Genetic Evaluation of Outbred Rats from the Breeder's Perspective
William J. White
Charles River Laboratories
Wilmington, MA
The origin of outbred laboratory rodents can be traced back to the 1500s. In the case of rats, populations of wild-caught animals were kept and bred to supply the blood sport of “rat baiting.” In doing so, small groups of rats from the much larger wild population were selected and bred, thereby providing a ready source for use in biomedical research in the 19th century. In the 1890s, a small number of these rats were brought to the United States from Germany to establish a laboratory-maintained research population. These initial animals were later randomly mated at a number of institutions with occasional infusions of a few animals from the wild and pet populations. During the early part of the 20th century, additional selection for a variety of traits, as well as the lack of a purposeful outbreeding system, likely caused a significant reduction in the individual genetic diversity of these noninbred rat stocks, which has continued until relatively recently.
POPULATION MANAGEMENT
Outbred population management was not seriously considered until the 1960s and even then was not widely applied to rodent production. Both commercially and academically, linking subpopulations and even starting new colonies did not include consideration of sampling error or population divergence. It was considered more important simply to begin breeding in a way that was not purposeful inbreeding with the hope that heterozygosity would be maintained. Unfortunately, the current interest in preserving and perhaps increasing heterozygosity in outbred populations, as well as addressing random genetic drift, is “historically”
hampered by the fixation of large amounts of the rat genome across all outbred stocks, compared with natural populations and perhaps humans.
SUPPLY AND DEMAND
Currently, approximately 75% of all rats and mice produced commercially (at least in the United States) are noninbred. Although inbred strains have the most prominent role in transgenic and knockout animal development, here too outbreds are still used for a number of applications. Worldwide, pharmaceutical and contract research organizations consume more than 70% of all commercially produced laboratory animals including rats. Because this demand will likely continue, we need to manage and genetically monitor outbred animals and, in particular, outbred rats correctly.
With proper management as the target, there are several things to remember. As mentioned by other speakers, random genetic drift occurs in outbred populations. Over time, two populations starting with equal gene frequencies of alleles arbitrarily designated as capital A and lower case a will undergo random genetic drift. Eventually, one population may develop an increasingly greater proportion of a single allele, A, and after many generations that allele may become fixed in that subpopulation while the other subpopulation may continue to segregate until such time as the proportion of one or the other of these alleles increases and also becomes fixed.
One of the goals of this meeting has been to consider how to use genetic monitoring and allele frequencies to assess subpopulations for the purpose of determining relatedness and presumably for management interdiction. At Charles River, as elsewhere, we began evaluating subpopulations of outbreds using biochemical markers. As expected, we have seen that many of them are monomorphic. We surveyed the three populations of Wistar Han rats, as shown in Figure 1 and Figure 2, using biochemical and immunologic markers. These populations were from unrelated commercial breeding facilities and represented different sources and dates of acquisition of breed stock. Although some differences do exist, there is striking similarity between the frequency of biochemical and immunologic phenotypes among all three populations. Existing differences are not consistent between subpopulations.
In trying to judge the similarity of two populations based on the distribution of a single marker, one can easily overlook contradictory information if all of the other makers are not considered. Even if a panel of markers is used, judgments regarding the similarity of populations will be limited by which markers are surveyed. The assumption that some standard panel of markers that can easily fingerprint populations for the purposes of authenticating them, as is done with inbred or F1 hybrid animals, does not consider the possibility that the distribution of phenotypes for any given marker can change over time even when comparing populations that are considered to be closely related.
|
Gene |
Source |
||||
|
Marker Type |
Designation |
Allele |
Pop. 1 (%) |
Pop. 2 (%) |
Pop. 3 (%) |
|
Biochemical |
ES-2 |
a |
78 |
80 |
80 |
|
d |
22 |
20 |
20 |
||
|
ES-6 |
a |
100 |
100 |
100 |
|
|
b |
0 |
0 |
0 |
||
|
ES-10 |
a |
100 |
80 |
100 |
|
|
b |
0 |
20 |
0 |
||
|
Pep-3 |
a |
100 |
100 |
100 |
|
|
b |
0 |
0 |
0 |
||
|
Hbb |
a |
29 |
30 |
40 |
|
|
b |
71 |
70 |
60 |
||
FIGURE 1 Wistar Han rat stocks: Partial genetic analysis of three populations using biochemical markers.
To interpret information gathered from phenotype assessment of outbred populations using biochemical, immunologic, or DNA makers, it is necessary to view this information as a whole and to consider not only similarities but also differences in the marker profiles. This inclusive information can best be obtained through the calculation of certain genetic monitoring statistics such as fixation index, estimate of polymorphism, conformity to Hardy-Weinberg equilibrium, and an estimate of average heterozygosity (Hartle 1988). Such computations yield a single number that considers all markers surveyed without unduly emphasizing a single marker, which may not be representative of the amount of divergence between two subpopulations if all possible markers are surveyed.
VARIABLES AFFECTING COMPARISONS OF SUBPOPULATIONS BY GENETIC MONITORING
Sample size is an important variable in any such analysis of subpopulations using an array of markers. Because outbred populations have considerable individual variation, small sample size may not truly yield a distribution reflective of the subpopulation, hence, 10 rats sampled may suggest one distribution whereas 100 rats may provide a different distribution and a better estimate of the subpopulation in question. It is also important to consider the “effective population number” and to make the appropriate corrections when analyzing data (Hartle 1988). The effective population number can be altered by age-related differences in reproductive rates as well as unequal numbers of males and females in a particular breeding scheme. It can also be affected by inequality of litter size due to production practices such as consolidation at birth or limitations placed on litters used for selecting future breeders. Unequal population numbers can also be an important variable inasmuch as population size can change generation to generation depending on production goals and other issues that may increase or decrease the population. Overlapping generations can also affect these analyses because the entire population does not progress to the next generation at exactly the same time.
CAUSES AND AMELIORATION OF GENETIC DIVERGENCE
There are three methods by which genetic divergence occurs. The first method is mutation, which can result from a number of physical/chemical processes. The chance of retention of any mutation in a population is relatively low. The second method is natural selection, which probably has a limited role in laboratory populations especially when rearing practices and the environment are relatively constant. The third method is unconscious selection wherein future breed selection is biased unconsciously by practices such as preferentially breeding good-tempered animals, animals with large litter size, or animals with lack of runted offspring. Such selection practices unconsciously favor phenotypes (and hence genotypes) that contribute disproportionately to the pool of future breeders.
The principal method for minimizing genetic divergence that occurs between geographically separated colonies (that is, subpopulations) is the trading of breed stock (migration) between colonies. Without migration, random genetic drift can be expected to cause at least moderate genetic divergence over time among outbred colonies derived from the same source. Migration of animals between colonies can be viewed as a form of genetic glue that holds colonies together and sets a limit on the amount of genetic divergence that occurs (Hartle 1988). Migration is not without its difficulties inasmuch as other factors such as the potential for microbiologic contamination of existing colonies must be considered in
the migration process. This risk can be minimized by indirect migration to and from a cesarean-derived isolator-maintained foundation colony.
Besides genetic divergence, colonies of outbred animals can differ significantly based on how and how many breeders were selected for the startup of the colony. This degree of difference is sometimes referred to as a genetic “bottleneck” or the founder effect. As can be seen in Figure 3, a comparison of the process of inbreeding with random mating of a colony beginning with 5 or 80 pairs of breeders, the amount of inadvertent inbreeding and hence fixation of alleles overtime can be influenced by the number of breeders selected to start the colony. If too few breeders are selected from the founder colony, the true allele frequencies in that colony will likely be misrepresented through sampling error in the newly established colony. This error can be magnified if a purposeful out-breeding system is not in place to adequately avoid inadvertent inbreeding and thus maintain the diversity of the population (White and Lee 1998). Random mating will not achieve this diversity, particularly if there is only a small number of breeders to choose from.
Another common error that produces nonrepresentative sampling is using entire litters as breed stock when forming a new colony. In addition, if a foundation colony that is linked to subpopulations by a regular migration process of breeders is not present, selection of a single production colony as a source of breed stock for a new colony may also misrepresent the genetic diversity present in all of the production colonies being maintained by an institution or breeder
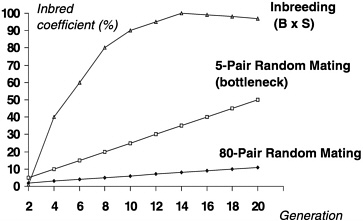
FIGURE 3 Coefficient of inbreeding with different colony size and mating systems.
(Figure 4). In the absence of a foundation colony, all of the production colonies must be sampled to develop a new production colony (Figure 5).
DEVELOPMENT OF A FOUNDATION COLONY-BASED OUTBRED PRODUCTION SYSTEM
The majority of commercially produced rodents are raised in barrier rooms that use very large breeding populations. The average rodent production room at our company is 2200 net square feet of floor area and will house approximately 60,000 rats that produce, in the case of outbreds, about 4000 rats for sale per week. In constructing our foundation colony-orientated outbred production system for CD rats, we began by identifying the 27 existing colonies of these animals worldwide and examining their stocking and production histories. To preserve the maximum amount of genetic diversity, we searched for colonies that had been separated for at least 5 to 10 years and found eight colonies that had been separated from each other (no infusion of new breed stock) for 12 to 22 years. We then selected 100 individuals (50 males and 50 females) from these eight colonies and placed them in a barrier room. Each group of 100 animals was designated as a separate line, and we began a circular-paired mating system from
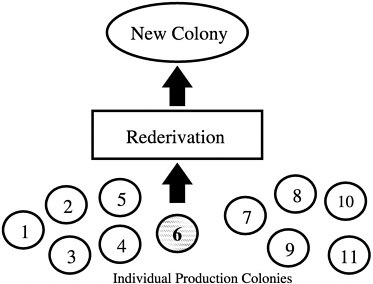
FIGURE 4 New colony set-up using only one of 11 colonies.
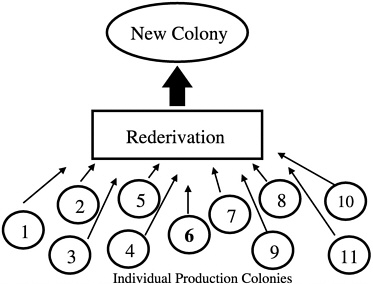
FIGURE 5 New colony set-up using animals from all colonies.
which we derived pregnant females for rederivation and subsequent selection of breed stock for population of new barrier rooms. We selected one pup per litter from 200 litters so as not to overrepresent any breeding pair (Figure 6). We repeated this process for each new breeding colony that we set up and began the process of closing breeding colonies and resetting them up with new stock using this system. We chose cesarean rederivation over embryo transfer rederivation because of the ease by which the process can be conducted in rats, which was important given the large number of pregnant females that had to be sampled.
The overall process to stock a barrier production room using a barrier room-maintained foundation colony was as follows. Pups were obtained by hysterotomy under aseptic conditions using a dual laminar flow hood technique. Rederived pups were aseptically transferred into 3-foot semirigid isolators and cross-fostered onto lactating females of defined flora status (Charles River Altered Schaedler Flora [CRASF]). Extensive health monitoring was conducted on both the environment and the foster mothers within the isolators throughout the course of the postpartum and weaning periods. Eight- to 12-week-old pups were packed into self-contained transport isolation shipping devices using aseptic technique and flown to the production site. Upon receipt at the production site, the integrity of the isolators was examined, and the shipping isolators were then
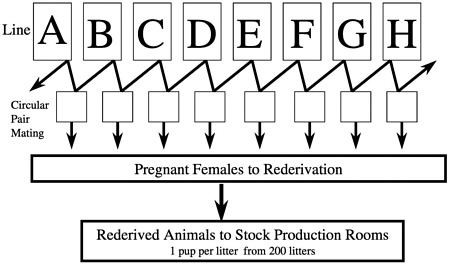
FIGURE 6 Crl:CD®(SD)IGS BR foundation colony system for producing stock for forward migration.
connected to a transfer isolator for unpacking. The animals were then transferred into the production room using a transfer isolation port built into the room that was connected to the transfer isolator using a plastic sleeve. All steps of the process were done using aseptic technique so that animals received into the barrier room retained their CRASF status. Transferred animals were used to start up the new production colony.
From a microbiologic standpoint, the maintenance of foundation colonies in a barrier production room that is entered by people poses a potential microbiologic risk especially if a regular forward migration process is conducted in addition to colony startups. To eliminate this risk, it was decided to rederive a colony of 250 breeding pairs representing the eight lines into 20 semirigid isolators. Each isolator contained up to 13 cages of breeding pairs, 12 cages holding future breed and stock, and two cages holding animals used for health monitoring of the isolator. In the case of the CD IGS foundation colony, the 20 isolators produce weanlings each week that are used for future breed replacement, forward migration, colony setup, and limited sale of overproduction for specialized customer use (Figure 7). In addition to the CD rat outbred foundation colony, there is a foundation colony for Wistar Han rats and CD-1 mice as well as foundation colonies for all of the inbred strains produced by the company. More than 200 isolators of varying sizes are used for foundation colony maintenance with another 1200 isolators being used for animal production, special animal services, and rederivation.
The 20 CD rat foundation colony isolators are maintained by a circular-paired mating system within each isolator and a migration system whereby fe
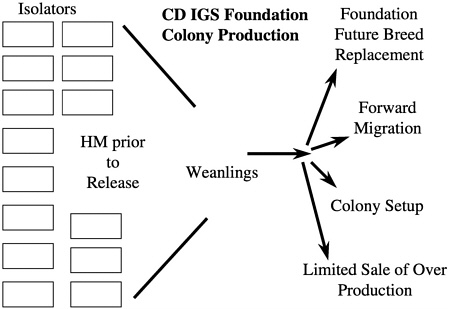
FIGURE 7 Schematic of foundation colony and its uses.
males from one isolator are migrated to another at designated times during the calendar year (Figure 8). These isolator transfers are linked to health monitoring, allowing isolators receiving transferred animals to be placed on quarantine hold until the results of health monitoring of that isolator have been obtained (Figure 9). This monitoring provides additional assurance that any change in the microbiologic status of the animals within an isolator can be detected before transfer of animals to new or existing colonies.
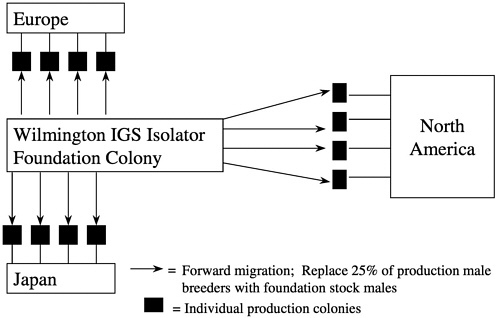
Forward migration of breed stock occurs from the foundation colony to the production colonies at 3-year intervals with 25% of the production male breeders being replaced in each colony by this migration process. Migrations are staggered over a 3-year period so that each year, one third of the colonies receive migrated animals (Figure 10). This process links all of the colonies to the foundation colony proactively making corrections without waiting for genetic monitoring to detect drift and a course of action to be undertaken.
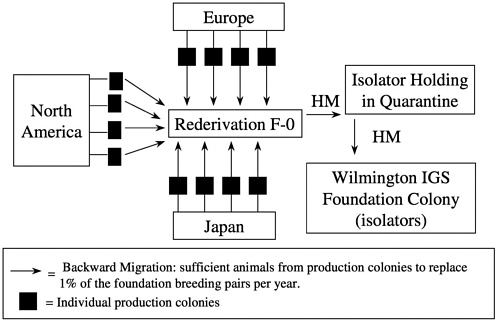
Unlike some migration systems, the genetic management of outbred foundation colonies by our company also includes backward migration (in-migration) at 5-year intervals. Each year, 1% of the foundation colony breeders is replaced by breeders brought back from the global network of production colonies of that stock. Animals received are mated, and the pregnant females undergo rederivation as previously described. Progeny are then held in extended quarantine during which time comprehensive health monitoring is conducted to ensure that
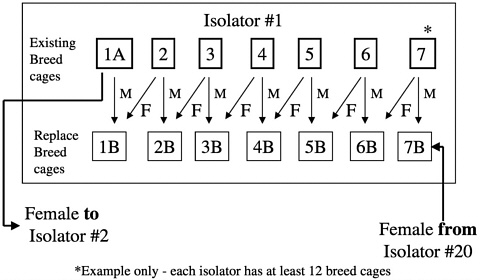
FIGURE 8 IGS outbred rat foundation colonies breed pair replacement.
the appropriate microbiologic state is maintained. Replacement breeders are then introduced into the foundation colony (Figure 11). This process of in-migration helps maintain genetic diversity in the foundation colony and ensures that the foundation colony reflects the variation found in the production colonies.
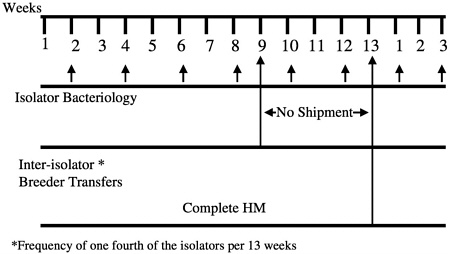
FIGURE 9 IGS foundation colony management environmental and health screening.
Within the individual barrier production rooms, we utilize a line-breeding system and rotate female breeders between lines. We have developed a number of operational procedures to minimize inadvertent inbreeding while ensuring consistency of production. Animals released for sale from production colonies are a mixture of all of the lines and hence a representative sampling of the entire colony. We refer to the overall genetic management system that uses an oubred foundation colony with forward and backward migration as the IGS system (White and Lee 1998).
CONCLUSION
I urge this group, as they consider methods for monitoring outbreds, to remember that “outbreds aren't inbreds.” We are not looking for authenticity when monitoring populations but for similarity. We are trying to preserve the great degree of individual variation in the population and hence should not expect individual uniformity as with inbred animals. Monitoring and management of outbred populations inevitably comes down to the issue of comparisons. It is unlikely that there is a single marker or set of markers, or a specific value in a population genetic statistic calculated from the frequencies of such markers, that can be used as an absolute cutoff in determining similarity of subpopulations. I urge the group that before undertaking such a determination, they include population geneticists in the deliberations.
So far, no one has discussed the role of cryopreservation in managing outbred populations. Some would argue that simply cryopreserving the foundation colony and using that for restarting or migration purposes would be the ultimate solution. Cryopreservation is important for preventing disastrous loss. One can argue, however, that cryopreservation is a selection method because only those embryos that can survive cryopreservation will survive into the reconstituted generation. It is unclear what traits, if any, might be linked to cryopreservation. If one is to maintain the diversity of an outbred population, some form of foundation colony of live animals is likely required even though cryopreserved animals from such a colony may be maintained as an insurance policy or be included in a backward migration program to provide some temporal refocusing of the foundation colony.
Overall, outbred rats will continue to play a role in research for the foreseeable future. The preservation of heterozygosity in outbred stocks and the linking of subpopulations are critical to the production of outbreds. Although genetic monitoring of outbreds can be used to compare subpopulations, such comparisons are relatively qualitative, are time sensitive, and cannot be depended on to completely quantify all genetic drift that has occurred. Proactive genetic management is the only practical way to ensure similarity of subpopulations.
REFERENCES
Hartle, D. L. 1988. A Primer of Population Genetics. 2nd edition. Sunderland, MA: Sinauer Associates, Inc. pp. 69-141.
White, W. J., and C. S. Lee 1998. The development and maintenance of the CRL:CD®(SD)IGS BR Rat Breeding System. In: Matsuzawa, T., and H. Inoue, editors. Biological Reference Data on CD(SD)IGS Rats. Yokohama, Japan: CD(SD)IGS Study Group. pp. 8-14.
QUESTIONS AND ANSWERS
DR. KAGIYAMA: Please restate why we need migration between sub-populations of outbred stocks.
DR. WHITE: If you maintain subpopulations without migration, you will experience genetic divergence. If you have an outbred stock such as Sprague Dawley or Wistar Han and you have 20 colonies (subpopulations), they are all going to drift independently unless you migrate animals between them to make them one functional colony. One could argue that you need only outward migration, but that argument is based on the assumption that whatever you call your reference colony will not vary significantly over time and will reflect all of the other subpopulations. We believe that some inward migration or back-migration is necessary to completely link the foundation with all of the production colonies (subpopulations), which are providing animals continually to the biomedical research community. It is the only practical thing you can do to proactively counteract genetic drift. By the time you obtain genetic monitoring data, the damage will have already been done. The practical approach is to be proactive in colony management and use genetic monitoring as a qualitative assessment of success.
DR. NOMURA: Please explain the idea of international genetic strains. After you mixed animals from different colonies to form the foundation colony, did you monitor them genetically? I would also like to see the actual genetic frequency data of the system you have theoretically described.
DR. WHITE: In developing the foundation colony, we made a decision to retain as much potential genetic diversity as existed in the production colonies at the time the foundation colony was formed. We looked for colonies that had been separated the longest without infusions of new breed stock, restarts of colonies, or the addition of animals into the breeding population by any other means. As you know, in commercial production, sometimes a colony will be developed and be in production for awhile and then be phased out because there is not sufficient demand for the animals being produced. Of the colonies available at that time, we were looking for ones that had been separated the longest, since they would have been more likely to have mutations or alleles that had become fixed, and thus, have made them different from other subpopulations of the same stock. In concept, this process is similar to construction of an F1 hybrid through a multiple F1 hybrid cross. We, in fact, did survey the individual colonies selected using
biochemical and immunologic markers; however, I did not bring those data with me today.
DR. SHEK: With respect to the markers for monitoring, we have biochemical and immunologic markers and are now looking at DNA fingerprinting and microsatellites for monitoring the IGS colonies. However, we and our consultants are not convinced that we currently have a set of markers that are extensive enough to do meaningful monitoring. We are continuing to look at minisatellites and trying to develop a more extensive set of microsatellites for comparing genetic divergence between colonies. If we use markers that are not highly polymorphic (for example, if we try to use microsatellites where there is a lot of band sharing), we will always get the results we want, which is that there is no divergence among our subpopulations because there is so much polymorphism there initially. This is an important point to consider even if biochemical markers are used. We are in the process of developing more effective DNA monitoring and results analysis techniques that should aid in comparison of genetic divergence between colonies.
DR. JACOB: One of the key points is how much monitoring is done. In my laboratory, if we are going to use a strain for something, we use a minimum of 600 markers. Hundreds are required for detecting diversity. At some point, you must characterize the amount of diversity to know how much diversity is there and to be able to construct the diversity in a population that you are seeking. You lose alleles every day you delay.
DR. SHEK: I disagree. Population geneticists have looked at this problem mathematically, and we are following their recommendations. Managing genetic divergence is definitely more important then the method or systems for monitoring it since monitoring only detects what has occurred and does nothing to counteract genetic divergence. Moreover, we are not trying to increase or decrease genetic diversity by artificial manipulation, but simply trying to preserve it.