7
Using Model Animals to Assess and Understand Developmental Toxicity
The recent advances in developmental biology described in Chapter 6 have established the central importance of a small number of highly conserved signal transduction pathways that mediate cell interactions crucial for animal physiology, reproduction, and development. It seems likely that many developmental toxicants might affect development by acting on those pathways. Application of the methods that have been so successful in elucidating them should now allow scientists to investigate that possibility and to determine the mechanisms by which developmental toxicants act. This chapter reviews the experimental approaches primarily responsible for the recent advances in knowledge about animal development and discusses how those approaches might be applied to developmental toxicology. Chapter 8 discusses how those approaches might lead to improved qualitative and quantitative risk assessment.
MODEL ORGANISMS AND THE GENETIC APPROACH
Single-Cell Organisms
Model organisms have been important throughout the study of modern biology. In the 1940s and 1950s, biochemical analysis of bacteria was important in working out the enzymatic pathways of metabolism. In the 1960s and 1970s, bacteria, especially Escherichia coli and its viruses (called phages), provided models for the new science of molecular biology and the elucidation of basic mechanisms for deoxyribonucleic acid (DNA) replication, transcription, and translation in prokaryotes. Since then, the budding yeast Saccharomyces cerevisiae and more recently the fission yeast Schizosaccharomyces pombe have
served as models for intensively investigating the molecular mechanisms of these and other functions unique to eukaryotic cells, such as the cdk-cyclin-based cell cycle, mitosis, meiosis, ribonucleic acid (RNA) splicing, regulation of chromatin structure, secretion, dynamics of the cytoskeleton, stress pathways, checkpoint pathways, and, to some degree, intercellular signaling and differentiation, the last two associated with yeast mating. Most of these cellular functions have been highly conserved during eukaryotic evolution, so that knowledge gained from yeast research is directly applicable to understanding human cell processes. However, understanding the interactions of cells and tissues in development and physiology of higher eukaryotes requires study of metazoans (i.e., multicellular animals). It should be appreciated, though, that as the processes are understood in metazoa, the components of each process can be introduced into yeast and the individual processes reconstituted there for further detailed study. For example, it has been found that a number of human cell-cycle proteins function well in the yeast cell cycle, when replacing the yeast cell’s components.
Utility of Model Animals
Much has been learned about human development and physiology through the study of model animals, a small set of diverse metazoans that have particular advantages for laboratory research. There are several reasons for their utility. Research on humans and other primates is expensive and limited by ethical considerations. The most commonly studied model animals are relatively inexpensive to maintain and are well suited for experimental manipulation. Most important, as outlined in Chapter 6, recent research has shown that there is a remarkable degree of similarity in the developmental mechanisms of all animals. Not only individual genes and proteins but also entire pathways of signaling and response and their functions in developing embryos appear highly conserved throughout evolution. This means that, although the embryology of simpler animals might appear superficially very different from that of humans, knowledge gained from those models can often be applied directly to understanding human developmental mechanisms.
On the other hand, there are important developmental and physiological attributes that can be investigated only in vertebrates, such as the adaptive immune system, or in mammals, such as placentation and lactation. Therefore, it is useful to study a representative range of model animals—from invertebrates that are only distantly related to humans but have particular experimental advantages, to rodents and other mammals that are less convenient but more closely related to humans.
Model Animals for Study of Development
For study of development, the currently most intensively investigated model animals, in order of increasing complexity, are the free-living soil roundworm
(nematode) Caenorhabditis elegans, the fruit fly Drosophila melanogaster, the frog Xenopus laevis, the zebrafish Danio rerio, the chick, and the laboratory mouse. Also particularly useful for certain investigations are sea urchin, sea slug (Aplysia), puffer fish, and a few mammals, including the rat. This set of model animals is somewhat different from those most widely used in the 1950s. Why have these species been chosen for recent intensive study? For four of them, the principal answer is genetics.
The genetic approach has become established in the last three decades as one of the most powerful tools for elucidating biological mechanisms. It allows researchers to compare wild type with a mutant phenotype and to identify new genes involved in controlling a biological process and to determine their functions in the organism. Genes that control important functions are identified by mutations that cause defects in those functions. These genes are then mapped, cloned, and identified at the molecular level so that the proteins they encode can be studied using methods of biochemistry and cell biology. This approach has proved to be extremely powerful, not only for basic research in model organisms but also for medical research on heritable human diseases. The approach was followed, for example, in the mapping, cloning, and subsequent study of the cystic fibrosis gene, the breast cancer susceptibility gene, and many others.
The four model animals chosen primarily on the basis of their convenience for genetic analysis are C. elegans, Drosophila, zebrafish, and mice. All are relatively small, easy to maintain in large populations in the laboratory, and have short generation times, which allow for rapid analysis of breeding experiments. The remaining animals are not well suited for classical genetic analysis, primarily because of much longer generation times, but have compensating advantages of convenience and manipulability or simplicity. Sea urchins, because of their reproductive properties, have been particularly valuable in studies of fertilization and gene regulation in early embryos. Aplysia are used in nerve growth and development studies. Puffer fish are useful for genomics because of their remarkably small genome size (400 megabases (Mb)) compared with most other vertebrates (about 3,500 Mb, including humans). The frog Xenopus has eggs and embryos that can be obtained in quantity and are relatively large (about 1 millimeter (mm) in diameter). The eggs and embryos are convenient for biochemical analysis as well as microsurgery and can easily be microinjected with cloned genes, RNAs, proteins, drugs, and so forth to study the developmental effects of those molecules. The embryos have been used in toxicant tests, such as the frog embryo teratogenesis assay–Xenopus (FETAX). FETAX is currently under consideration for validation (Bantle et al. 1996; NIEHS 1998). Chick embryos, more closely related to mammalian embryos, are readily accessible for observation and microsurgery (unlike those of mice, which develop in the uterus) and are convenient for tissue transplantation experiments. Putative developmental toxicants can be added directly to the embryo, thereby bypassing the modifying effects of maternal metabolism and selective transfer by the placenta. Rat, rabbit, and
guinea pig have long been standard systems for physiological and toxicological investigation. However, because of the power of genetic analysis, the four genetically tractable model animals (C. elegans, Drosophila, zebrafish, and mouse) have become mainstays of recent research in developmental biology and, for the same reason, are also likely to be particularly valuable in emerging approaches to developmental toxicology. These systems are described in more detail below, following a brief review of methods in genetic analysis.
Rationale and Strategy of the Genetic Approach
Genetic analysis has a powerful advantage in that it can “dissect” functionally and define the important components of any biological process without knowing anything about the process in advance—simply by isolating mutations that affect it, using those mutations to define the genes that control the process, and then cloning and characterizing those genes and their gene products, thereby revealing molecular mechanisms. Over the past two decades this approach has been successfully applied to many aspects of animal development, as indicated in Chapter 6. It can also be applied to elucidating the mechanisms of action of developmental toxicants. The general steps in the standard genetic approach, described below, are sometimes referred to as “forward genetics” (going from the mutant phenotype to the gene) in contrast to the more recently developed methods of “reverse genetics” (going from the gene back to a phenotype) made possible by molecular biology and genomics (see Chapter 5 for some of the genomic methods). Although the terms forward and reverse genetics are now generally accepted, it should be noted that the term “reverse genetics” has had a history of use in earlier medical genetics literature to describe the progression from mapping of a heritable disease state to cloning of the responsible gene (called “forward genetics” elsewhere).
Forward Genetics
The steps in this approach are as follows:
-
Choose a defective phenotype of interest (e.g., failure to develop a particular structure or increased sensitivity to a toxicant) that is specific and selectable or easily recognizable.
-
Using mutagenized populations, carry out a saturation screen for mutants with the defective phenotype (i.e., a screen large enough so that mutations are likely to be found in every gene required in development of the normal phenotype).
-
Use classical genetic analysis of these mutations to define the genes they represent by genetic mapping and complementation tests and to determine their null phenotypes (i.e., the effects of complete loss of gene function). The incisive-
-
ness of studying null mutants is worth mentioning in the context of developmental toxicology. Their phenotypes match the toxicologist’s ideal of what the “perfect” toxicant would generate for observation if it completely inhibited just one target component of the organism.
-
If possible, establish the order of function of the identified genes by constructing double mutants to determine which of two distinguishable phenotypes takes precedence (epistasis test).
-
Identify additional modifier genes by using suppressor and enhancer screens in a sensitized genetic background for secondary mutations that make the defective phenotype of an existing mutant less or more severe.
-
Using fine-structure genetic mapping and positional cloning, obtain genomic clones of each gene for molecular analysis and verify their identities by demonstrating that each gene in the corresponding mutant animal carries a DNA sequence alteration.
-
From suitable complementary (c) DNA libraries of cloned cDNA copies of the animal’s messenger (m) RNA population, isolate cDNAs corresponding to each gene, sequence them to determine the predicted amino acid sequence of each encoded protein, and carry out a similarity search, comparing those sequences with the sequences available in databases, which often can be used to discern motifs and reveal the functional class to which a protein belongs. (Function was initially deduced for the class from other kinds of studies—biochemical, cellular, developmental, and physiological.)
-
Determine when and where the mRNA and the protein encoded by each gene are found during development by using, respectively, nucleic acid probes and antibodies made to fusion proteins. A faster but sometimes less reliable alternative is to make reporter constructs, which carry the promoter region of the cloned gene fused to a gene encoding a reporter protein that can be detected by its activity (e.g., the E. coli β-galactosidase gene lacZ) or fluorescence (e.g., green fluorescent protein (GFP)). Embryos into which such a construct has been introduced (by DNA transformation) can be observed at various stages to determine when and in what cells and tissues the promoter is active. Generally (but not always), an active provider will reflect the expression pattern of the normal gene.
-
Supplement that information with genetic mosaic analysis, by producing animals in which only certain cells or tissues are mutant, to discover where a gene must normally function and whether its functions are cell autonomous (i.e., intracellular) or cell nonautonomous (i.e., intercellular).
-
Isolate and biochemically analyze proteins encoded by the mutationally identified genes to study further the function of the proteins.
All these steps are not always carried out. The most important and difficult step, once mutants have been obtained, has been positional cloning of the gene. However, shortcuts are becoming available with the accumulation of genomic mapping and sequence information and the development of new technologies
(see following sections). For example, if a mutation defining a gene of interest has been mapped to a region of the genome for which the entire DNA sequence is known, the “candidate gene” approach can be used to identify it. Computer analysis of the genomic sequence can predict which sequences in the region represent coding sequences and open-reading frames (ORFs) of genes and what proteins these DNA sequences encode. It is then often possible to guess one or a few most likely candidate genes and confirm that one of these is correct by sequencing one (or preferably more) mutant allele and finding the responsible sequence alteration(s) or by expressing the candidate gene to see if its encoded product reverts the mutant phenotype back to wild type.
A new method called genomic mismatch scanning (GMS), using DNA microchip technology, will allow more rapid identification of the candidate gene and the mutational lesion in one step. Oligonucleotides representing the entire sequences of all candidate genes in the region to be tested, as well as all possible single base-change mutational variants of each sequence, are synthesized and fixed in an indexed array on a microchip (see description of the method in Chapter 5). The chip is then annealed to differently labeled probes from nonmutant and mutant forms of the cloned gene. By comparing these patterns, both the correct candidate gene and the nature of the mutational lesion can be determined.
Reverse Genetics
With the increasing availability of genomic sequence information, the following somewhat different approach is becoming more useful for studying biological processes, especially in organisms such as mammals, for which the forward genetic approach is difficult. It is called reverse genetics, because it starts with a cloned gene of potential interest. The cloned gene is then used to obtain animals with defects in the gene or its expression for functional analysis. The steps in this approach are as follows:
-
Identify a gene of interest from its sequence (e.g., the mouse homolog of a developmentally important gene in Drosophila) and obtain a clone of the gene by standard methods based on sequence similarity (such as screening a mouse library (collection) of genomic DNA clones with the cloned Drosophila gene).
-
Determine its expression pattern (as described above) for clues to its function.
-
Inactivate the gene (often referred to as “knocking out,” “targeted inactivation,” or “homologous recombination” of the gene) and observe the phenotypic consequences for more definitive information on function. This can be done either transiently, by injection of an antisense or double-stranded mRNA that specifically prevents gene expression, or permanently (preferable, but requiring considerably more effort), by generating animals that carry a null mutation in the gene. In the nematode C. elegans, the double-stranded mRNA method works
-
particularly well (described in more detail below). In the mouse, mutations can be obtained efficiently by targeted recombination of mutant DNA constructs introduced into the germ line (described in more detail below). In flies (Drosophila) and nematodes (C. elegans), the desired mutant individual can be screened from a large population after random transposon insertion or chemical mutagenesis.
-
Once mutations are obtained, they can be subjected to any of the genetic analyses described above.
Again, emerging technologies, such as microchips carrying ordered arrays of cDNAs to allow rapid analysis of how a mutation affects mRNA populations, will accelerate and enhance the above approaches.
Extrapolation to Humans
For many genes identified by forward or reverse genetics in model animals such as the mouse, and particularly for genes relevant to human disease states, the next step is to isolate and characterize the corresponding (orthologous) gene in humans. Several recent developments have simplified the task of cloning human homologs for molecular analysis. Extensive and detailed maps of molecular markers are now available for many areas of the human genome, and rapid progress is being made on the remainder in connection with the Human Genome Project. Comparison of mouse and human maps demonstrate extensive linkage conservation (synteny) between the two genomes (i.e., the arrangement of orthologous genes has been conserved over large regions from the last common ancestor). Considerable linkage conservation is found even between fish and mammals. As ancestral species diverged hundreds of millions of years ago and evolved into present-day species, local gene order in most instances has been maintained while large blocks of contiguous genes have been rearranged. For example, genes A-B-C-D-E found on mouse chromosome 12 might be found as A-B-C-D-E or, in reverse order, as E-D-C-B-A on human chromosome 7. As a result, if the chromosomal location of a gene responsible for a trait in the mouse is known, it is now possible to predict quite accurately the chromosomal location of its ortholog in humans (see Web site at http://www.informatics.jax.org, under mammalian homology and comparative maps). This approach will also be useful in defining human genes that affect responses to developmental toxicants (e.g., the genes for various enzymes that metabolize exogenous chemicals).
There also are large libraries (expressed sequence tag (EST) libraries) of sequences representing pieces of mRNAs transcribed from genes at various times and tissues in the human (see description of EST methods in Chapter 5). Transcripts from almost 90% of all human genes are estimated to be sequenced and present in these libraries. The transcripts are of great value for isolating the human homologs of genes and gene products that have been well characterized in other organisms.
THE MAJOR MODEL ANIMALS FOR GENETIC ANALYSIS
The four genetically tractable model animals, C. elegans, Drosophila, zebrafish, and mouse, are useful for somewhat different reasons. Relevant characteristics of each are described briefly below, along with some of their experimental advantages and disadvantages (see also Tables 7-1 and 7-3). The potential utility of each animal for identifying and investigating mechanisms of developmental toxicants is discussed later in this chapter.
The Nematode Caenorhabditis elegans
History, Biology, and Genetics
Caenorhabditis elegans is a roundworm found commonly in soils all over the world. It has become widely exploited as a model animal largely because of the early efforts of Brenner (1974), who recognized its experimental advantages and pioneered its genetic analysis. The adult is about 1-mm long, just visible to the naked eye. It feeds on bacteria, such as the common bacterium E. coli, and is easy to grow and breed on agar plates in the laboratory. C. elegans is one of the simplest animals known, with a small fixed number of somatic cells: 959 in the adult hermaphrodite and 1,031 in the adult male. It is transparent throughout the life cycle, so that its entire development can be analyzed in living animals with the light microscope. Its generation time is only 3 days, and development is rapid (Figure 7-1). Embryogenesis is complete by 14 hours after fertilization. The first-stage (L1) larva hatches from the egg, growing and molting through three larval stages (L2, L3, and L4) as its reproductive system develops before the final molt to adulthood. Adult males make sperm and can mate with hermaphrodites, making genetic crosses possible. The hermaphrodites are essentially females but produce some sperm during late larval development and can self-fertilize, which simplifies genetic analysis. C. elegans has a genome size of about 100 megabases (Mb) packaged into six small chromosomes, including five autosomes and a sex (X) chromosome (hermaphrodites have two and males one). Extensive genetic and physical maps have been constructed, and its genome has recently become the first in a metazoan to be completely sequenced under the auspices of the Human Genome Model Organisms Project (C. elegans Sequencing Consortium 1998). The genome includes about 19,000 genes.
Because of its transparency and the invariance of cell-division patterns throughout C. elegans development, it has been possible to describe embryonic and larval development completely at the cellular level. By observation of developing animals using Nomarski microscopy, Sulston and coworkers were able to define all the larval cell lineages (Sulston and Horvitz 1977) and later the entire embryonic cell lineage (Sulston et al. 1983), so that the ancestry of every cell in the adult organism is now known. Perturbation of normal development by laser
TABLE 7-1 Comparison of Four Model Animals for Genetic Analysis and Humans As a Reference
|
Animal |
Adult Size (cm) |
Genome Size (Mb) |
Period of Organogenesis (d) |
Generation Time (wk) |
Experimental Advantages |
|
Nematode (Caenorhabdits elegans) |
0.1 |
97 |
0.2-0.4 |
0.4 |
Convenient forward and reverse genetics, complete genome sequence known, complete description of development available, simplicity, transparency |
|
Fruit fly (Drosophila melanogaster) |
0.4 |
180 |
0.5-1 |
2 |
Most convenient forward genetics, many genetically defined signaling pathways known, extensive knowledge of development |
|
Zebrafish (Danio rerio) |
3 |
1,700 |
1-4 |
12 |
Vertebrate, good forward genetics, transparency, external, well-studied development, accessible to test chemicals in water |
|
Mouse (Mus musculus) |
6 |
3,000 |
6-15 |
10 |
Placental mammal, closest model to humans, good forward and reverse genetics, well-studied development |
|
For comparison: |
|
||||
|
Human |
170 |
3,500 |
14-60 |
27 yr (1,400 wk) |
|
|
Abbreviations: cm, centimeter; d, day; Mb, megabase; wk, week; yr, year. |
|||||
ablation of specific cells has provided information on inductive cell interactions during embryogenesis and larval growth. This knowledge has been extremely useful in analyzing the genetic control of cell-fate determination and the roles of cell signaling pathways by using genetic approaches, as described further below. For more comprehensive reviews on current knowledge of C. elegans, see Wood et al. (1988) and Riddle et al. (1997).
Transgenic Technologies
DNA Transformation. Cloned genes can be reintroduced into the C. elegans genome by injection of DNA into the syncytial region of the hermaphrodite gonad (Mello et al. 1991). The injected DNA recombines to form large replicating extrachromosomal arrays, which become incorporated into developing oocytes
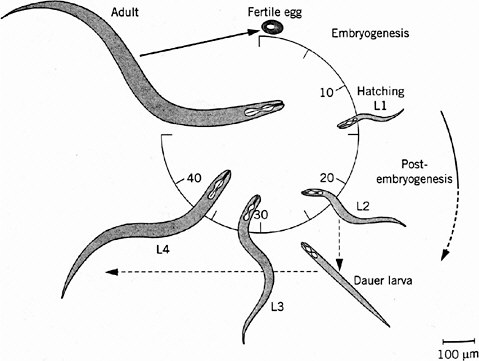
FIGURE 7-1 Life cycle of Caenorhabditis elegans. The numbers 10 through 40 indicate hours after fertilization of the egg. L1 through L4 indicate larval stages, each ending in a molt, a shedding of the tough cuticle. The dauer larva is a diapause stage entered when food (usually bacteria) is in short supply. Source: Wood (1999). Reprinted with permission from Encyclopedia of Molecular Biology; copyright 1999, John Wiley & Sons.
and then embryos. These arrays can be transmitted to most cells of the resulting animals and through the germ line to their progeny. Although the genes on such arrays are present in high and somewhat variable copy number, they are efficiently expressed and can be useful for many types of investigations, such as transformation rescue in positional cloning and analysis of a cloned gene’s expression patterns using lacZ or GFP reporter-gene constructs. From transmitting lines, more stable integrated lines can be obtained in which the array has inserted randomly into a chromosomal locus, allowing various gene-trapping technologies for identifying loci with tissue-specific expression patterns. Targeted insertion of transgenes by homologous recombination has not yet been achieved. Reverse genetics using targeted gene disruption is therefore difficult but can be accomplished by random transposon insertion (Plasterk 1995) or deletion mutagenesis followed by appropriate screens, or it can be accomplished by RNA-mediated gene interference (RNAi), as discussed next.
RNAi. A powerful tool for reverse genetic analysis has been provided by the discovery that introduction of double-stranded mRNA for a particular gene into C. elegans will specifically inactivate that gene, resulting in loss-of-function phenotypes that generally mimic the gene’s null phenotype for at least a generation or two (Fire et al. 1998). Although the mechanism of this inactivation, referred to as RNAi, is not yet understood and gene expression in some tissues is more susceptible to inactivation than expression in other tissues (Montgomery et al. 1999), it is clear that RNAi will be extremely useful for rapid functional tests of genes identified by genome sequencing as potentially important, for example, in development or in responses to environmental toxicants. Moreover, recent results indicate that the technique is applicable to Drosophila (Kennerdell and Carthew 1998) and perhaps to other organisms as well.
Signaling Pathways in Development
Most of the progress in understanding C. elegans development has come from application of forward genetics as described above, combined with laser ablation experiments to identify required cell interactions. A variety of inductive events, which in C. elegans can be analyzed at the single-cell level, are mediated by signaling pathways that are still under investigation. However, it is already clear that nematode development uses most of the pathways described in Chapter 6, often in developmental contexts similar to those found in more complex metazoans. Two exceptions are the Hedgehog and cytokine signaling pathways, which C. elegans appears to lack (Ruvkun and Hobert 1998).
The Fruit Fly Drosophila
History, Biology, and Genetics
Drosophila melanogaster is the common fruit fly found worldwide in orchards, where adult flies lay eggs on rotting fruit. Since the beginning of this century, fruit flies have been cultured in the laboratory in half-pint milk bottles and more recently in shell vials and plastic tubes by using a solid food, typically composed of agar, cornmeal, dried yeast, and molasses. At 25°C the life cycle takes approximately 2 weeks. Embryogenesis and the first two larval stages require 1 day each; the third larval stage, 2 days; and the pupal stage, 4-5 days (Figure 7-2). Two-day-old adults begin to lay eggs. Because of the short life cycle, ease of rearing in large populations, and the many diverse phenotypes readily visible under a simple dissecting microscope, many mutations have been accumulated in the organism since its initial use by T. H. Morgan and his associates at Columbia University in the 1920s. The study of fly genetics has been instrumental in many classic discoveries in eukaryotic genetics, such as linkage, gene mapping, recombination frequency, and chromosomal aberrations. Discov-
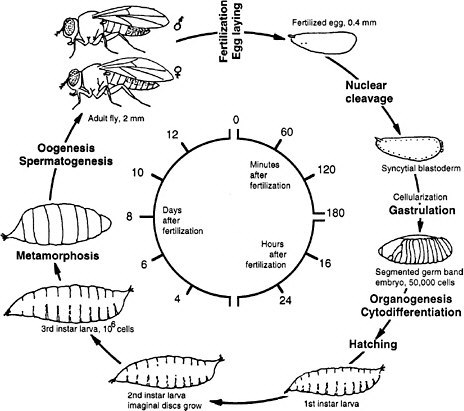
FIGURE 7-2 Life cycle of Drosophila melanogaster. The larva hatches 1 day after the egg is fertilized. First, second, and third instar are larval stages, each ending with a molt. During pupation most of the larval tissues are destroyed and replaced by adult tissues derived from the imaginal discs that were growing in the larva. Times are given for the life cycle at 25°C. Source: Adapted from Wolpert et al. (1998).
ery of the giant polytene salivary gland chromosomes in the 1930s provided a cytological basis for those genetic theorems and thus made Drosophila a key organism for genetic analysis.
As discussed in Chapter 6, the use of fruit flies for developmental studies awaited the saturation screens for lethal and female sterile mutations. These screens were conducted in the late 1970s and 1980s and led to the discovery of cascades of gene functions responsible for the organization of the egg and early pattern formation in the embryo. The advent of recombinant DNA and cloning quickly led to the isolation and sequencing of key genes, which affect the regional specification of body parts. Such genes were defined by the homeotic mutations studied by E. B. Lewis. These studies led to the startling discovery in 1983 that sequences of amino acids coded for by homeotic genes (the homeobox
DNA sequence, and the homeodomain protein sequence) were conserved not only in different homeotic genes in flies but also across the whole animal kingdom, including humans. This conservation of genetic structure and function has become the cornerstone of modern developmental biology, forming the basis for the usefulness of model organisms in understanding human developmental mechanisms.
D. melanogaster has a genome size of approximately 180 Mb, a third of which is centric heterochromatin (regions rich in simple sequence repeats that remain condensed during interphase). The 120 Mb of euchromatin (unique sequence, decondensed during interphase) are located on two large autosomes, one dot chromosome, and paired XY sex chromosomes. The 120 Mb of euchromatic DNA have now been sequenced (Adams et al. 2000), and are estimated to encode approximately 13,600 genes, somewhat fewer than the C. elegans genome but with comparable functional diversity. The polytene chromosomes of Drosophila provide a cytogenetic map of the euchromatic portion of the genome, and by means of in situ hybridization to those large chromosomes, molecular markers have been identified within most subdivisions of the map.
Transgenic Technologies
One of the principal tools for Drosophila research is the availability of a transposon, called the P-element, which can be used as a vehicle for introducing nearly any genetic construct into the Drosophila genome at high efficiency via a relatively easy process of transformation. In addition, flies possessing single P-elements, containing dominant markers, such as the bacterial β-galactosidase gene, can be used as mutagens to disrupt coding sequences and as markers of the disrupted gene for cloning. Similarly, P-elements lacking a strong promoter for the expression of β-galactosidase can insert adjacent to enhancer elements that activate the enzyme in an enhancer-specific manner and thus identify potential new genes for study. By a combination of saturation screens and newer insertional mutagenesis experiments, it is possible to accumulate large sets of mutations of known genes and of related gene functions. Among reverse genetic technologies, for determining the function of a gene identified only by its nucleotide sequence, is insertional mutagenesis using transposable elements or RNAi, which has recently been shown to be effective in Drosophila as well as C. elegans.
Signaling Pathways in Development
One of the most useful outcomes of the genetic analysis of D. melanogaster development has been the identification of developmental pathways that are conserved in most organisms. Initially, these sets of genes were identified because of their similar phenotypes. As indicated above, using epistasis relationships, genes could be put into developmental sequence. More recently, the innovative use of a
sensitized genetic background has led to the identification of additional components in those pathways. These new components have been difficult to discover, because they are either stored in the egg and thus show a maternal perdurance or they are used in multiple pathways, the consequence being that their mutant phenotype diverges from that seen in any one pathway or is generally lethal. In this section, the committee will describe sensitized mutants in some detail, because it believes that they will be useful in the future for toxicant assays.
The screen used by Simon et al. (1991) has set the pattern for many subsequent screens (and the committee will later draw attention to the potential use of this strategy for assaying toxicants). The Sevenless gene encodes a tyrosine kinase receptor that is required specifically for the formation of rhabdomere 7, one of eight light-receptor cells in each ommatidium of the Drosophila compound eye. The authors conducted a genetic screen on flies bearing weak, temperature-sensitive mutations in the Sevenless gene. They screened for genes for which inactivation of one copy caused the Sevenless phenotype. Hence, even if such genes are needed in multiple tissues, the activity of the remaining intact gene would suffice in all but the eye. Increases or decreases in gene activity in the tyrosine kinase pathway would affect the intermediate eye phenotype of the fly strain used. The screen detected a series of genes functioning downstream of the receptor, including homologs of Ras, Raf1, and a guanine nucleotide exchange factor (Figure 7-3). The dose-dependence of the Ras mutation suggested that a small (two-fold) reduction downstream should modify the rough-eye phenotype imparted by the mutant Ras and do so only in the eye. The suppressors so discovered are the Drosophila homologs of components of the MAP kinase pathway. These are used by many tissues, so their mutations would be recessive lethal. This study provided the basis for integrating the downstream effector pathway for many tyrosine kinase receptor functions.
Since this initial screen, screens using sensitized genetic backgrounds have become commonplace in developmental genetics of flies. For example, the initial set of downstream functions of the decapentaplegic pathway (a TGFβ signaling pathway), now called SMADs, was discovered in a screen for mutations that enhanced the phenotype of a weak Dpp allele (Raftery et al. 1995). Mutations in the Notch pathway have identified additional components functioning downstream of the receptor (Xu and Artavanis-Tsakonas 1990).
A major difficulty in identifying components of these critical signaling pathways is that they play essential roles in many developmental processes; hence, mutations in the genes involved are lethal. Genetic approaches have been developed in Drosophila to circumvent that phenomenon. The site-specific recombinase system from yeast (FRT-FRP system, Golic and Lindquist 1989) has been particularly useful (Figure 7-4). In the case of essential genes that are expressed during oogenesis, germ-line clones that are homozygous for a lethal mutation are made in the background of an ovary expressing a dominant female sterile mutation. These clones, lacking the dominant female sterile mutation, are
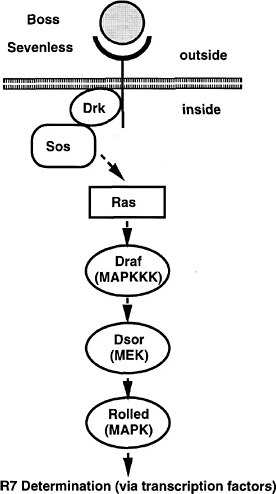
FIGURE 7-3 The signal transduction pathway by which the development of the R7 photoreceptor cell is induced in the eye of Drosophila. This pathway is a receptor tyrosine kinase pathway. Many of the components of the pathway were first discovered in Drosophila by doing selections in mutants in which the pathway was operating close to threshold due to a component of reduced activity. These “sensitized strains” revealed secondary mutations easily, and might be useful for detecting toxicant effects on development. BOSS, the ligand presented on the surface of neighboring cells R2 and R5. SEV, the transmembrane receptor that binds the BOSS ligand protein from an adjacent cell. DRK, an adapter protein binding to the phosphorylated cytoplasmic tail of the receptor. SOS, a GTP:GDP exchange factor protein activated when binding to DRK. RAS, a small G protein active when GTP is bound. DRAF, a protein kinase that phosphorylates DSOR, which in turn phosphorylates ROLLED, which in turn phosphorylates a transcription factor, which then activates specific gene expression involved in R7 determination. MARPKKK, mitogen-activated protein kinase kinase kinase; MEK, mitogen-activated protein kinase/extracellular signal-related kinase kinase; MAPK, mitogen-activated protein kinase. See text for details. Source: Adapted from Simon et al. (1991).
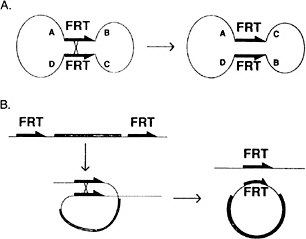
FIGURE 7-4 The FLP recombination method for removing DNA sequences from the genome at specific times and places in the animal to inactivate or activate specific genes. This method is now widely used in Drosophila and in mice. (Panel A): A circular piece of DNA is shown, containing two FRT sites in opposite 5′-3′ orientation. When crossover occurs, the gene order ABCD changes to ACBD. (Panel B): Two FRT sites are in the genome in the same orientation. When cross-over occurs, a circle of DNA is looped out carrying the equivalent of one FRT site. The other remains in the genome. See text for details. Source: Golic and Lindquist (1989). Reprinted with permission from Cell Press, copyright 1989.
able in many instances to produce mature eggs lacking the gene product (Perrimon et al. 1989; Chou and Perrimon 1996). This provides the opportunity to determine the phenotype of embryos lacking the function of the gene in question. Similarly, somatic clones can be produced in embryos or larvae heterozygous for various mutations. These somatic clones, induced at high frequency by the FRT-FRP system, will be homozygous for the new mutations and show a phenotype (Xu and Rubin 1993). Thanks to these powerful methods, components of several of the signal transduction pathways have been identified first in Drosophila and C. elegans and later confirmed in vertebrate cell systems.
The Zebrafish
History, Biology, and Genetics
The zebrafish is a relative newcomer to the list of model animals. It has become important as the first vertebrate to be subjected to large-scale genetic screens, which were shown to be feasible by C. Nüsslein-Volhard and others in the 1980s (Haffter and Nüsslein-Volhard 1996). Such screens are possible be-
cause, unlike other vertebrate model systems, adult zebrafish can be reared in large numbers (each is 5-cm long) at reasonable expense (although zebrafish are more expensive than C. elegans and Drosophila), and they are fecund, laying hundreds of eggs at regular intervals. The embryos are permeable to, and in fact, bioconcentrate many chemicals added exogenously in the water. The effects on development might be assayed simply and visually, although such tests have not been done systematically. Most important for assaying effects, the embryo is transparent and develops rapidly (128 cells develop 3-4 hours after fertilization), so all organs are visible and established during a few days. The organs, including heart, vessels, kidney, and liver, are nearly identical to those in the early human embryo. The zebrafish generation time is about 12 weeks. Its genome, carried on 26 chromosomes, is small for a vertebrate, 1,700 Mb (about half the size of the human genome).
Large-scale mutagenesis screens in zebrafish identified genes involved in development of vertebrate-specific body plans and tissues. One such chordate feature is the transient embryonic “backbone,” the notochord. The notochord generates signals (e.g., proteins of the Hedgehog family) that pattern embryonic development of adjacent tissues, including the nervous system and muscles. The neural crest is found only in vertebrates. The migratory population of neural-crest cells emanates from the neural tube and disperses widely, contributing to neural ganglia, pigmentation, jaw structures, and the major blood vessels from the heart. Other important vertebrate organ systems, without close cognates in invertebrate model genetic organisms, include the bony skeleton, an endothelial lined vascular system, a chambered heart, and gut derivatives such as the pancreas and liver, and the kidneys.
Organogenesis in Development
Individual mutants from large-scale screens of zebrafish were found to have perturbations in organogenesis and in other aspects of development that are highly informative. For example, the notochord is ablated entirely in some mutants, and in others, the notochord is structurally present but particular notochord signals are absent. Neural-crest derivatives, such as neurons, craniofacial structures, and melanocytes (pigment cells), are affected by mutations. Different modules of organ form or function are selectively removed by individual mutations. For example, the Pandora mutation eliminates the heart ventricle; Slo Mo causes a slow heart rate. Those mutations, therefore, provide an entrance point to specific pathways of organogenesis. In some cases, the phenotypes resemble congenital disorders, such as aortic coarctation (as noted in Chapter 2, cardiac defects are the most common of live-born human developmental defects). Others have phenotypes that are common in the adult, such as heart failure. Current screens are pinpointing even more subtle phenotypes, in part by inclusion of molecular probes to reveal particular cell populations. For example, a large number of mutants
have been isolated as ones affected in the projection of retinal nerves to the tectum of the brain.
Large-scale efforts are under way in many laboratories to clone the mutant genes. Genetic and physical maps, crucial for positional cloning, are being constructed. Additionally, a large-scale EST project is under way, along with development of methodologies to map the ESTs. Because there is extensive synteny (conservation of chromosomal gene order over short distances) even between zebrafish and mammals, the dense EST maps of mouse and human should suggest candidate genes once the map position of an EST is established. The large insert libraries needed for positional cloning are now available. One important advantage of zebrafish for positional cloning, compared with the mouse, is the ease and thrift of scoring thousands of embryos in mapping crosses. The large numbers greatly enhance genetic resolution, thereby delimiting the chromosomal region in which to search for a mutant gene. With current maps, and some guess-work, more than 30 zebrafish mutant genes have been located and identified as of late 1999. One mutant gene had not been previously described in any other species and is critical to normal endoderm (gut) differentiation. Other mutant genes are related to known genes in other organisms and have refined the understanding of how signaling pathways pattern the early vertebrate embryo.
The Mouse
Biology and Genetics
Although lower vertebrate and nonvertebrate organisms are valid model systems for studying many aspects of cell and molecular processes that are shared by widely disparate organisms, certain characteristics are restricted to mammals. These characteristics include placentation, intrauterine development, lactation, and aspects of immunology and carcinogenesis. To study these characteristics, only a mammalian model is ultimately appropriate. The laboratory mouse provides a small, tractable, and genetically well-characterized model, in spite of minor differences among mammals in the details of development and metabolism.
Many different mammalian models have been used for different aspects of biomedical research, but among mammals, the mouse is perhaps the most versatile and best studied. Among the advantages, mice are among the smallest mammals and have a short generation time of around 10 weeks. They are prolific breeders and their reproductive cycles are easily monitored for the timing of pregnancies. They have been bred in captivity for biomedical research for nearly a century and are docile animals. All these features add up to a great practical benefit in cost efficiency when large numbers of animals are required for research (e.g., in genetic and toxicological studies). As many as 3,000 pups (and up to 5 generations per year) can be raised per year per square meter of area in an approved animal facility, assuming cage racks are 5-6 shelves high (Silver 1995).
Recent advances in genomic research in humans and mice have reinforced the mouse as a model genetic system. They have closely related genomes of approximately the same size (about 3,000 Mb) and probably diverged from a common ancestor 80 million years ago. Counterparts of most human genes can be found in the mouse. In terms of the genomic structure, large segments of chromosomes containing the rank order of hundreds to thousands of genes have been preserved virtually intact (synteny) between the mouse and human, facilitating the application of forward and reverse genetic techniques. Laboratory mice present a vast resource of defined genetic strains, including inbred and recombinant inbred strains with characterized allelic differences that can serve as models for human genetic polymorphisms. There is a growing resource of naturally occurring and induced mutants (including a large variety of knock-out null mutants) that are commercially available, easily obtained, and easily maintained. Furthermore, the embryos of mice are accessible to embryological and genetic manipulation and have been widely used in the development of transgenic technologies.
Transgenic Technologies
Manipulating the mammalian genome has become a commonplace experimental procedure during the past two decades, and transgenic animals have been widely used in many research areas. “Transgenic” is a term that was originally coined to describe animals that had a foreign or “trans” gene inserted at random into their genome by experimental means. Its use has been broadened as more sophisticated techniques for altering the genome have been developed, and it can now be used to include any animal whose genome has been altered by addition of genetic material or by alteration of existing genes by gene targeting. Transgenic techniques, which were first devised in the 1975-1985 period, have been applied to a variety of experimental animals and agricultural animals, although by far the most common mammalian subject of gene manipulation remains the laboratory mouse.
The transgenic approach, whereby genes can be isolated, altered, and then returned to the animal, has provided a new means to investigate experimentally the function of genes and their regulation in different tissues and at different times during development. As it became clear that foreign genes could indeed function after insertion into the genome of a host, and that transgene expression was, to some extent, under experimental control, the practical uses of transgenic animals began to emerge, including uses in toxicology.
Although the first successful transgenics were made using a viral vector to deliver DNA to mammalian embryos through viral infection, the direct microinjection of cloned genes into the pronucleus (haploid nucleus) of a fertilized egg has proved to be the more versatile method and has been used widely. The principle is simple: a gene of interest is cloned, with or without regulatory elements, and is microinjected into the pronucleus. One or more copies of the gene will
integrate into a chromosome at random, either immediately or after one or two cleavage divisions. Because integration is a rare event, individual eggs rarely contain more than one integration site. If the transgene enters the germ line, it subsequently behaves as a Mendelian gene in meiosis.
Table 7-2 summarizes transgenic technologies commonly used in experimental mice, and those technologies are described in detail below.
Overexpression and Misexpression. A common use of transgenics is to overexpress a given gene either in the tissue where it is normally expressed or at ectopic sites (where it is not normally expressed). A transgene is constructed that includes the coding region of the gene and its own regulatory elements or those of another gene that will drive expression constitutively, inducibly, or ectopically. Some of the earliest transgenic experiments were ones in which transgenic mice were produced with a human-growth-hormone gene driven by the inducible metallothionein promoter. The use of a human gene provided a means of distinguishing the expression of the endogenous mouse-growth-hormone gene from the expression of the transgene.
Promoter and Enhancer Analysis. To identify the regulatory elements of a given gene, a series of transgenes can be constructed containing ever increasing amounts of the 5′ regulatory region linked to an unrelated gene with an assayable gene product (a reporter gene) or the gene itself. Then, a series of transgenic animals is produced with those constructs and assayed for the expression of the reporter gene or gene product. In this way, regulation of levels of expression and tissue specificity of expression can be assigned to specific positions in the regulatory region of the gene under study. This has been widely done, for example, in the analysis of the expression of the 39 Hox genes of the mouse.
Antisense Transgenes. Transgenes can be constructed to contain antisense sequences. If expression of an antisense transgene is directed to the tissues where the endogenous gene of complementary coding sequence is being expressed, antisense RNA transcribed from the transgene can hybridize to the endogenous mRNA and reduce its translation. This is useful for assessing the function of the endogenous protein.
Gene Trapping. A serendipitous means of finding new genes includes the phenomenon of insertional mutagenesis, where, by chance, a transgene inserts into a chromosomal gene causing a mutation that results in an unexpected mutant phenotype. A similar procedure was discussed above for P-element insertion in Drosophila. Another means of screening for new genes in the mouse is to make transgenics using a promoterless reporter gene. Successful expression of the reporter transgene will indicate its intergration near an endogenous promoter, and the expression pattern can provide information about the endogenous gene that
TABLE 7-2 Transgenic Technologies Commonly Used in Laboratory Mice
|
Method |
Purpose |
DNA Construct |
Desired Effect |
Variations |
|
DNA microinjection into zygotic pronucleus |
Overexpression |
Gene of interest with strong promoter |
Higher than normal expression |
Tissue specific, ubiquitous or inducible promoter |
|
Misexpression |
Gene of interest with ectopic promoter |
Ectopic expression |
Temporal or spatial misexpression |
|
|
Promoter and enhancer analysis |
Regulatory regions driving reporter gene expression |
Mapping of regulatory elements |
Various reporter genes (e.g., lacZ), green fluorescent protein, luciferase |
|
|
Antisense |
Endogenous promoter with antisense gene |
Reduced expression of endogenous gene |
|
|
|
Insertional mutagenesis |
Any |
Phenotype associated with serendipitous disruption of unknown gene |
|
|
|
Gene trapping |
Promoterless reporter gene |
Expression of reporter gene indicating expression of endogenous gene |
Various reporter genes (e.g., lacZ), green fluorescent protein, luciferase |
|
|
Targeted mutagenesis via homologous recombination in embryonic stem cells |
|
|||
|
DNA electroporation into embryonic stem cells |
Mutations |
Homologous genomic regions flanking selectable marker |
Mutation of endogenous gene |
Insertion into or deletion of endogenous genomic sequences to produce null (knockout), hypomorph, or dominant negative mutations |
|
Method |
Purpose |
DNA Construct |
Desired Effect |
Variations |
|
|
Expression reporting |
Homologous genomic regions flanking in-frame reporter gene and selectable marker |
Disruption of endogenous gene and expression of reporter gene under control of the endogenous gene’s promoter |
Various reporter genes (e.g., lacZ), green fluorescent protein, luciferase |
|
Targeted mutagenesis via homologous recombination in embryonic stem cells |
|
|||
|
DNA electroporation into embryonic stem cells |
Point mutations |
Homologous genomic regions with desired point mutation flanking lox-P-flanked selectable marker |
Replacement of homologous region and removal of selectable marker |
Different recombinase systems possible |
|
Conditional mutations |
Homologous genomic regions containing lox-P site in intron, flanking lox-P-flanked selectable marker |
Produce mutation by removing regions flanked by lox-P sites at desired time |
Mutation produced at different times or in specific tissues |
|
|
Conditional restoration |
Homologous genomic regions with lox-P-flanked selectable marker |
Restore mutated gene function by removing selectable marker at desired time |
Function restored at different times or in specific tissues |
|
|
Knockin |
Homologous genomic regions of gene 1 flanking coding regions of gene 2 |
Replacement of coding region of gene 1 with that of gene 2 to misexpress gene 2 in expression domain of gene 1 |
|
|
has been “trapped” by the inserted transgene. Cloning of the insertion site then leads to the identification of the chromosomal gene.
Embryonic Stem Cell; Mediated Gene Targeting. A major limitation of the DNA microinjection method for making transgenic animals is that it does not allow the targeted alteration of endogenous genes. A technically more complicated method of gene targeting in embryonic stem (ES) cells provides the means for accomplishing that alteration, particularly for the directed inactivation of genes. All the knock-out mutants for signal-transduction components, which were discussed in Chapter 6, were produced by this method. The method, worked out in 1980, is simple in principle. A gene-targeting construct is made that incorporates a selectable marker flanked by cloned sequences of the gene to be targeted. It is introduced, usually by electroporation, into ES cells in vitro. ES cells are obtained from the inner cell mass of a mouse embryo at the blastocyst stage. The cells can be cultured in a Petri dish in artificial medium, where they proliferate for tens to hundreds of generations. Random integration of the targeting construct DNA into a chromosomal site of the ES cell will occur as a rare event (1 in 1,000 cells), and even more rarely (1 in 1 million cells), a double-crossover event of recombination will occur between the homologous DNA of the construct and the endogenous gene, the result being that the selectable marker is inserted into the endogenous allele. Cells that have incorporated the transgene are selected using the selectable marker. They are cloned, and the cell clones that have undergone homologous recombination are distinguished from random integrants by analysis of the DNA.
Cells with the desired genetic change are then introduced into early embryos to produce chimeric pups (i.e., ones containing both normal cells and genetically altered cells). The gene alteration is recovered in the subsequent generation if, and only if, the ES cells, which would be heterozygous, contribute to the pup’s germ cells. With further mating, homozygotes can be obtained for study. Although technically demanding, the flexibility and precision of this method ensures its widespread application. A glance at recent databases indicates almost exponential growth in the number of reported mutations produced this way during the past 10 years (more than 1,000 published at last count). The majority of these were intended as loss-of-function mutations (null or knockout mutations or targeted deletions), although gene targeting is increasingly being used to produce other types of mutations (such as more subtle mutational alterations of a resident gene, sometimes referred to as “knockin” mutations). Some of the possible types of mutations are listed below.
Knockout or Null Mutations. Gene targeting has been most commonly used to disrupt the function of an endogenous gene. The simplest means of accomplishing this is to make a targeting construct with the selectable marker, such as neomycin resistance (the selection cassette), in a position that will disrupt gene tran-
scription or translation. This can be accomplished either by inserting the cassette in a critical region or by replacing a portion of the endogenous gene with the selection cassette producing a deletion. The selection cassette usually has its own promoter and polyadenylation sequences. Note that such a mutation cannot properly be called a knockout, or assumed to be a null mutation, until evidence has been obtained that no gene product is produced in the mutant. Expression of a partial gene product might result in a phenotype other than the null phenotype.
Expression Reporting. A variation on a simple knockout construct can provide information on the expression pattern of the targeted gene. For this purpose, a reporter gene (e.g., the E. coli gene lacZ or the jellyfish gene for GFP) is used without a promoter. The targeting construct is made in such a way that the reporter gene is in frame and is driven off the targeted gene’s promoter following homologous recombination. In this way, the reporter-gene product will be produced in all cells that would normally express the targeted gene. Usually, the reporter gene is studied in a heterozygote so that the other allele provides a functional gene product.
Point Mutations. Various schemes have been devised to produce mutations that are more subtle than complete loss of function. These usually involve a targeting construct that replaces the endogenous sequence with a homologous sequence containing a point (or other type) mutation. The trick is then to remove the selectable marker. This is most commonly done by using the bacterial Cre recombinase system, which is similar to the FLP recombinase system described in Figure 7-4. In the targeting construct, the selectable marker is made to be flanked by lox-P sites, short DNA sequences that are the substrate for Cre recombinase. Correctly targeted cell clones are then transiently transfected with the Cre gene, whose encoded product causes recombination between the lox-P sites, popping out the intervening DNA. The targeted allele is then left with a point mutation and a single lox-P site, which, if located in an intron, has no effect on the targeted gene.
Conditional Mutations. The Cre recombinase system (similar to the FLP recombinase system) has proved to be extremely versatile in gene-targeting schemes. The greatest potential is perhaps in the production of so-called conditional and tissue-specific mutations. The targeting construct is similar to that described above for making point mutations. The difference, however, is that a third lox-P site is introduced into a neutral position in the endogenous gene, in addition to the lox-P-flanked selectable marker. Then, following transient Cre expression, some cells will be recovered in which the lox-P-flanked selectable marker has been removed but two lox-P sites still remain. Any further expression of Cre will result in the removal of the intervening DNA, producing a deletion that can be planned to result in a null mutation. Provided the construct has been engineered so that the two remaining lox-P sites do not interfere with endogenous gene expression, normal mice can then be made with this ES cell line following
removal of the lox-P-flanked selectable marker. The final step is the removal of the lox-P-flanked DNA in specific tissues of the whole animal, which is accomplished by breeding the gene-targeted mice with transgenic mice expressing Cre, as a transgene, in specific tissues.
A variation on this scheme can be used to restore normal expression of a mutated gene. In this case, a lox-P-flanked selectable marker is inserted into a gene, by gene targeting, to disrupt its function. Function can be restored in specific tissues in the resulting mutant mice by mating with transgenics expressing tissue-specific Cre in order to remove the inserted deleterious DNA, leaving behind a single lox-P site in a noncritical position.
Limitations and Pitfalls of Transgenic Technologies
Variations in the design of any transgene or gene-targeting construct, as well as local features of the integration or target site, will affect the outcome of transgenic experiments. Integration of a transgene is random. Thus, its expression might be affected unpredictably by other promoters or enhancers at its integration site, or the transgene might be entirely silenced by integration into a transcriptionally inactive chromosomal region. This has been termed the “neighborhood effect.” Expression might also be influenced by epigenetic phenomena, such as methylation. Another chance event that will alter the intended experimental outcome, but which can be exploited, is insertional mutagenesis, described above. If the transgene happens to integrate in a position that causes the disruption of an endogenous gene, the experiment might be more informative about the endogenous gene than the transgene.
With gene targeting, a possible complication can arise if the selectable marker, which is essentially a foreign transgene, remains in the genome. Either this gene or its promoter could potentially affect expression of the targeted gene or neighboring genes. In gene-targeting experiments that involve deletions, it is possible to unknowingly remove cryptic regulatory regions located within introns and thus, potentially, to affect expression of nearby genes.
Phenotypic effects observed following any mutational change, whether through a transgene, gene targeting, or a naturally occurring mutation, are subject to what are called “genetic background effects.” The mutant phenotype might vary in different animals depending on what other genes that animal possesses (its genetic background). For example, a mutation might show a different phenotype in two inbred strains if those strains carry different alleles in other genes that directly or indirectly modify the phenotype of the mutant gene. These background effects will be largely unpredictable, but this situation provides material to identify and isolate modifier genes and thus to increase understanding of genetic pathways. The judicious use of inbred strains, in which the mice are theoretically 98% genetically identical, allows these background effects to be studied (e.g., see Lander and Schork 1994).
Animal-Cloning Technology
Recently, it has become possible to produce small numbers of genetically identical mice by the procedure of fusing individual cumulus cells from an adult female into individual enucleated eggs, thereby providing each egg with a diploid nucleus. This Cumulina family is in its third generation of transfers (Wakayama et al. 1998). Recently, mice have been cloned from fibroblasts derived from adult tail snips (Wakayama and Yanagimachi 1999a,b; Wakayama et al. 1999). In testing situations in which genetic variability is a problem, such clones could provide a uniform population.
POSSIBLE APPLICATIONS OF MODEL ANIMAL RESEARCH TO DEVELOPMENTAL TOXICOLOGY
Are Simple Toxicological Tests Possible?
Using the New Knowledge
The new knowledge gained from model animal research should be applicable to developmental toxicology in at least three important ways:
-
In developing more effective assays to test for environmental toxicants.
-
In assessing the risks of known toxicants.
-
In investigating toxicological mechanisms, the understanding of which will allow development of new therapeutic approaches to toxicant-induced defects.
In keeping with the third charge to the committee to evaluate how this information might be used to improve qualitative and quantitative risk assessment, this section deals primarily with possible new model organism approaches to toxicant detection and to the analysis of the mechanism of action of toxicants on developmental processes. The committee will draw upon these new approaches in Chapters 8 and 9 in proposing a multilevel, multidisciplinary strategy to improve developmental toxicity assessment.
Learning from the Ames Test
Ideally, scientists would like to have an inexpensive test system analogous to the Ames test, which is used, and sometimes misused, for detecting potential carcinogens. The Ames test was based on the assumption that many carcinogens are mutagens and that most mutagens are carcinogens. This test uses sensitized bacteria to measure the mutagenic activity of test samples. It is inexpensive, rapid, and suitable for testing many compounds under many conditions.
Is a similar test possible for developmental toxicants? Probably not. Whereas carcinogens act in a limited number of ways, primarily by inducing mutations in the DNA of somatic cells, developmental toxicants probably act by a large variety of mechanisms involving many aspects of development. Attempts to use very simple metazoans, such as hydra, to test for general effects on development have proved to be unsuccessful, because the results obtained were not interpretable as predictive of mammalian responses. On the other hand, the rodent assays, which are now considered the most predictive of human developmental responses, are expensive and slow and hence suitable for assaying only a small number of compounds. As currently performed, they also might detect only gross effects.
From the knowledge now being gained about developmental mechanisms, it seems possible that many developmental toxicants (those that defy the drug-metabolizing defenses of the animal) will prove to act by perturbing the signaling pathways involved in the many inductive interactions between cells and tissues. (As previously emphasized, signaling pathways appear to be highly conserved among most animal phyla.) However, this hypothesis remains largely untested. Do known developmental toxicants affect signaling pathways, and if so, is this how they cause developmental defects? Pursuit of these questions is a search of mechanisms of developmental toxicity. Using the simple and relatively inexpensive animal model systems amenable to genetics, scientists should be able to answer these questions. If the answers are yes, as is the committee’s hypothesis, it should be possible to design evaluation approaches for potential developmental toxicants with the use of animals, having sensitized genetic backgrounds and reporter-gene outputs, to detect effects on specific signaling pathways, as described in further detail below. Results will have to be used with caution, so that false positives are not overinterpreted. However, a judiciously applied battery of such tests could represent a major advance in developmental toxicity testing.
Sensitized Genetic Models for Testing of Specific Pathways
Some mutations essentially shut down a pathway by completely inactivating a component. Others that produce less inactivation cause no visible phenotype, although they bring the pathway close to a threshold of function and, therefore, render it sensitive to changes of activity of other components of the pathway—changes that by themselves might be asymptomatic. In signaling pathways, such sensitization can be accomplished either by raising or by diminishing the level of activity of a particular component, depending on its activating or inhibiting contribution. Change in the activity of a second component of this pathway due to mutation would cause the threshold to be crossed to altered function and phenotypic consequences. Hence, depending on how the assay is established, a phenotype might be enhanced or suppressed by perturbing a second element. In attempting to define a pathway, genetic screens for new dominant mutations that enhance or suppress the pathway-defective phenotype in a mutagenized, geneti-
cally sensitized strain of the test animal can be used to identify genes for new pathway components. Because the sensitization can often be designed to affect the pathway only in a particular tissue, this approach can succeed, even if a pathway is used in many places and many stages of development and null mutations cause phenotypes too pleiotropic to be interpretable.
Such sensitized strains should also be useful for identifying toxicants that modify the activity of a pathway component. Advantages of testing on animals having a tissue-specific sensitized pathway include the following:
-
The chemical’s effect can be assigned to a pathway without knowing the particular target protein, or even all elements, of the pathway.
-
Biologically relevant thresholds of effect can be sought, because low doses should suffice.
-
Phenotypes are more readily and reliably assessed, because they are revealed in a tissue-specific manner.
A variety of sensitized models and other approaches to assaying effects of known and potential developmental toxicants on specific pathways should be possible in the test animals considered here. All the model animal systems provide opportunities for developing methods of toxicity assessment and for investigating toxicological mechanisms (these opportunities are discussed in greater detail in Chapter 8). (Questions will be considered below on extrapolation to humans and the differences between animals and humans in uptake and metabolism of toxicants and in developmental processes.) Although the readouts of the assays involve scoring the development of various invertebrate organs, these readouts are chosen because they are likely to reveal effects of toxicants on conserved signaling pathways, and not because the organs are like mammalian organs. A relevant point is that human polymorphisms of signaling components might sensitize certain individuals to the detrimental effects of environmental agents.
Test animals for which genetic manipulations are difficult are not mentioned in the following section. For example, the FETAX test makes use of the frog Xenopus laevis. Although much has been learned about the development of Xenopus by mRNA injections into the egg, assays of cDNA libraries, and in situ hybridization, the organism is not yet amenable to easy genetic manipulation, and transgenesis procedures are in the early stages of use (Kroll and Amaya 1996). Thus, the committee does not believe that it equals the genetic model organisms for use in sensitive and ultimately informative assays of toxicants.
Caenorhabditis elegans
Suitability for Developmental Toxicology
Advantages of C. elegans include its low maintenance cost in the laboratory and properties mentioned above: its facility of genetic analysis, including rapid
reverse genetics using RNAi, its anatomical and developmental simplicity, and its transparency throughout the life cycle, allowing visualization of internal phenotypes at the cellular level and expression of fluorescent reporters, such as GFP, in living specimens. One possible disadvantage is that, as a soil organism, it might have evolved resistances to some chemicals that can act as developmental toxicants in higher animals. These differences could in principle be characterized and genetically modified. Another possible disadvantage is that its collagenous cuticle might be impermeable to many test compounds. However, since larvae and adults constantly ingest materials from their environment, compounds that are not rapidly degraded should enter the animal through the gut.
Assays with Sensitized Pathways
Many of the signaling pathways described in Chapter 6 have now been demonstrated to function in C. elegans development. Of particular potential utility for toxicological applications are pathways important for postembryonic development but not essential for viability up to that point, so they can be assayed in living animals. A few examples of such pathways follow. For some, sensitized strains are already available; for the others, they can be easily constructed.
Receptor Tyrosine Kinase (RTK) Pathways. One of the best-studied signaling pathways is not essential for viability in C. elegans but does mediate the induction of the hermaphrodite vulva in the hypodermis. The gonadal anchor cell releases an epidermal-growth-factor-like signal to nearby hypodermal cells that receive it via an appropriate RTK and the downstream components of a typical Ras signaling pathway. Defects in this pathway lead to an easily visible lack of a vulva (and, hence, inability to lay eggs) or to multiple vulva-like structures (Sternberg and Horvitz 1991). This organogenesis operates in the last larval stage, when the animal feeds actively. Sensitized strains for screens for enhancer and suppressor mutations are already available and could be used to test for effects of toxicants on pathway function.
Transforming-Growth-Factor (TGF) β Pathways. Also nonessential for larval viability are two distinct pathways responding to different TGFβ-superfamily ligands, which interact via receptor serine and threonine kinases with typical downstream Smad protein components. One is involved in controlling development of C. elegans larvae into a diapause form (the dauer larva) under adverse conditions, and the other is in control of body size and patterning of the tail of the male (Riddle and Albert 1997; Padgett et al. 1998; Suzuki et al. 1999).
Notch and Delta Pathways. Pathways involving the Notch-like receptor LIN-12 and a Delta-like ligand, also nonessential for larval viability, affect postembryonic gonadal and vulval development. Another pathway involving the Notch
homolog GLP-1 and a Delta-like ligand controls germ-line proliferation in the hermaphrodite. However, since GLP-1 signaling is also essential for early inductions in the embryo, further genetic modification would have to be carried out in order to use the GLP-1 variant of the pathway.
Wnt Pathways. Several recently discovered pathways involving WNT-like ligands and the homologous receptors appear to be important for establishing cell polarities and resulting patterning processes throughout C. elegans development; however, some of these pathways appear to be required only postembryonically and are nonessential for viability (Wood 1998).
Stress Pathways. GFP reporters have been made for various heat-shock proteins of the cytosolic unfolded protein pathway. Because C. elegans is transparent, transgenic animals carrying such reporters could provide a convenient readout of stress-pathway activation in response to toxicants.
Apoptosis Pathway. Much of the current fundamental knowledge of cell-death control was worked out in C. elegans. The pathway of interacting gene products that controls apoptosis during normal C. elegans development is well defined (Metzstein et al. 1998). It is considerably simpler than the pathways that are emerging in mammals, as is true for the simple model organisms in general; therefore, all aspects of the mammalian mechanisms are not represented in C. elegans. For example, there is only one cysteine protease, CED-3, in C. elegans, and there are at least 10 in humans (Salvesen 1999). Nevertheless, the control pathways are fundamentally similar. Sensitized pathways that can be produced by mutations in C. elegans should prove useful in testing for toxicants that cause developmental defects by way of apoptosis. As noted in Chapter 6, a variety of toxicants increase apoptosis in affected rodent embryos.
Behavioral Development Pathways. Although the behavioral repertoire of C. elegans is limited, the neural and molecular bases for several behaviors are well understood in the context of the completely mapped connectivity of its simple nervous system, which includes only 302 neurons. Because these behaviors are easily scored in the laboratory, assays for abnormal development or function of neuronal signaling pathways could provide simple, inexpensive, and useful screens for neurotoxins and other toxicants affecting development.
Genetic analyses have identified over 100 genes required for development of animals with normal movement. The “uncoordinated” (UNC) phenotypes resulting from mutations in these genes can be the consequence of either neuronal or muscular defects (Moerman and Fire 1997; Ruvkun 1997). Assays for toxicant effects on movement could therefore detect interference with the normal development and function of both muscles and the neurons that control them. Moreover,
a skilled observer can distinguish many different UNC phenotypes associated with specific defects that are genetically and often physiologically understood. Therefore, comparison of toxicant-induced abnormal movement to behavior of known mutants could rapidly provide initial evidence on the point of action of the toxicant.
More specialized assays could be developed based on the animal’s chemosensory abilities. C. elegans normally senses and responds appropriately to a variety of ionic and volatile chemoattractants (e.g., Na+, K+, ethanol, and ketones) and chemorepellants (e.g., acid pH, Cu2+, octanol, and benzaldehyde) in their environment (Bargmann and Mori 1997). These responses are mediated in the head by chemosensory neurons, which send information for control of movement via the major anterior ganglion, called the nerve ring, and the ventral and dorsal nerve cords. Chemoreception involves a large family of 7-membrane-pass cell-surface receptors, which are divergent from chemoreceptors in vertebrates. However, the neurotransmitter receptors (both ligand-gated channels and G-protein linked), neurotransmitter synthesis and release pathways, and G-protein-linked second-messenger pathways involved in chemoresponses are highly conserved between C. elegans and mammals (Bargmann 1998). Extensive genetic and neuron ablation studies have shown that defects in the development of these signaling pathways lead to abnormal chemosensory responses, which are easily and inexpensively assayed on agar plates in the laboratory.
Another potentially informative assay system could be based on the C. elegans dauer pathway. The dauer (enduring) larva is a state of diapause that allows a population to survive periods of limited food and overcrowding. Under such conditions, the animals produce a pheromone that can be detected by sensory neurons in the head. Presence of the pheromone induces molting of L2 larvae (see Figure 7-1) to an alternative form of the L3 larva called the dauer larva or simply dauer, which is resistant to dessication, does not feed, and has a low metabolic rate and an increased lifespan of several months compared with the normal lifespan of about 2 weeks. This response is mediated by a signaling pathway that is clearly homologous to the metabolically crucial pathway of insulin signaling in mammals. Genetic analysis has identified components of the pathway that are required for dauer formation under starvation conditions, as well as components required for preventing dauer formation when food is available (Riddle and Albert 1997). Defective function of either class of components is easily assayed in the laboratory.
Not only could such assays serve as the basis for testing toxicant effects on nervous system development and function, but also the extensive knowledge of the cells and molecules involved should make it possible to rapidly identify the components affected by a toxicant that causes behavioral defects.
Testing for Pathway Readouts Using Functional Genomics Technology
A limitation of the above approach is that only genetically well-defined pathways can be tested for toxicological responses, and only one pathway can be tested at a time. More global tests will soon be possible that take advantage of the complete genomic sequence of the nematode. (As discussed in Chapter 5, functional genomics refers to the profiling of all transcription from the genome.) The targets of most signaling pathways in development are transcription factors; thus, the end result of the signal is a change in the pattern of transcripts synthesized by responding cells. DNA microchips carrying ordered arrays of cDNAs representing all the C. elegans expressed genes will soon be available, allowing rapid comparisons of mRNA populations from different stages of development and from wild-type and mutant animals at the same stage. As data from such tests accumulate, it should be possible to define “fingerprints” of the mRNA changes that result from perturbation of specific signaling pathways. Once these fingerprints are defined, comparison of animals treated versus not treated with a developmental toxicant could indicate whether a particular pathway or pathways are being affected. Such DNA microchips with arrays for detecting 6,000 mRNAs have been successfully used to detect changes of gene expression in yeast cells in mating circumstances or not (Spellman et al. 1998) and in cultured human cells exposed or unexposed to serum (Iyer et al. 1999).
RNAi Testing of Candidate Genes Affecting Toxicant Responses
Several genes known to encode proteins affecting toxicant responses in mammals have homologs in C. elegans. Examples are the genes for the aryl hydrocarbon receptor (AHR) and the AHR nuclear translocator ARNT (Powell-Coffman et al. 1998), whose normal functions are not yet fully understood in any animal. As an approach toward understanding the role of such gene products, their normal functions can be conveniently investigated in C. elegans by reverse genetics using the RNAi technique described above.
Drosophila
Suitability for Developmental Toxicology
Both the low cost of rearing flies and the ease of constructing special stocks are special virtues of Drosophila for the testing of potentially dangerous chemicals. One difficulty, however, will be to determine the effective dosage of the chemical being tested and the most effective route of delivery. Development of the eye and wing takes place during pupal development, although most of the growth of the eye and wing primordia (imaginal discs) occurs in the larva. The pupal case is relatively impermeable to anything except very lipophilic compounds. These compounds can be dissolved in acetone and applied directly to the
pupal case. For example, juvenile hormone, an isoprenoid, readily passes through the cuticle. However, water-soluble test chemicals would have to be included in the food fed to 3- and 4-day-old larvae (third instar) or injected into pupae or flies (see later discussion of successful cases).
Assays with Sensitized Pathways
Because Drosophila has already shown the utility of sensitized screens for detecting new components in developmental pathways, there is little doubt that appropriate genetic stocks can be developed that would be useful for identifying potential toxic compounds. Such compounds would formally act like secondary mutations in reducing or increasing the activity of some other pathway component. In cases in which the sensitized strains are vigorous, as was the case for the sevenless mutants in the Simon et al. (1991) screen, mutant flies can be used without further genetic manipulations. In many instances, however, the sensitized strains are weak due to the fact that the signaling pathway is used for multiple functions, resulting in reduced viability. In these cases, the FRT-FRP system for producing clones of cells (see Figure 7-4), homozygous for sensitizing mutations, will be extremely useful. Careful construction of strains will also provide for “twin clones,” one for the homozygous mutation and the other for clones homozygous for the wild-type genes. This twin clone will provide a record of the loss of the mutant clone in these instances where the homozygous mutant cells become inviable due to chemical treatment.
The compound eye and the wing are two excellent organs for the analysis of potential toxic chemicals. Both are nonvital organs under laboratory conditions, and indeed eyeless and wingless flies are viable and fertile. Both organs are easily scored for developmental effects. Both have a relatively large precursor population so that the frequency of clones is relatively high, if the FRT-FRP system is used. Finally, many of the major signaling pathways function in establishing the regular pattern. A few examples will illustrate these features.
Ras Signaling in Eye Development. Earlier in this chapter, the committee described an example of a screen that uses a sensitized genetic background to identify genes in the receptor tyrosine kinase pathway involved in Drosophilia eye development (Simon et al. 1991). The use of sensitized systems to reveal chemical modifiers has in fact been well demonstrated for the Drosophila eye. Peptidomimetics that block isoprenylation interfere with RAS membrane binding and activity and, as shown in Figure 7-5, when injected into flies, can abrogate the abnormal phenotype of the activating RAS val12 mutation (Kauffmann et al. 1995). The same agents block activated Ras-induced tumors in mice, confirming cross-species relevance of the assay used as a chemical and genetic indicator.
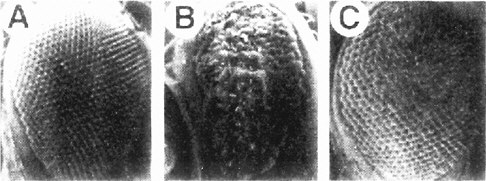
FIGURE 7-5 Effects of peptidomimetics on eye development in Drosophila. (Panel A) Normal compound eye with 800 ommatidia, each a bundle of seven photoreceptor cells and a bristle. (Panel B) The eye of a RAS val12 mutant in which the receptor tyrosine kinase (the sevenless pathway) is overactive. Note the disarrangement of ommatidia. (Panel C) A pupa was injected with a peptidomimetic that blocks isoprenylation, hence interfering with Ras membrane binding and activity. This reduces the activity of the pathway toward normal. The eye is more normal in development. (Kauffmann et al. 1995).
Transforming-Growth-Factor (TGF) β Pathways. The Dpp gene in Drosophila has been instrumental for identifying downstream components of the mammalian TGFβ cascade, such as the SMAD genes (Chanut and Heberlein 1997). Dpp is required in both the eye and the wing for normal development, and there are specific alleles of Dpp that strongly affect each of those organs. In addition, stronger alleles could be used in combination with a clonal analysis.
Hedgehog Pathways. Although Hh functions in the eye, its effect is not easily seen in small clones, presumably because of the diffusible nature of the HH protein (Heberlein et al. 1995). Effects might be more visible in wing clones, or it might be useful to develop sensitized systems for one of the downstream functions of the Hedgehog pathway, such as protein kinase A or the transcription factor cubitus interruptus (equivalent to the GLI oncogene in mammals).
Notch Pathways. Notch is required both for eye development and formation of the sensory components along the wing edge (Baker and Yu 1997). A wide variety of mutations specifically affecting the wing are available; therefore, clonal analysis should be useful.
Wnt Pathways. The Wingless ligand is required for establishing pattern in the wing and legs, and for limiting the eye domain. Clonal analysis of sensitized mutations should be efficient in detecting components of this widely used pathway.
Fibroblast-Growth-Factor (FGF) Pathways. The FGF pathway, which is a subset of the RTK pathways, is not used in either the eye or the wing in Drosophila, but it is required for proper tracheal branching, a process that is readily visible in living larvae by a variety of techniques. Consequently, specific interference with this pathway might also be detectable (Klambt et al. 1992). The tracheae are branched airways of the respiratory system of Drosophila. Their development has interesting similarities to angiogenesis in vertebrates, which involves FGF signaling.
Stress Pathways. Much of the original characterization of these pathways (e.g., the heat-shock response) was done in Drosophila. For convenient scoring of chemical effects, a variety of transgeneic strains have been constructed with reporter genes that are activated under conditions of stress.
Cell-Cycle Control. Drosophila might also be useful for the analysis of compounds that interfere with control of the cell cycle or that activate various checkpoint control pathways. Compounds stored in the egg during oogenesis support nearly all of the critical cell divisions required to achieve the free-living larval stage. During the larval stages, growth of the larva occurs by polyploidization and cell enlargement rather than by cell division. The only dividing cells in the larva are the precursors of the adult structures, and they increase in number prior to metamorphosis in the pupal stage. Most loss-of-function mutations in genes that are required for the cell cycle result in larval lethality, often at the larval and pupal boundary (metamorphosis), when the absence of adult precursor cells becomes critical. Consequently, it should be possible to test various chemicals for their ability to thwart the growth of adult precursor cells.
It could be argued that toxicant effects of the cell cycle are better studied in defined cultured conditions with, for example, culture-adapted differentiated cells. Indeed, cell lines are available, or could be prepared, with one or the other of the 17 intercellular signaling pathways functioning in culture, and these would be valuable for assessments of toxicant effects (e.g., effects on activin’s action as an erythroid differentiation factor in human erythroleukemic cell lines). The advantage of a proliferating developing system, such as the Drosophila imaginal discs described above, is that several signaling pathways simultaneously influence proliferation, in their full complexity of signal release via the Golgi and signal modification in the extracellular space, in addition to signal reception and transduction. Thus, the initial net cast to find toxicant effects might be wider than that in a cell culture system. Still, the latter could be very useful in focused identification and analysis of toxicant effects.
Possible Assays of Behavioral Development in Drosophila
Because of complex courtship behavior, feeding habits, and biological
rhythms, flies have become an excellent organism for identifying genes involved directly in behavior. Some of the major discoveries of genes controlling specific behavior traits have been accomplished in Drosophila. For example, the recent identification of genes controlling circadean rhythms in mammals (Takahashi 1995) owes its origin to the genetic screens that revealed the per locus in Drosophila (Konopka and Benzer 1971). Subsequent Drosophila screens have disclosed other components of the molecular clock—such as Timeless, Cycle, and Clock—which are also conserved in other organisms for photo-period regulation. Similarly, genetic screens in Drosophila have identified the mutants, such as Dunce, Turnip, Cabbage, Amnesiac, and Rutabaga that affect associative learning (Dubnau and Tully 1998). These genes have been shown to be part of the adenyl cyclase pathway.
Possibly the most elaborate behavioral patterns of Drosophila involve courtship behavior, which includes a series of species-specific activities essential for successful mating (Hall 1998). Starting from these successful pioneering studies, many of which were initiated in the laboratory of S. Benzer, Drosophila has become a useful organism for exploring the genetic basis for alcoholism (Bellen 1998), susceptibility to drugs (McClung and Hirsh 1999), aging (Lin et al. 1998), and other neurobehavioral traits. Because of the simplicity of its life cycle and rearing needs, the fly will continue to be an important first step in the identification of genes controlling a wide variety of behaviors. It should also be extremely useful for detecting deleterious effect of toxicants on specific behaviors, and as described above, the tests could be done on sensitized strains.
Zebrafish
The zebrafish is an appropriate model organism in which to test potentially toxic chemicals for several reasons. First, the zebrafish is a vertebrate and therefore of more direct relevance to the role of pathways in development of vertebrate-specific tissues and organs, such as the neural crest (Kelsh et al.1996; Schilling et al. 1996) and parts of the heart (Stainier et al. 1996), which are affected in many congenital anomalies. Second, chemicals can be added directly to the water (Stainier and Fishman 1992). Third, the zebrafish embryo is transparent, so tissue and organ development can readily be assayed. Viable, preferably dominant, mutations would be needed as the sensitized target to make such screens achievable on a large scale. Some viable pigmentation mutations have different heterozygous and homozygous phenotypes, indicating a dose responsiveness to the defect (Haffter et al.1996), and therefore are candidates, but these mutations have not been cloned, so the affected pathways remain to be determined. The potential is great for devising assays for chemical effects on development.
The zebrafish embryo has been used for toxicological assays (Ensenbach and Nagel 1995, 1997; Henry et al. 1997; Mizell and Romig 1997); however, it has not been developed for sensitized assays. Such assays are now feasible and would
be advantageous. Also, there are now a number of mutants for components of signaling pathways (e.g., BMP2, Nodal, and Cripto).
One of the original hopes for the study of zebrafish was that it would permit the genetic dissection of behaviors, including learning. In principle, the transparency of the embryo offers the opportunity for simultaneous assay of neuronal activity. Several behaviors are established early during development. For example, the embryo becomes motile and will respond to touch within the first day of postfertilization (Saint-Amant and Drapeau 1998), and eyes follow a striped drum (termed the optokinetic response) by 3 days (Brockerhoff et al. 1995). Screens have already identified genes that modify these activities (Brockerhoff et al. 1995; Granato et al. 1996). Mutations that perturb locomotory behavior have been shown to affect a variety of sites, including receptors, the CNS, or muscle (Granato et al. 1996). Rhythmic activity, such as circadian rhythms (Cahill et al. 1998) and cardiac pacemaking (Baker et al. 1997), are embryonic in their time of onset and can be dissected by genetics (Baker et al. 1997). Whether these behaviors may be modified is not known, but modification would provide a means to garner genes for learning, addiction, and memory. The effects of chemicals on the development of these behaviors remain to be examined, but the availability of specific behavioral assays is at hand.
A radiation hybrid map of the zebrafish genome has been completed, the map coverage being 81.9% of the genome. The map is based on a panel of 94 radiation hybrids (Geisler et al. 1999). A large-scale insertional mutagenesis screen in the zebrafish, with the goal of isolating about 1,000 embryonic mutations, is under way (Amsterdam et al. 1999). This approach is similar to the Nobel Prize-winning approach of isolating a large number of Drosophilia mutants—described in an earlier chapter.
Mouse
Transgenic Animals and Developmental Toxicology
Mice and rats have long been the mammals of choice for toxicological tests. Their advantage over the previously discussed model organisms is their similarity, as mammals, to humans. An advantage of mice over rats is the advanced state of the procedures for genetic manipulation and the large number of mutant and inbred strains already available. The disadvantage of mice and rats, compared with the other model organisms presented above, is their expense. To use them to test tens of thousands of compounds under a range of exposure conditions or in combinations is not feasible. Thus far, little use has been made of sensitized strains and reporter strains to improve toxicant detection and to learn more about mechanisms of toxicity.
Furthermore, little analysis has been done on the differences between mice and humans that lower the validity of cross-species extrapolations. There are
likely to be two main kinds of differences: (1) differences in the steps by which the active or activated toxicant is introduced to the embryo or fetus, and (2) differences in the components of developmental processes, making the toxicant have more effect or less effect on developmental function and resulting in a more severe or less severe developmental defect. Enough is now known about absorption, distribution, metabolism, and excretion, especially regarding drug-metabolizing enzymes (DMEs), to make systematic comparisons between humans and rodents possible. Many components of signaling pathways and genetic regulation, which are central to development, are also now known in test animals and humans, and those components could be compared systematically as well.
As summarized by Malakoff (2000), the use of mice in biomedical research and drug testing is expected to increase dramatically in the next few years, especially as the sequencing of the mouse genome approaches completion, and the increased use of mice will create opportunities in developmental toxicology.
Gene-Deletion Transgenics for the Study of DMEs
Only within the last 5 years have transgenic methodologies been brought to bear on the reciprocal relationship between the animal’s genetic constitution and its susceptibility to developmental toxicants. A particularly important and approachable set of genes are those encoding enzymes that metabolize exogenous chemicals. As discussed in Chapter 5, there are two major categories, the oxidizing enzymes (mostly P450 proteins) and the conjugation enzymes. There might be tens of these enzymes that metabolize most chemicals and hundreds more that metabolize a few chemicals each. In addition, several known transcription factors activate the expression of genes encoding these enzymes, and some of these factors themselves bind exogenous chemicals.
Much remains to be learned about the role of the various enzymes in generating and removing active toxicants, and the gene-deletion approach in the mouse has great advantages. At least in some cases the knockout mice are viable and fertile. One pioneering example in which gene-deletion transgenics was used concerns the dioxin-inducible mouse gene battery (Nebert and Duffy 1997), a group of genes believed to play an important role in developmental toxicity. Much more of this analysis can and should be done, both to understand the role of these enzymes in potentiation and detoxification and to define human and mouse differences for better-informed extrapolations of animal-test data.
Overexpression Transgenics for the Study of DMEs
Another approach to the study of developmental toxicity using transgenic animals is to overexpress, or ectopically express, a gene of interest in the embryo and fetus. For example, one could ask whether overexpression or ectopic expression of a gene encoding a particular oxidizing or conjugating enzyme either sensitizes or protects the embryo and fetus from the adverse effects of developmental
toxicants. Expression of transgenes can be driven by ubiquitous, constitutive promoters, such as β-actin, to induce expression in most, if not all, cells of the embryo. Alternatively, transgene expression can be limited to specific tissues by using tissue-specific promoters. Finally, transgene expression can be limited to specific stages of development and specific tissues by using one of several inducible expression systems such as the Cre-recombinase described above.
Transgenes as Biomarkers for the Activation of Stress, Checkpoint, and Apoptosis Pathways
Transgenic mice might also play a role in testing drugs and chemicals for potential developmental toxicity (see further discussion in Chapters 8 and 9). As scientists learn more about normal development and about the mechanisms of developmental toxicity, in part from studies using gene deletion or overexpression approaches, sufficient information should become available to construct transgenic mouse lines designed to contain biomarkers of the animal’s toxic response to a chemical. For example, a transgenic mouse could be constructed that contained a transgene consisting of the heat-shock promoter linked to an appropriate reporter gene, as has been done in Drosophila. This reporter, in turn, could be used to determine whether specific drugs and chemicals induce a stress response, which is often associated with developmental toxicity. Such a biomarker transgene could be used for in vivo developmental toxicity studies, or cells from appropriate tissues could be used for in vitro testing. Other stress and checkpoint pathways could be connected to reporter genes as well, as could components of apoptosis.
Signaling Pathways as Developmental Targets of Toxicants
As noted above, the mouse is at present the animal of choice for the selective knockout or replacement of genes. A large number of genes encoding components of many signaling pathways have now been eliminated, one at a time, as summarized in Chapter 6 (Tables 6-4 and 6-5), and the phenotypes of the single-gene homozygous null mutants have been examined. Surprisingly, many of these mutants achieve advanced development, living past birth, and some reach fertile adulthood. Many have discrete developmental defects. The current explanation of the viability of these mutants is that vertebrates have large, partially diversified families of genes for most signaling components, and the genes, which are expressed at different times and places in the embryo and fetus, encode proteins of partially redundant function. Defects tend to occur at times and places where there is no overlap of expression. A number of double and triple mutants have been made as well, and they have more severe effects.
Sensitized test mice can certainly be prepared for the testing of toxicant effects on development (i.e., mice with a particular pathway operating at or near the limiting rate in a designated developing tissue). Animals can also be prepared with lacZ or GFP reporter genes to give enhanced readout of effects on pathways.
Assays of Altered Behavioral Development in the Mouse
As noted in Chapter 2, developmental defects include functional as well as structural defects, but functional defects are less well diagnosed and less well understood. Behavioral defects comprise a large category of functional defects. The testing of environmental agents in animals for behavioral defects as an outcome of prenatal exposure is sometimes done, but not routinely. If an agent is suspected to have neurotoxic effects, a developmental neurotoxicity battery is required, and the suspected agents are mostly pesticides, fungicides, and rodenticides (see Makris et al. 1998; EPA 1998f).
In the developmental neurotoxicity tests, pregnant rodents are treated with the agent from day 6 after conception until 10 days after birth to include transfer of the chemical in the milk, and then the pups are tested in various ways from day 4 to day 60. Tests include a functional observational battery (FOB) and specialized behavioral tests.
The FOB includes cage-side observations of arousal; autonomic signs; convulsion; tremors; gait; mobility; posture; rearing; stereotypy; responses to touch, approach, tail pinch, and clicks; foot splaying; grip strength; righting reflex; body temperature; and body weight. The specialized tests include those of motor function, sensory function, and cognitive function. Motor-function tests involve tests of grip strength, swimming endurance, use of the suspension rod and rotorod, discriminative motor function, gait, righting reflex, and various ratings of spasms or tremors. Sensory-function evaluations include discrimination conditioning and reflex modification related to auditory, visual, somatosensory, and olfactory inputs and pain sensitivity. Cognitive-function tests include habituation of the startle reflex and classical conditioning related to the nictitating membrane, flavor aversion, passive avoidance, and olfactory conditioning.
Twelve chemicals, mostly pesticides, were tested by the above protocol, reviewed retrospectively as a group, and various developmental neurotoxic effects were distinguishable (Makris et al. 1998). Concordance was found between structural alterations of the nervous system and behavioral defects. The difficulties of interpreting the developmental causes of behavioral defects were discussed.
In the effort to assess functional defects, the committee favors greater inclusion of behavioral tests with other tests of toxicants. It would be useful to calibrate these tests with mice of known genetic defects affecting aspects of neurodevelopment. Null knockout mutants in homozygous and heterozygous condition could be used. The importance of modifier genes in the genetic background of each transgenic mouse line cannot be underestimated. It would also be useful to calibrate the tests with animals treated with doses of toxicants just below the level giving detectable structural defects in CNS development.
TABLE 7-3 Advantages and Disadvantages of Four Model Animals for Toxicant Testing
|
Organism |
Advantages |
Disadvantages |
|
C. elegans |
Relatively inexpensive, short life cycle, small size, large populations, extensive information on genetics, genomics known |
Some signaling pathways are apparently not represented (e.g., Hedgehog); short life cycle, so test compounds might persist long enough to complicate the study of sensitive developmental periods and of primary versus secondary effects |
|
Drosophila |
Relatively inexpensive, short life cycle, small size, large populations, extensive information on genetics, genomics known |
During pupal development organism is inaccessible to toxicants; short life cycle, so test compounds might persist long enough to complicate the study of sensitive developmental periods and of primary versus secondary effects |
|
Zebrafish |
Simple vertebrate, transparent, external embryos, best organogenesis model |
Relatively expensive, long life cycle, genetics and genomics research in progress |
|
Mouse |
Mammalian model, most similar to humans, targeted gene replacement possible |
Relatively expensive, long life cycle, embryonic development in utero and not as easily accessible for manipulation as other model animals |
Comparative Utility of Model Organisms
In summary, each of the four genetically tractable model organisms—C. elegans, Drosophila, zebrafish, and mouse—offers promise for development of tests to identify potential developmental toxicants, and each has advantages and disadvantages, as summarized in Table 7-3.
A useful approach, which the committee will present more fully in Chapter 8, might be to develop the more-rapid and inexpensive systems, namely, C. elegans and Drosophila, for preliminary large-scale assays, which can be followed up by more definitive tests using the vertebrate models, zebrafish and mouse, which, although slower and more costly, are more likely to give results relevant to humans.
GENE EXPRESSION AS DETERMINED BY IN VITRO AND ENGINEERED CELL TECHNOLOGIES
Mammalian cells in culture, as an inexpensive and fast complement to whole-animal (e.g., mouse and rat) studies, can be used for a wide variety of specific
purposes related to mechanisms of developmental toxicity and polymorphic variation in susceptibility of toxicants. The incisive use of cultured cells has been enhanced greatly by molecular methods in which genes of choice (from any organism including humans) can be introduced into such cells, for example, to express variant forms of DMEs and to investigate the relation of function to allelic variation.
Human allelic variants code for gene products different from those encoded by the wild-type allele, and it is important to characterize and, if possible, quantify any functional alterations. The fundamental techniques available for studying gene expression include (1) expression in vitro where DNA from the allelic variant or wild-type allele is transcribed to the mRNA and then translated into a functional, active protein in a cell-free extract; (2) high-level expression in cells from which proteins can be purified with relative ease; and (3) expression in eukaryotic cell culture in which the cell or the cell’s DNA has been modified (i.e., genetically engineered). These techniques rely on the gene (or at least the cDNA-encoded “coding region” responsible for generating the amino acid sequence of the protein). The cDNA encoding the allele being studied must first be cloned into an appropriate plasmid vector that includes regulatory sequences that drive and terminate transcription and a selectable marker gene. As outlined below, each of the aforementioned expression strategies is used by the investigator to meet specific needs. In general, these in vitro and cell-culture systems are very useful, because they are relatively quick, efficient, inexpensive, and use simple technologies when compared with the development of mouse lines or other animal-model systems.
There is a major shortcoming, however, in the in vitro and cell-culture expression systems. Single-nucleotide polymorphisms (SNPs) outside the amino-acid-determining region of the gene (e.g., splice junctions; promoter sequence; 5′ and 3′ untranslated regions; and enhancers upstream, downstream, and inside the gene) can have striking effects on expression of the gene under study, and the effects would generally not be realized, characterized, or quantitated by the in vitro and cell-culture expression systems (Nebert 1999).
Expression In Vitro
There are both reticulocyte lysate and wheat-germ lysate combined transcription and translation kits now commercially available to assess the interaction of the protein under study with another purified protein. To use these systems, a plasmid containing the cDNA of interest and a radiolabeled amino acid are added to the lysate. Although the protein of interest is not expressed at high levels relative to the total amount of proteins in the lysate, it is the only radiolabeled protein in the reaction mixture.
High-Level Expression in Cells
Proteins or enzymes of interest can be expressed at very high levels, which can be advantageous for studying certain functional alterations. The most commonly used systems include plasmid-transformed bacteria (Oudenampsen et al. 1990), baculovirus-infected insect cells (Kost and Condreay 1999), and vaccinia-virus-infected mammalian cells (Chakrabarti et al. 1985; Eckert and Merchlinsky 1999). All three systems allow high-level expression of cloned genes. In general, bacterial expression is the easiest to use. However, both the baculovirus-infected and the vaccinia-virus-infected expression systems are eukaryotic, and it might be particularly important to use them if post-translational processing or intracellular accessory factors are needed in the production of a functional gene product. Moreover, proteins expressed in any of the above systems can be given “tag sequences” to allow for rapid, specific purification.
Transient and Stable Transfections of Mammalian Cells in Culture
Yeast (Oeda et al. 1985), African green monkey kidney fibroblast COS cells (Zuber et al.1986), and vaccinia virus (Battula et al. 1987) were among the earliest expression systems in vivo. They have been successfully used to study DMEs. Allelic DNA can be transfected (passed into the cell) by microinjection or by chemical-DNA aggregation methods including calcium phosphate precipitation and liposome-mediated transfection. By using such methods, followed by antibiotic treatment to isolate cells that house the plasmid containing the selectable marker gene, cells can be transfected either transiently or stably. In transient transfections, expression of the gene is generated from extrachromosomal copies of the transfected plasmid and persists until the expression plasmid is degraded or diluted by cell passage. In general, 5% to 50% of all cells in culture contain the incoming gene, the DNA is not stably “integrated” in the cell’s genome, and the transfected cells contain many copies of the new genetic material. In contrast, for stable transfections, the incoming DNA is integrated (albeit randomly) into the cell’s genome. Cells expressing the gene under study are initially selected on the basis of co-expression of a gene that provides antibiotic resistance. After antibiotic selection, continued cellular propagation in the presence of the antibiotic will ensure that the gene of interest is expressed in a more or less permanent (i.e., stable) fashion—remaining after the cells are passaged on through many additional generations. The copy number of integrated genes is highly variable and can range from one to several dozen, and the genes are generally arranged in tandem (head to tail) at an “integration site” that normally cannot be directed, or controlled for, on an experimental basis.
For better gene expression in culture, one or more introns (even when they are artificially inserted) have been found to enhance gene activity in cultured cells (Palmiter et al. 1991); alternatively, small genes with fewer than 10 and
usually fewer than 5 exons and introns can often be transfected transiently or stably into cells in culture as the complete gene. Dozens of promoters have been studied over the past 15 years. They include the Drosophila heat-shock promoter (Hsp), the mouse mammary tumor virus long-terminal repeat (MMTV LTR) having a glucocorticoid response element (GRE), enhancer sequences from simian virus 40 (SV40) and herpes simplex virus type 1 (HSV-1), human cytomegalovirus (hCMV), thymidine kinase (Tk), and locus control regions of the metallothionein gene (LTR-MT) displaying variable potencies in driving transcription (Blackwood and Kadonaga 1998; Makrides 1999).
Both transient and stable expression of genes in mammalian cells have many advantages. First, genes are expressed in a native environment so post-translational modifications and subcellular targeting are authentic. Second, many transformed mammalian cell types are available for transfection. This allows for the selection of cell types—with the proper intracellular accessory factors or proteins—for enzyme activities that most closely resemble their in vivo counterparts. Finally, expression of gene products can be controlled by an increasing number of eukaryotic promoters. Thus, the levels of expression, including inducible expression, can be controlled.
Generations of cell lines that stably express DME genes represent the new generation of pharmacological and toxicological test systems (Langenbach et al. 1992). Expression of stably transfected DME genes in Epstein-Barr virus (EBV)-transformed human B-lymphoblastoid cell lines has provided cell lines that scientists can use to study and categorize numerous foreign compounds. Those drugs or chemicals can be classified according to the intermediates formed by way of the different metabolic pathways. More than a dozen EBV-transformed human B-lymphoblastoid cell lines are already commercially available (Crespi and Miller 1999). They contain anywhere from 1 to 12 human P450 and other DME genes (i.e., cDNAs), which retain their substrate specificity with regard to the metabolism of particular drugs or classes of pharmaceutical agents.
Some of these cell lines are already being used by pharmaceutical companies to determine whether a newly developed drug is metabolized by a particular DME. If, for example, the new drug is shown to be a CYP2D6 substrate, it is already known that humans differ by 10-fold to more than 30-fold in the activity of that enzyme (see Chapter 5). Poor metabolizers (the PM phenotype, which comprises 6% to 10% of Caucasian populations) would thus be expected to metabolize the new drug more slowly than extensive metabolizers (EM phenotype) and much more slowly than ultra-metabolizers (UM phenotype). If the parent drug is toxic, the PM individual would be at increased risk; if the CYP2D6-mediated metabolite is toxic, then the EM and especially the UM individual would be at increased risk. Dosage of the drug can therefore be adjusted to the patient’s genotype before the physician prescribes the new drug. For drugs already on the market, molecular epidemiologists can search for possible associations between reported differences in birth defects or other developmental problems and genotype of the
mother (or child, fetus, or embryo)—keeping in mind the range of drug doses that might have been given.
During the next decade, dozens more of the DME genes (alone and in combination) are likely to be expressed in the human B-lymphoblastoid cell lines or in other similar stably transfected cell backgrounds to determine which enzymes are responsible for either detoxification or metabolic potentiation of a particular drug under study. This information will be useful in the future of developmental pharmacology and toxicology, as well as molecular epidemiology.
SUMMARY
The committee has evaluated the state of the science for elucidating mechanisms of developmental toxicity and concludes that such elucidation, although not yet realized, can be achieved in the next decade for the following simple reasons:
-
Developmental biology has reached the molecular level of mechanistic explanations.
-
The accumulation of new and relevant information about vertebrate development is rapid (assisted greatly by research on model organisms, such as Drosophila and C. elegans).
-
The accumulation of genome sequence data for humans, mouse, rat, and Drosophila is rapid, adding to that already available for C. elegans, yeast, and many prokaryotes. Information on human polymorphisms and rare variants and their disease relatedness is increasing rapidly, as are data on the ever-increasing library of mouse mutants.
-
The methods are powerful and widely applicable, and the species barriers to comparative study have been greatly reduced in the past few years.
The committee begins in this chapter, and continues in the next, the third charge to evaluate how this information can be used to improve qualitative and quantitative risk assessment. In this chapter, the committee summarizes some of the techniques for modifying model organisms, including the mouse, for effective use in assays evaluating agents for potential developmental toxicity and for elucidating mechanisms of toxicity. The committee concludes that the methods and background knowledge are at hand to make incisive comparisons of humans and model animals so that the extrapolation of results from model animals to humans can be more accurate and useful for risk assessment.













































