After reaching consensus on the need for a New Taxonomy, the Committee deliberated extensively on the question “How do we get there?” In this context, “there” refers to successful creation of a system for acquiring and analyzing information relating the molecular profiles and health histories of large numbers of individuals. In Chapter 3, we describe the properties we would expect a Knowledge Network of Disease and the New Taxonomy to have and the type of Information Commons that would be needed to create them. However, we also emphasized that these resources will forever remain “works in progress.” As information technology, basic science, health research, and medicine undergo successive waves of change, both the content and structure of the New Taxonomy and Information Commons are expected to evolve, likely in directions that are presently impossible to envision. Consider, by analogy, early attempts to conceptualize the world-wide web compared to the use of the internet today. The Committee’s view is that we presently lack the infrastructure required to produce a dramatically improved disease taxonomy. Rather, we propose a path forward to develop the infrastructure and research system needed to create the Knowledge Network of Disease that we believe would be an essential underpinning of a molecularly-based taxonomy. We also address the sustainability of this initiative. Just as public leadership and investment played essential roles in bringing the world-wide web into existence, we believe such investment will be critical if we are to achieve a grand synthesis of data-intensive biology and medicine. However, we also recognize that, just as the world-wide web needed to pay its own way before it could truly flourish, the Knowledge Network and its underlying Information Commons will need to do the same.
The Committee believes that initiatives will be required in three areas to exploit the wealth of information now emerging on molecular mechanisms of
disease by creating a dynamic and comprehensive, yet practical and widely-used, Knowledge Network:
- Design of appropriate strategies to collect and integrate disease-relevant information. The Information Commons would be developed by linking molecular data to patient information on a massive scale. Creating a system for establishing this linkage for increasing numbers of individuals—and making the resulting data widely available to researchers—is the key step in moving toward a Knowledge Network and New Taxonomy. Such coupled data can be generated in several ways—including the modest-scale, targeted molecular studies on patient materials that dominate current practice. However, the most direct and effective discovery paradigm involves observational studies that seek to relate molecular data to complete patient medical records available as by-products of routine health care. Effective follow-up of the most promising hypotheses generated through such studies will require laboratory-based biological investigations designed to seek explanations at the biochemical or physiological levels.
- Implementation of pilot studies to establish a practical framework to discover relationships between and among molecular and other patient-specific data, patient diagnoses, and clinical outcomes. The new discovery model will involve the mining of large sets of patient data acquired during the ordinary course of health care. This is a novel, largely untested discovery approach. Pilot studies designed to identify and overcome obstacles to successful implementation of this approach will be required before a set of “best practices” can emerge.
- Gradual elimination of institutional, cultural, and regulatory barriers to widespread sharing of the molecular profiles and health histories of individuals, while still protecting patients’ rights. The sharing of data about individual patients among multiple parties—including patients, physicians, insurance companies, the pharmaceutical industry, and academic research groups—will be essential. Current policies on consent, confidentiality, data protection and ownership, health cost reimbursement, and intellectual-property will need to be modified to ensure the free flow of research data between all stakeholders without compromising patient interests.
A NEW DISCOVERY MODEL FOR DISEASE RESEARCH
The current model for relating molecular data to diagnoses and clinical outcomes typically involves abstracting clinical data for a modest number of patients from a clinical to a research setting, then attempting to draw correlations between the abstracted clinical data and molecular data such as genetic
polymorphisms, gene-expression levels, and metabolomic profiles. When discoveries are judged definitive and potentially useful, an effort is made to return this information to the clinical setting—for example, as a genetic or genomic diagnostic test. This model creates a large gulf between the point of discovery and the point of care with many opportunities for mis- and even non-communication between key stakeholders. For example, there have been approximately ten times more genome-wide association studies (GWASs) performed on individuals of European ancestry than other groups (Need and Goldstein 2009). The current model also fails to exploit the wealth of molecular data that are likely to be generated routinely in the future as personalized genomics and perhaps other personalized “omics” become routine in clinical settings. Perhaps most seriously, the current discovery model offers no path toward economically sustainable integration of data-intensive biology with medicine.
The Committee views it as both desirable and ultimately inevitable that this discovery model be fundamentally transformed. Instead of moving clinical data and patient samples to research groups to allow analysis, the molecular data of patients should instead be directly available to researchers and healthcare providers. The Committee recognizes that this is a radical departure from current practice and one that faces significant challenges, nonetheless, because we believe this new discovery model would have dramatic benefits, we believe that aggressive steps should be taken to implement it.
The changes in science, information technology, medicine and social attitudes—as discussed in Chapter 2—provides the opportunity to implement this model. Indeed, there are concrete instances of research initiatives already underway that substantiate the Committee’s belief that a special effort to implement its core recommendations can be achieved. In addition to the eMERGE Consortium discussed in Chapter 2, an excellent example is a collaboration between Kaiser-Permanente Northern California and the University of California at San Francisco (UCSF). Kaiser members were asked to participate in a study that would allow genetic and other molecular data to be compared with their full electronic health records. The study has faced major hurdles, and required more than ten years to progress from its conceptualization to large-scale acquisition of genetic data. A pivotal challenge was to build trust between Kaiser’s members, management, and oversight groups such as the relevant Institutional Review Boards. While all parties recognized it was essential that the Kaiser members who were being asked to “opt in” to the research study be fully aware of its aims, the outreach infrastructure required to educate members had to be created nearly from scratch. A second major challenge was acquiring funding to cover the cost of generating extensive molecular data that lacked direct and immediate relevance to patient care—a responsibility that Kaiser itself could not be expected to take on given the pressure to constrain health-care costs. Moreover, changing perceptions about what constitutes appropriate informed consent required costly and time-consuming reconsenting of the participants.
Nonetheless, the ability of committed investigators—working within strongly supportive institutions—to overcome these obstacles has been impressive: nearly 200,000 Kaiser members have joined the study and large-scale data collection is now underway.
The pioneering UCSF-Kaiser study makes clear that a discovery model based on direct use of patient data is possible, even as its implementation faces significant hurdles. In order to address and resolve these hurdles, the Committee envisions the design of several targeted pilot studies. These studies would probe key aspects of this new research paradigm and demonstrate to healthcare providers the value of a molecularly informed taxonomy of disease. By demonstrating value for patients, the pilot studies will seek to lay the groundwork for a sustainable discovery model in which relevant clinically validated molecular data are routinely generated at the “point of care” because they meet the commonly accepted risk-benefit criteria that apply to all clinical test results.
PILOT STUDIES SHOULD DRAW UPON OBSERVATIONAL STUDIES
As emphasized above, the Committee believes that much of the initial work necessary to develop the Information Commons should take the form of observational studies. In this context, what we mean by observational studies is that, although molecular and other patient-specific data would be collected from individuals in the normal course of health care, no changes in the treatment of the individuals would be contingent on the data collected. This approach to discovery is already in use today, although most current initiatives draw in a very limited range of clinical data. Notably, many GWASs have compared the genetic make-ups of individuals who receive a diagnosis of a disease to those who do not (McCarthy et al. 2008). For example, GWASs comparing individuals with and without a diagnosis of Crohn’s disease securely identified a number of gene variants that implicate autophagy in the pathophysiology of Crohn’s disease while similar comparisons for Age-Related Macular Degeneration implicated complement factor H (McCarthy et al. 2008; Ryu et al. 2010). In other instances, clinically relevant genotype-phenotype correlations have been discovered in the course of observational studies performed during randomized clinical trials. For example, a randomized clinical trial was performed to compare the efficacy of different formulations of interferon alpha in the treatment of chronic infection with hepatitis C. A subsequent observational study used a GWAS to identify variation near the IL28B gene as strongly correlated with response to treatment (Ge et al. 2009). Tests for the genetic variants identified in this study are already in widespread clinical use (PRNewswire 2011; Scripps Health 2011).
The enrollment of individuals in these studies had no bearing on their diagnoses, treatments, or in most cases, anything else in their lives. The goal of these observational studies was simply to ask the question “Are there gene
variants in the general population that are associated with who ends up with a particular diagnosis or experiences a particular treatment response?”
While observational studies will be primary tools used to develop hypotheses about new and clinically useful ways to group patients, the findings emerging from such studies will need confirmation and investigation using other approaches. For example, there are likely to be a great many ways to classify patients based on molecular data, and only some will have clinical utility. In general, clinical utility will need to be evaluated using randomized clinical trials.
Observational studies will also need to be followed by functional studies that seek to determine the mechanistic basis of observed molecular associations with clinical outcomes. An example of this type of combined discovery path is the identification of BCL11 as a modifier of the severity of sickle cell disease. Initially implicated in this role in GWAS studies, the biological basis of the association was quickly determined by focused analyses that established that BCL11A acts as a repressor of fetal hemoglobin. It is the persistence of fetal hemoglobin into adulthood in patients with particular variants at the BCL11 locus that ameliorates the symptoms of sickle cell disease (Sankaran et al. 2008). We anticipate that laboratory-based research of this sort will be essential to elucidate the underlying reasons for observed associations between molecular data and clinical outcomes and that these mechanistic insights will play an essential part in establishing the Knowledge Network and guiding its use.
The Committee envisions pilot studies that would:
- Be of a sufficient size, as well as scientific and organizational complexity, to reveal on the basis of actual experience the most significant barriers to the development of point-of-care discovery efforts.
- Address one or more unmet medical needs for which deeper biological understanding of a disorder would likely lead to near-term changes in treatment paradigms and health outcomes.
- Include the generation and analysis of a range of molecular-data types potentially including, but not limited to genomic data (sequence and expression), metabolomic data, proteomic data, and/or microbiome data.
- Be led by an organization charged with delivering health care with strong partnerships with researchers.
- Be supported by research funding to establish a “proof of principle.”
- Involve partnerships with a broad array of stakeholders, both public and private, including health-care providers, patients, payers, and scientists with expertise in genomics, epidemiology, social science, and molecular biology.
- Seek to remove barriers to data sharing and provide an ethical and legal framework for protecting and respecting individual rights.
- Develop IT networks of sufficient scale to allow assembly analysis and sharing of the integrated datasets.
- Draw on laboratory research to assess the biological underpinnings of associations between molecular data and clinical outcomes.
- Establish validation standards for clinical, evidence-based decision-making.
Below, we outline two example pilot studies; the first, “The Million American Genomes Initiative”, is selected to pilot the use of one of the key layers of ‘omic information that is “ready to go”. This pilot project would help to populate the Information Commons with relevant data and facilitate learning how to establish connections with other layers. By focusing on health care recipients in diverse states of health and disease, this project would also help evaluate the new discovery paradigm by allowing correlations to be made between germline sequences and a vast range of phenotypes. The second “Metabolomic Profiles in Type 2 Diabetes” is disease specific and is designed to ensure the early introduction of a different ‘omic layer (metabolomics) into the Information Commons and to pilot evaluation of more targeted questions in the new discovery paradigm.
EXAMPLE PILOT STUDY 1:
THE MILLION AMERICAN GENOMES INITIATIVE (MAGI)
A natural pilot study that would contribute to the development of the Information Commons and Knowledge Network of Disease would involve the sequencing of the genomes of one million or more individuals and the establishment of appropriate infrastructure for drawing correlations between the sequence data and the medical histories of these individuals. In focusing on a pilot study involving complete sequence data, we do not intend to elevate sequence data above other data in their importance to the Knowledge Network. Instead, this proposal recognizes that sequencing methods are “ready to go,” or nearly so, for very-large-scale implementation and the acquisition of such data in a point-of-care setting would, of necessity, require addressing key challenges related to informed consent, protection of data, data storage, and data analysis that will be common to all types of data. This proposal also recognizes that sequencing on this scale will inevitably be undertaken in the near future in an effort to make connections between human-genome-sequence data and common diseases. We view it as important to the development of the Knowledge Network that this effort be grounded in the new discovery model, which would make possible systematic comparisons of the molecular data with electronic medical records, now and into the future: that is, the study design should allow correlations between genotypes determined now and health outcomes that occur years or decades later.
The sequencing of one million genomes would include a sufficient range of individuals with different health outcomes and sufficient statistical power to detect associations. For example, amoxicillin-clavulanic acid is a widely used antibiotic that causes severe liver injury in one out of approximately 15,000 exposures. In a one-million-patient sample we would expect to include many individuals with this—and other similarly rare—adverse drug reactions and other medical conditions. It is also essential that the sample size be large enough to build a concrete picture of the distribution of gene variants in individuals free of specific diagnoses.
EXAMPLE PILOT STUDY 2:
METABOLOMIC PROFILES IN TYPE 2 DIABETES
Recent metabolomic profiling of blood samples from individuals who subsequently developed type 2 diabetes showed marked differences in the characteristics of branched-chain amino acids sampled from blood draws (Wang et al. 2011). These early analyses suggest the potential of metabolomic analyses to help identify those individuals at most risk of developing diabetes, and in particular, may help to elucidate the physiological steps involved in the transition between insulin-resistant pre-diabetes and full-blown diabetes. We therefore envision a pilot project focused on understanding this transition using metabolomic profiles in blood. This work would begin with targeted quantitative metabolomic studies transitioning toward more comprehensive metabolomic profiles over time. Such an effort, combined with knowledge gained from Pilot 1 and research from other layers of the Information Commons (such as the microbiome and exposome) could contribute substantially to strategies to delay or prevent the development of type 2 diabetes.
ANTICIPATED OUTCOMES OF THE PILOT STUDIES
The pilot studies are intended to lead to new connections between genetic or metabolomic variation and disease subclassifications, often with implications for disease management and prevention. More importantly, they will provide the lessons necessary to facilitate a more rapid transition in the way molecular data are used. For example, pilot projects of sufficient scope and scale could lead to the development of new discovery models, including those in which patient groups self-organize in recognition of shared clinical features and then pursue efforts to generate relevant molecular data. Such an initiative also would permit many logistical, ethical, and bioinformatic challenges to be addressed in ways that would benefit future efforts and lead toward the sustainable implementation of point-of-care discovery efforts.
Research to develop a Knowledge Network of Disease will need to resolve complex ethical and policy challenges including consent, confidentiality, return of individual results to patients, and oversight (Cambon-Thomsen et al. 2007; Greely 2007; Hall et al. 2010).
The Committee’s vision of a Knowledge Network of Disease and its associated benefits for future patients will become a reality only if the public supports a new balance between research access to materials and clinical data and respect for the values and preferences of donors. Ultimately, there should be no dichotomy between “patient data or materials” and “those who benefit from this research.” The patients who are giving their materials and data for research would also receive the benefits of research leading to a Knowledge Network and the resulting new molecularly-based taxonomy.
How might these ethical and policy challenges be resolved so that the pilot studies described previously might be carried out? The Committee recommends that an appropriate federal agency initiate a process to assess the privacy issues associated with the research required to create the Knowledge Network and Information Commons. Because these issues have been studied extensively, this process need not start from scratch. However, in practical terms, investigators who wish to participate in the pilot studies discussed above—and the Institutional Review Boards who must approve their human-subjects protocols—will need specific guidance on the range of informed-consent processes appropriate for these projects. Subject to the constraints of current law and prevailing ethical standards, the Committee encourages as much flexibility as possible in the guidance provided. As much as possible, on-the-ground experience in pilot projects carried out in diverse health-care settings, rather than top-down dictates, should govern the emergence of best practices in this sensitive area, whose handling will have a make-or-break influence on the entire Information Commons/Knowledge Network/New Taxonomy initiative. Inclusion of healthcare providers and other stakeholders outside the academic community will be essential.
An approach to these issues might include:
- Intensive dialog about the benefits of an Information Commons containing individual-centric data about health and disease. This dialog should include researchers and the public, patient representatives, and disease advocacy groups. Reaching out to communities that have been suspicious of research because of historical abuses would strengthen trust. At the workshop the Committee convened, we heard patient advocates and public representatives argue forcefully that more trans-
parency regarding research and more collaboration among researchers, research institutions, and the public would facilitate research. For example, when constructively engaged, advocacy groups have advanced biomedical research by helping to design studies that are attractive to patients, publicized the projects, helped to recruit participants, and raised money to help pay for the research (Giusti 2011; Patients LikeMe.com 2011).
- Exploration of approaches to informed consent that would allow patients to give broad consent for future studies whose details remain unspecified. Once provided with concise, understandable information on how their data and biological materials would be used for research, many patients are willing to consent provided they are treated as true partners in an activity that will provide broad public benefit (IOM 2010a; Trinidad et al. 2011). On the other hand, some patients will object generally to the research use of “leftover” specimens originally collected for clinical purposes or, more narrowly, object to their use in certain types of research. These concerns must be carefully addressed. Current approaches to informed consent for research rely on long, complex consent forms that may deter participation while doing little to help participants understand the nature of the research. As noted below, the Health Insurance Portability and Accountability Act (HIPAA) requires authorization or waiver for each specific research study: common interpretations of this requirement are so restrictive that investigators and Institutional Review Boards thwart or substantially delay research of the type that will be needed to develop the Information Commons.
- Strong public representation and input on oversight and governance. Public participation in biobanks and research projects would build trust (Levy et al. 2010) and help resolve issues that arise in the course of research, such as whether to offer to return individual research results to persons whose biological materials are analyzed (Beskow et al. 2010a) As noted earlier, the gray areas around the potential that researchers may have a “duty to inform” participants of clinically relevant results need to be clarified.
The HIPAA required the federal government to develop regulations for protecting the privacy of personal health information. The HIPAA privacy regulations, which are intended to protect patient privacy, inhibit research that requires widespread sharing and multi-purpose use of data on individual patients in several ways (IOM 2009): First, rich molecular data about an individual (particularly whole-genome sequencing) could be considered a unique biological identifier under HIPAA, even if overt identifiers are removed. Although a waiver of authorization to use identifiable health information may
be granted under certain circumstances, many health-care organizations are reluctant to participate. Secondly, because HIPAA does not allow authorization for unspecified future research or for several projects at one time, authorization must be given for each specific use of patient data. Thirdly, requirements for “accounting” to patients for research uses of data are burdensome and discourage data sharing. These regulations are strong deterrents to the kinds of pilot projects envisaged in this report.
The Committee found a need to re-interpret—or perhaps reformulate—HIPAA regulations, and is in agreement with the 2009 IOM report “Beyond the HIPAA Privacy Rule: Enhancing Privacy, Improving Health Through Research,” which found that the HIPAA privacy rule fails to protect privacy as intended (IOM 2009), and, as currently implemented, impedes important health research and imposes burdensome administrative requirements (IOM 2009). This IOM report concluded that stricter security would be a better approach to protect privacy than requiring patient authorization to use identifiable data for research. It recommended that much research based on existing materials and data be exempted from an amended HIPAA privacy rule (IOM 2009). For example, the Committee suggested that researchers be allowed to work with “secure, trusted, non-conflicted intermediaries that could develop a protocol, or key,” for linking identifiable data from different sources (IOM 2009). A biobank might serve as a trusted intermediary for the pilot projects described above, giving researchers only data and materials without overt identifiers but retaining a key to coded samples so they could update clinical information or re-contact patients or donors when appropriate. Furthermore, the IOM report recommended that “researchers, institutions, and organizations that store personally identifiable data should establish strong security safeguards and set limits on access to data” (IOM 2009). These precautions might include, for example, requirements for physical security of data and provisions in materials and data-transfer agreements that forbid researchers who receive de-identified data from trying to re-identify patients or donors or to contact them directly.
Furthermore, new approaches to informed consent are being proposed and tested. Some examples include: (1) incorporating highly specific patient preferences regarding use of their personal health information data (PCAST 2010), (2) using a short form for informed consent for participating in biobanks, with additional supplemental information for participants who desire more information (Beskow et al. 2010b), (3) de-identified data-based, opt-out model used by Vanderbilt and i2b2 (Pulley et al. 2010), and (4) consent for whole genome sequencing and study of all phenotypes, coupled with respect for individualized preferences regarding the return of clinically validated results (Biesecker et al. 2009). The Committee envisages that best practices and ultimately consensus standards will emerge from the different models of consent and return of clinically significant results to participants.
To accelerate the development of new tests and products based on a Knowledge Network of Disease, precompetitive collaboration between nonprofits and industry and among different for-profit companies would be desirable (IOM 2010b, 2011). The research needed to build the Information Commons, which will require projects involving vast amounts of data from large numbers of patients, will proceed more efficiently if such collaborations can be developed both between academia and industry and among for-profit companies that have historically been competitors (Altshuler et al. 2010).
These collaborations could include developing common standards and database formats and building infrastructure to facilitate data sharing. Consortia might be organized to share upstream research findings widely that have no immediate market potential but are critical to downstream product development. Examples of such upstream research include the identification and validation of biomarkers and predictors of adverse drug reactions. To build a flourishing culture of precompetitive collaboration, drug companies will need to overcome their reluctance to share all data from completed clinical trials, not just the selected data relevant to regulatory proceedings. Finally, and most significantly, guidelines for intellectual property need to be clarified and concerns about loss of intellectual-property rights addressed. Precompetitive collaborations will only emerge if individuals and organizations have incentives to join them (Vargas et al. 2010). The Committee believes that without such incentives, it will prove difficult or impossible to collect the new information that must be acquired before precision medicine, with its attendant benefits in improved health outcomes and reduced health-care costs, can become a widespread reality.
Similar principles apply whether the collaborations involve commercial entities or are confined to academia. To encourage the collection of materials and data, organizations and researchers who collect them should have first access to their use for research, while still ensuring their timely availability to others. The Committee does not envision the desirability or need, in the context of the research required to populate the Information Commons with data and derive a Knowledge Network from it, for the instant-data-release model adopted during the Human Genome Project. However, it does believe that timely, unrestricted access to datasets by researchers with no connections to the investigators who created them will be essential. The cost of populating the Information Commons with data precludes extensive redundancy in publicly financed research projects. At the same time, the size and complexity of these datasets—as well as the need for diverse, competitive inputs to their analysis—precludes giving any one group prolonged control over them. They must be regarded as public resources available for widespread and diverse research into ways to improve health care and to increase the efficiency of health care delivery.
Because the Committee is skeptical that one-size-fits-all policies can accommodate the conflicting values associated with incentivizing researchers and insuring adequate access to data, it believes that pilot projects of increasing scope and scale should put substantial emphasis on addressing the challenges associated with data sharing, rather than focusing exclusively on data collection and analysis.
COMPETITION AND SHARING IN THE HEALTH-CARE SYSTEM
A distinct and critical question is whether payers, such as health insurance companies, will provide access to their vast databases of patient and outcomes data and whether they will be willing to integrate these data with data from other companies and researchers with the goal of creating Knowledge Networks such as those described in Chapter 3. On one hand, these organizations recognize the potential value and cost saving that could emerge from such an effort. On the other hand there are considerable impediments. One of the main impediments is cultural: many of these organizations view their data as a propriety asset to be used in efforts to generate competitive advantages relative to other organizations. For example, large health-care systems and insurance providers are interested in developing decision-support tools for physicians that would cut down on the substantial waste caused by misdiagnosis or inappropriate treatment decisions. Integration of biological data, patient data, and outcomes information into Knowledge Networks that aggregate data from many sources could dramatically accelerate such efforts. However, if the data and the research results are shared, it would undermine one type of competitive advantage that large data providers might otherwise have. In this way, there is a tension between the sharing that would be good for the health-care system as a whole and the short-term competitive instincts of individual providers and payers.
Apart from the culture of competition there are other impediments related to cost pressures. Cost pressures within the health-care system are such that providers and payers are unlikely to be willing to invest substantially (or in some cases, at all) in the collection of biological data for research purposes. Over the long term, once such data have been shown to yield clinically useful information, it will become justifiable to expend health-care resources on the collection of actionable data, just as is presently done for standard diagnostic tests. However, until such data are shown to be clinically useful, it is unrealistic to expect that the Information Commons will become populated by biological data (such as genome sequences) acquired from providers and payers. Similarly, the information technology challenges associated with integration of large datasets and new disease classification systems are substantial. For example, Aetna is currently engaged in a multi-year effort to update its information technology systems to support the planned conversion to the ICD-10 coding standards. This effort alone will cost tens of millions of dollars. While the goals
of integrating datasets and changing classification systems are achievable in principle, they will be beyond the technical capacity of all but the largest and most technologically sophisticated providers and payers. Thus, the transition to non-proprietary Knowledge Networks into which all data would be deposited would have to involve strong incentives for payers and providers. This may mean that the government will ultimately need to require participation in such Knowledge Networks for reimbursement of health care expenses. At an even more fundamental level, the longstanding issue of equity in access to a sufficiently advanced level of health care should also be addressed if the data in the Knowledge Network is to adequately represent the diversity of our society.
Decision-making based on a Knowledge Network of Disease and the New Taxonomy, which will incorporate a multitude of parameters, will represent a significant adjustment in the practical work of the primary care physician. Given the demands on the time of physicians and other care-givers in the present health-care environment, few are likely to have the time or to feel qualified to interpret the results of “omics”-scale analyses of their patients. The importance of this issue will escalate over time as the Knowledge Network and its linked molecular-based taxonomy evolve into a system whose sheer complexity greatly exceeds current approaches to disease classification.
One concern is that the infusion of large molecular datasets into clinical records will reinforce a tendency many perceive as already crediting genetic and other molecular findings with more weight than they deserve. In extreme cases, this cultural bias has enabled the promoting and marketing of “omic” tests with no clinical value whatsoever (Kolata 2011). In other cases genetic or “omic” tests with real value in specific contexts may be over-interpreted and thereby occlude consideration of other relevant clinical data. To develop the Knowledge Network of Disease and the New Taxonomy that will be derived from it, health-care providers will need to develop much greater literacy in the interpretation and application of molecular data.
To meet these challenges, health-care providers will require both decision-support systems and new training paradigms. The decision-support systems will need to provide useful information about the propensity of patients to develop disease, facilitate a correct diagnosis, guide selection of the most appropriate strategies for disease prevention or treatment, inform the patient about the prognosis and management of the disease, and provide the opportunity for both physicians and patients to access more detailed information about the disease on an “as interested” or “as needed” basis. Whenever possible, such decision-support systems should enable shared decision-making by patients and
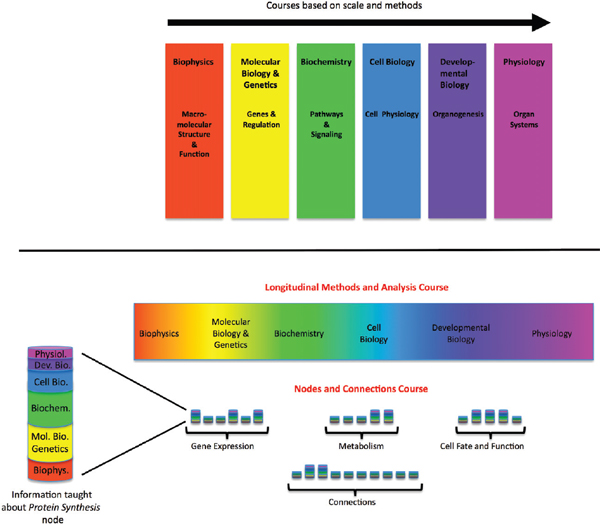
FIGURE 4-1 Curriculum for biomedical graduate program—proposed new model.
The current model of the first-year curriculum in a typical biomedical graduate program (top) and an alternative model (bottom). The multicolored bars in the nodes and connections course represent fundamental principles and essential facts about each key process integrated across scales.
SOURCE: Modified with permission from Lorsch 2011.
their care-givers. Such systems should be readily updatable as more information is acquired about disease classification, the ability of particular test results to predict disease development, progression, or response to treatment, and the success of particular disease-prevention and management strategies.
In order to prepare physicians for the use of a comprehensive, dynamically changing Knowledge Network, biomedical education will need to adjust. Lorsch and Nichols (2011) recently proposed that graduate and medical life sciences curricula would significantly benefit from a major shift away from the current discipline-specific model to a vertically integrated nodes-and-connections framework (see Figure 4-1). This model is not the only possible way of reorganizing instruction to reflect new knowledge about molecular processes, but it demonstrates how the development of a molecularly-based taxonomy, and the underlying Knowledge Network of Disease, could lead to major changes in education, while preparing students pursuing research careers to function in a scientific landscape that increasingly requires multidisciplinary approaches to solve biomedical problems (NRC 2009; MIT 2011). It also would give future physicians a more holistic view of biological processes, which reflects what will be required to fulfill the promises of genomics and personalized medicine (Ashley et al. 2010; Wiener et al. 2010).
The teaching model proposed by Lorsch and Nichols very closely mirrors the properties of the Knowledge Network of Disease described in Chapter 3. In this teaching model a given topic—for example, gene expression—would be taught in a vertically integrated fashion, with essential information all the way from the atomic to the whole-organism scale discussed. Adjusting teaching strategies to reflect the biological reality of the material has the potential to create significant synergies. Students may retain more knowledge of basic science when this information is directly connected to medicine. The enhanced ability to use the New Taxonomy in medical practice and research would reinforce the student’s conception of biology. Although it is beyond the scope of this report to suggest detailed reforms of the medical-school curriculum, the Committee would like to emphasize that full realization of the power of the Knowledge Network of Disease and the New Taxonomy derived from it would almost certainly require a major shift in educational strategy.
















