5
The Importance of Data
Data are critical for characterization, calibration, verification, validation, and assessment of models for predicting the long-term structural durability and performance of materials in extreme environments. Without adequate data to verify and assess them, many models would have no purpose. An accurate structural durability model requires a database that consists of several master pedigrees of material systems with a uniform data set that represents a multitude of test conditions and several applications.
DATA AND MODELING
The Federal Aviation Administration (FAA) has been intent for several years on encouraging the sharing of data on composite properties and developing standards for these properties and designations. The goal, of course, is to ensure safe and efficient deployment of composite technologies for existing and future aircraft. FAA’s safety and certification initiatives were started in 1999 to address issues associated with the growing use of composites for the construction of planes. A large part of this effort is to coordinate the groups that update policies, issue advisory circulars, offer training, and furnish detailed background to support standardized composite properties and engineering practices.
One of the main efforts in this area is maintaining MIL-HDBK-17, which has included standardization and shared databases in PMCs for many years—the first PMC data set was approved in 1990. Sharing of standards, guidelines, and data is intended to achieve a number of goals, including (1) the control and stabilization of raw material, which is needed for continued safe and reliable use of PMCs in aircraft products; (2) the expansion of applications for PMCs in other industries to drive materials supplier developments; and (3) the promotion of consistent engineering practices. Overall, this effort aims to share composite databases and specifications throughout industry, thereby improving the efficiency of suppliers, users, and regulators.
This ongoing effort demonstrates the need for consistency in data collection and use and the difficulties this poses. Ideally, models should be universal, accounting for all materials and every possible damage mechanism or combination of mechanisms and every possible environment and condition of application. Because this is nearly impossible, a more practical approach would be to create a central database that would allow a model developer to select a PMC material system and its associated data based on the application and the anticipated damage mechanisms. The development of such a resource would clearly require an interdisciplinary team such as that described in the recommendation on teaming (see Chapter 3).
Finding: A certain amount of data is critical for modeling and life prediction.
However, not all data that are measured or can be measured are important to modeling and life prediction. The data needed should be intelligently selected for relevance and efficiency. This means, for
example, that experimentalists should not measure short-beam shear strength simply because they have always done so and there happens to be a test standard for it. A database should include properties because they support promising models or because they can be used to validate (or repudiate) promising models. Making use exclusively (or nearly exclusively) of existing ASTM or industry standards without considering their relevance is a recipe for disaster.
Data as defined today are expensive to gather, difficult to harmonize, and, many times, irrelevant to a specific model. Furthermore, data tend to become outdated and must be refreshed and improved. This calls for an overhaul of the way we collect, use, tally, and communicate data. A word has been recently coined that encompasses these concepts: informatics.
INFORMATICS AS A DISCIPLINE
Informatics is a relatively new field that is fast impacting the other fields it touches. A working definition of the discipline is the research, development, or application of computational tools and approaches for expanding the use of data, including those required to acquire, store, organize, archive, analyze, or visualize the data. It is important to note, however, that the field did not appear in academia fully formed but took many years to overcome academic inertia and gain the acceptance of researchers in the disciplines involved.
Biomedical Informatics
Biomedical informatics applies computer science, decision science, cognitive science, and organizational theory to the management of biological and medical information. The biological community did not universally agree on the importance of developing information resources and tools, even those that are today some of the most visible—GenBank, model organism databases, and the Human Genome Project. In retrospect, however, each of these efforts has revolutionized the accessibility and applicability of data and analysis tools to biologists, and many say these resources have created new perspectives for how biological research is done and interpreted. Extending these centralized data and analysis resource concepts to PMCs would enable researchers to make connections and draw conclusions about PMC mechanism-property relations that cannot be made with the current decentralized state of data, analysis, and algorithms in the field.
Physicians, clinical researchers, and biologists have long needed to organize and sift through large quantities of data. Since the 1970s, biomedical professionals have joined with computer and information scientists to find new ways of dealing with their data, and the field of biomedical informatics, particularly the subfield of bioinformatics, has exploded in the last 10 years. Biomedical informatics research has been able to capture significant amounts of funding, primarily from the National Institutes of Health, and there are now biomedical informatics laboratories at every institution doing major biological research. According to gradschools.com, 62 schools in the United States and another 29 outside of the United States currently have medical informatics or bioinformatics M.S. or Ph.D. programs.
The emergence of this academic discipline has opened up new areas of research, drawing in scientists who wish to make new discoveries by mining existing data. Biomedical informaticists have made basic research discoveries by creating and then applying computational tools to find subtle patterns in vast amounts of data that were previously inaccessible to biologists. In one example, computational biologists were able to analyze the genome of an intriguing and poorly understood microorganism to find a DNA sequence—unrelated to that of any other known organism—that was critical for understanding how the microorganism, an enzyme, replicated its DNA. This enzyme has been exploited in the development of the polymerase chain reaction and resulted in a Nobel prize for its achiever.
As part of their research, bioinformaticists are constantly developing new databases and computational tools and publishing them on the Internet, providing a tremendous service for biological scientists. In general, informatics researchers generate algorithms to extract valuable information from massive quantities of raw data. When the informaticists provide these tools on the Internet, the larger community of biological scientists can use them for applications specific to their research, such as comparing gene sequences of various organisms to determine their evolutionary relationships.
Of the many bioinformatics databases, Genbank may be the most widely known and has set a number of standards for such resources. Researchers have come to routinely place data in this community archive for the common good, knowing that it can be freely used by anyone.1 In addition, all leading journals have adopted a policy that requires sequences to be deposited in the public databases and the corresponding access numbers to be cited in published articles. All publicly funded laboratories now consider it de rigueur to contribute sequence data to Genbank within 24 hours of its generation, even if there is no accompanying research paper.2
Genbank provided a centralized way to store and access this data, which now contains over 44 billion gene pairs. In part because of this tremendous resource, and in part because GenBank does not have an experimental division, GenBank cannot thoroughly verify the submitted data, much of which has never been experimentally validated. As a result, there are duplicate, conflicting, and erroneous entries in the database.
As with data from any other source, biological researchers must keep the nature of the submission and the curation process in mind when using data that are not their own. To some extent, such problems exist in all databases, even local, lab-based data, but the curation and semantic consistency of data are harder to assure as the size and number of sources of data increase. The resources allocated to database management, submission requirements, curation efforts, validation methods, and the nature of the included data, as well as the priority assigned to them, all affect reliability.
Biomedical Informatics as a Model
The materials science and engineering community may be able to learn much from the example set by biomedical informatics. Because most materials scientists are not biologists as well, they are not likely to see parallels between the two disciplines. For example, a materials scientist might believe that a gene sequence is always a gene sequence, just as a biologist might believe that a polymer is always a polymer, although neither is true.
In fact, gene sequences vary with many of the same factors of measurement and external conditions that PMCs do. Just as a gene sequence is identical only for clones, wet strength after exposure may be identical only for two adjacent coupons. Every person’s gene sequence differs; every 1,000 bases on average is different from one person to another. This variance in the gene sequence is sufficient to cause variations in body chemistry and performance, but it is very difficult to unravel cause and effect.
This is similar to the difficulty of determining how small changes in a polymer's structure due to processing may change the way its properties evolve during environmental exposure. The chemical structure of a particular polymer set as a reference is analogous to the setting of a map of a particular gene as a reference. Untangling how small variations in the sequence interact with other equally variable genes and with environmental variables is a huge challenge for a biologist. Similarly, understanding how a small change in polymer structure interacts with processing and environment changes to evolve properties is a huge challenge for the materials scientist.
Given these parallel challenges, some existing bioinformatics databases may serve as a model for the proposed polymer composites database. One salient example is dictyBase, at <http://dictybase.org>, which contains information on the Dictyostelium genome and functional genomics. It is a relatively new database, so at its inception the biology community understood the value of a model organism database and readily accepted the concept. The Dictyostelium research community is relatively small, with fewer than 200 laboratories actively focused on Dictyostelium as a research organism. However, in the first 18 months that dictyBase was available, 20,000 distinct computers in more than 30 countries requested pages, and many of those requests came from students in high school and undergraduate lab courses. It also provides more than 5,000 pages per week to people looking at the Dictyostelium genome and
|
1 |
J. McEntyre and D.J. Lipman. 2001. GenBank—a model community resource? Nature webdebates. Available at <http://www.nature.com/nature/debates/e-access/Articles/lipman.html>. Accessed February 2005. |
|
2 |
Further, the Director of the National Institutes of Health (NIH) recently issued a public access policy requiring that NIH-funded investigators whose research was supported in whole or in part with direct costs from NIH submit to the PubMed Central database an electronic version of the final manuscript upon acceptance for publication. Although the current policy allows the paper's posting to be delayed up to a year after publication, the effect of this policy on open-source access to data is expected to be profound. |
associated research data. These data do not include spurious hits from search engines and the like and represent intentional usage of the site.3 Other databases of similar size are the Saccharomyces Genome Database, at <http://www.yeastgenome.org>, and the Drosophila Genome Database, at <http://www.flybase.org>.
The popularity of these sites would seem to augur well for proposing a research project to develop a database for polymer composite materials. Today, more than 40 universities have laboratories focused on the development, characterization, environmental performance, or modeling and simulation of PMCs. More than 200 companies are listed in the Composites Sourcebook4 as buying and selling PMCs or their components (in addition to the five main defense integrators). Many other universities teach courses on these topics, and the number of companies in the supply chain with interest in such data could be much greater.
It takes three curators, two programmers, and between 10 and 20 percent salary support for the project leaders to run dictyBase. Currently, this cost is $450,000 annually. Because of the relative maturity of the program codes, there were no additional costs to set up the database. The dictyBase is based on modern architecture and automation and as such is also less expensive to run than some older systems.
Operating a database, however, is more than a matter of software. The curators are responsible for reading the papers, reviewing data, coupling sequence data to function, annotating gene function and literature, and generally interrelating information and capturing the quality of the evidence within the database. The programmers are responsible for the hardware and the software, for continuously uncovering better ways to enter, retrieve, and visualize the data, and to develop user-friendly analytical tools for the research community.
While collecting the data is difficult but important, merely having it in a spreadsheet is not sufficient. More is needed than simply dollars, although funding is important. Each successful bioinformatics database started with an agency’s or a research group’s vision for responding to a specific scientific need. Whether the database was initiated by a proposal from a principle investigator (or group of investigators) or a call for proposals from a funding agency, it is crucial to support researchers who are committed to devoting time, energy, and dollars to develop a community resource. Success in such an endeavor requires dedication and the attention of someone who has solid experience in accumulating and working with the data and who is now willing to branch out into informatics to provide this kind of service to the community.
Other examples of public bioscience databases include these:
-
Protein Data Bank (PDB), at <http://www.rcsb.org>.
-
National Center for Biotechnology Information (NCBI), at <http://www.ncbi.nlm.nih.gov>.
-
National Institute of Standards and Technology (NIST) databases, at <http://www.nist.gov/srd>.
-
Tetrahymena Genome Database (TGD), at <http://www.ciliate.org>.
PARALLELS FOR MATERIALS DATA
In biomedical informatics, the raw data being stored might be a gene sequence and related details on specific organism function (for example, raw data on gene X along with data on blood pressure, on heart rate, and on various biochemical pathways and their responses to stress) (Figure 5-1). This is a vastly complex issue because a single function is regulated by many genes, and there is not a one-to-one correspondence of cause and effect. In addition, the history of the organism, the environment, and other stressors all feed into the organism’s response.
An analogous issue in materials is the relationship between a material's structure and its properties: The raw data may include details of the material’s composition and structure; related information may be data on properties, response to environmental stressors, chemical changes that occur with certain exposures, time to failure under various conditions, and many others. Again, this is a vastly complex issue. Any property, such as strength, is regulated by many details of the chemical composition
|
3 |
Personal communication, W.A. Kibbe, dictyBase, Northwestern University. February 2005. |
|
4 |
Composites Sourcebook, Ray Publishing. Available at <http://www.compositesworld.com/sb/>. Accessed February 2005. |
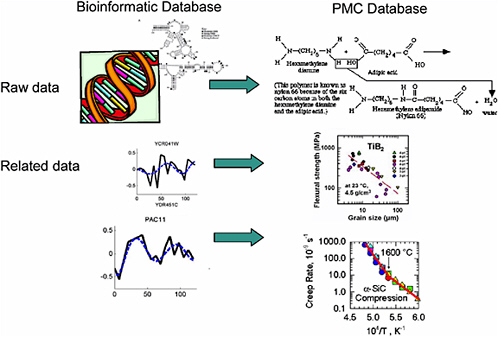
FIGURE 5-1 Parallels between bioinformatics data and materials data.
and microstructure of the specimen. Thus, the same type of algorithms that are being studied and used in bioinformatics to make connections in the vast sea of biological data can be explored and developed in the context of material informatics to learn about the mechanisms underlying a material’s response.
Biomedical databases can also provide examples for the materials community in the area of protected information. As export-controlled data are guarded to maintain competitiveness, patient data must be kept confidential for personal privacy reasons. In some biomedical databases (see Box 5-1), only certain people may access patient-specific results, while summaries of multiparticipant studies can be more widely distributed.
The key point for materials data is that a comprehensive tool—data coordinated with models—is far more useful than a typical handbook.
Recommendation: Mission agencies should offer sustained support to develop and maintain comprehensive information on PMC properties in a new materials informatics initiative. A steering committee on the needs of the PMC community should be formed to ensure that this effort is effectively targeted. This committee should be responsible for developing guidelines and overseeing operations for a national PMC informatics initiative.
The members of a steering committee should be a multidisciplinary team made up of design, structural, mechanical, and fabrication engineers and organic/physical polymer chemists, as outlined in the recommendation on teaming. It should have subcommittees to direct the activities for each class of material being considered for database inclusion. For such a committee to be effective and maintain its neutral position, it must have sustained funding from several sources.
|
BOX 5-1 Bioscientists developed the first public bioscience databanks in the 1970s and have been depositing data into free, Internet-based repositories since the inception of the Internet. In general, development and maintenance of a major databank cost between $1 million and $10 million per year, and this money most often comes from the National Institutes of Health (NIH). Depending on its size and complexity, a database may be maintained by a collaborative group spanning multiple universities, a government entity such as the National Center for Biotechnology Information (NCBI), or a single research laboratory. A relatively new and still fairly small example is the Pharmacogenetics and Pharmacogenomics Knowledge Base (PharmGKB). In April 2000, the NIH initiated a collaboration with Stanford University and granted it $9 million, spread over 5 years, to develop and maintain this database. Unveiled in February 2001, PharmGKB serves as a resource for clinicians, clinical researchers, and basic researchers to help them understand the relationship between human genetics and patients’ responses to various disease treatments. When queried with the name of a drug, disease, or gene, PharmGKB offers users results of clinical drug studies along with lists of related literature references and gene sequences. The results are coded with icons to help searchers to rapidly find the type of information they need. Unlike most bioscience databases, personal privacy issues have forced PharmGKB to restrict access to some of the data. Only scientists affiliated with registered projects may access clinical trial data with individual identifiers. However, anyone may view the pooled and summary data. Another atypical aspect of PharmGKB is that data submissions on human clinical studies come only from registered users. Many of the researchers who submit their data are required to do so as a condition of their funding from NIH/National Institute of General Medical Sciences. Most bioscience databanks allow submissions from anyone, and the scientific curators verify the data. Very often, scientific journals require that researchers submit their data to the databanks as a condition of publication. In other cases, such as the databases maintained by the National Institute of Standards and Technology, staff members search current literature and compile relevant information. The PharmGKB project director, Teri Klein, says that one of the biggest challenges has been encouraging investigators to share their information. She says that while biologists are accustomed to sharing many types of data, the concept of sharing the type of data included in the PharmGKB database is new and is not as widely accepted. One of the frequently underestimated costs of databases is that of staffing. To maintain and improve the database, the PharmGKB project currently employs 13 people. There are 10 full-time staff for the project and 3 part-time—the principle investigator, the project director, and an administrative assistant. Three curators verify submissions and search for publications containing relevant unsubmitted data; five software developers and two IT specialists deal with the data. The heavy presence of computational staff reflects the complexities of data representation and manipulation and the importance of providing a consistent architecture and analysis environment to the community. |






