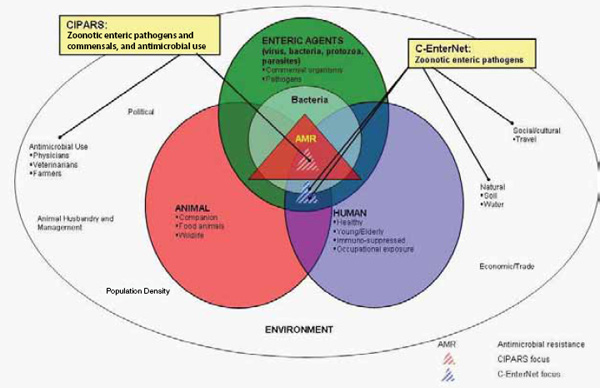
In the summer of 2011 Germany experienced one of the largest outbreaks of a food-borne infection caused by enterohaemorrhagic Escherichia coli (EHEC) with the serotype O104:H4. A large number of cases with bloody diarrhea and haemolytic uraemic syndrome (HUS) occurred. Never before was such a high rate of HUS cases observed in an outbreak caused by a food-borne pathogen. The events in Germany caused by EHEC O104:H4 in the summer of 2011 show dramatically how rapidly an infectious agent is able to develop into a major health threat for a whole country. The outbreak caused widespread concern among the population, turning soon into fear. People expecting safe and healthy food felt threatened. It changed the eating habits of the majority of the population, and it had enormous economic consequences, particularly for farmers producing salad ingredients. It resulted in a large number of seriously ill patients and in a substantial number of deaths. The burden of disease and the economic consequences
![]()
1 For the HUS investigation team of the Robert Koch Institute.
2 Robert Koch Institute, Nordufer 20, 13353 Berlin, Germany (BurgerR@rki.de).
have made it a tragedy for many. It is important to analyse this outbreak scientifically in order to learn from this unique event and to be prepared for comparable infections in the future. In particular, all the steps regarding detection of cases, diagnostic procedures, identification of vehicle and origin, and infection control measures, all the way to therapy, should be reflected carefully. Usually, even experienced physicians encounter only a few cases of EHEC-induced HUS in adults in their whole career. Therefore, the large number of cases in Germany represents a valuable source of information for future epidemics.
This manuscript summarises the work of the HUS investigation team of the Robert Koch Institute (RKI) and gives an overview of the work done by the colleagues in the Department of Infectious Disease Epidemiology at the RKI (G. Krause, C. Frank, D. Werber, K. Stark, and U. Buchholz), the Department for Infectious Diseases (M. Mielke and A. Fruth), and the RKI-Consultant Laboratory for HUS/EHEC at the University of Münster (H. Karch). Many additional colleagues were involved.
Epidemic Profile and Development of the Outbreak
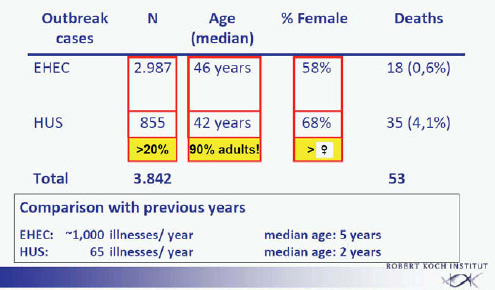
The extent of the outbreak becomes apparent by comparison with the average annual numbers of EHEC cases or HUS in Germany. In previous years about 1,000 patients per year were identified, with a median age of about 5 years. Of these patients about 70 per year developed HUS, with a median age of about 2 years (Frank et al., 2011a). In the outbreak from May to September 2011, approximately 3,000 EHEC cases were observed with a median age of 46 years, 58 percent of those patients were female, and 18 deaths were observed among the EHEC patients (0.6 percent). An additional 855 EHEC patients who developed HUS were identified (Frank et al., 2011b). This represents more than 20 percent of the total number of patients (3,842). The large majority of these patients were adults, the average age was 42 years, 68 percent of the HUS cases were female, and 35 deaths were observed among the HUS patients (4.1 percent). The total death toll was 53 patients (Figure A1-1).
Analysis of the incidence of HUS by the likely county of infection revealed that northern Germany was mainly affected. The same is true for cases with travel history; also for these patients the county of residence at the time of infection was northern Germany. Most cases were observed in the states of Schleswig-Holstein, Mecklenburg-Western Pomerania, Hamburg, Bremen, and Lower Saxony. Later in the epidemic, cases were found in all of the 16 German states. The incidence in the five northern German states varied from 1.8 to 10 cases per 100,000 persons. All other states had incidence rates with less than 1 case per 100,000 persons (Frank et al., 2011b; Wadl et al., 2011).
A substantial number of EHEC or HUS cases occurred also internationally during this time, particularly in the European Union, but also a few cases in the United States and Canada. Particularly affected was Sweden with 35 EHEC
and 18 HUS cases including one fatality, Denmark with 15 EHEC and 10 HUS cases, and France with 10 EHEC and 8 HUS cases. Single cases were found in 12 additional European countries. In the United States 2 EHEC and 4 HUS cases were identified with one fatality, and Canada had a single EHEC case. An epidemiological analysis revealed that—with two exceptions—all cases in this outbreak of EHEC or HUS found internationally were directly or indirectly associated with a visit to Germany during the weeks of the outbreak. Most of these patients visited northern Germany for a shorter or longer period of time during the peak of the outbreak.
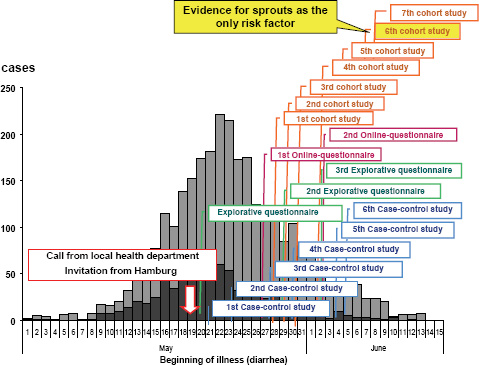
The RKI was notified about the outbreak by a phone call from the local health authority of the state of Hamburg on May 19, 2011. Immediately (i.e., the next day), the RKI sent a substantial team of experts to Hamburg in support of the local colleagues. The subsequent epidemiological analysis revealed in retrospect that the outbreak had in fact started at the beginning of May and reached the peak of cases on May 22, 2011 (Figure A1-2). Thus, there was an obvious and substantial notification delay (Altmann et al., 2011). Up to the moment of notifying the RKI, a large proportion of the infections had already occurred. After May 22 both the reported number of EHEC gastroenteritis and the number of HUS cases decreased (Wadl et al., 2011).
The team of epidemiological specialists sent to Hamburg started right away with initial explorative interviews. The team size was enlarged in the next days
SOURCE: Robert Koch Institute.
(up to 15 members), and a substantial number of case-control studies, additional explorative interviews, and cohort studies were started. As early as May 21 (i.e., 2 days after the RKI was notified), the first qualitative evidence for the role of vegetables was obtained. Raw milk products or products from raw meat, which frequently represent a source of infections with EHEC, had already been ruled out as the origin of infection in this outbreak. On May 22 the corresponding information was submitted to the European Early Warning and Response System and to the World Health Organization. Local public health authorities were warned, and initial interviews were given to the German press. During the next few days, information was provided on the website and in a series of press conferences and interviews. On May 25 (i.e., 5 days after the outbreak), the pathogen was identified from patient samples as EHEC O104:H4 by the RKI-Consulting Laboratory for HUS in Münster and the National Reference Centre laboratory for bacterial enteric pathogens at the RKI (Buchholz et al., 2011).
After a number of telephone conferences, the RKI together with the Federal Institute for Risk Assessment and the Federal Office of Consumer Protection
and Food Safety conducted a press conference advising on food consumption. Advice was given not to consume raw tomatoes, cucumbers, and salad in northern Germany. This recommendation was based on the increased risk of illness after consumption of these raw salads in northern Germany. Unfortunately, the majority of the press reported this advice as warning against salad from northern Germany.
Once the magnitude of the outbreak became apparent, the RKI immediately established a website providing all details about the infectious agent, updated as they developed, both for the medical specialists and microbiological laboratories in Germany and abroad and for the general public. Data sheets on the infectious agent and frequently asked questions, sometimes updated several times a day, proved to be an important source of information.
After mid-June 2011 only single cases of HUS occurred. On July 26 the RKI declared in a press conference the end of the outbreak because no new cases clearly associated with the outbreak had been reported for 3 consecutive weeks since the last newly reported illness on July 4.
Identification of the Infection Vehicle
In addition to the explorative interviews and case-control studies, cohort studies in disease clusters proved to be particularly helpful. Beginning on June 1, more than 30 cohorts were investigated in order to identify the vehicle of infections and to identify further cases. Particularly useful were cohort studies of travel groups that included international visitors or tourist groups from abroad. Here a close cooperation with foreign health authorities was instrumental. For a number of travel groups the length of stay, the particular location, and food consumption could be reconstructed in detail. Also, cluster analysis of patients associated with food consumption in different restaurant-associated outbreaks provided information. An analysis of billing data of guests at an affected canteen provided further data. In these studies a detailed investigation was performed using ordering information and additional details documenting the consumption as revealed by the corresponding bills. The most substantial evidence regarding the vehicle of infection was obtained by a so-called recipe-based restaurant cohort study ( Buchholz et al., 2011).
Sprouts as the Responsible Vehicle of Infection
In the course of the epidemiological analysis it became obvious that patient memory is not a reliable source of information. This proved to be particularly true because in these EHEC/HUS patients not only symptoms of gastrointestinal infection and impaired kidney function were observed but also major neurological symptoms, preventing reliable interviews. Therefore, the recipe-based restaurant cohort study was designed to obtain information independent of a functioning patient memory (Figure A1-3).
| Table 3. Relative Risk of Infection Associated with Sprouts and Other Raw Food Items in Univariable Analysis. | |||||||
| Food Item | Total Subjects Evaluted no. |
Subjects Exposed (Percent of Cohort) | Cases among Subjects Exposed (Attack Rate) no.(%) |
Subjects Not Exposed (Percent of Cohort) | Cases among Subjects Not Exposed (Attack Rate) | Relative Risk(95%) CI | P Value |
| Sprouts | 152 | 115(76) | 31(27) | 37(24) | 0 | 14.23(2.55-∝) | 0.001 |
| Tematoes | 152 | 50(33) | 14(28) | 102(67) | 0 | 1.68(0.77-3.62) | 0.18 |
| Cucumbers | 152 | 50(33) | 14(28) | 102(67) | 0 | 1.68(0.77-3.62) | 0.18 |
| Chinese cabbage | 152 | 45(30) | 13(29) | 107(70) | 18(17) | 1.72(0.77-3.71) | 0.17 |
| Radicchio | 152 | 45(30) | 13(29) | 107(70) | 18(17) | 1.72(0.77-3.71) | 0.17 |
| Lattuce | 152 | 45(30) | 13(29) | 107(70) | 18(17) | 1.72(0.77-3.71) | 0.17 |
SOURCE: Taken from Buchholz et al., NEJM, 365, 1763 (2011).
Ten cohorts with a total of 168 guests of a given restaurant in the city of Lübeck in Schleswig-Holstein were identified. All persons had dinner at the same restaurant between May 12 and 16. Eighteen percent of the guests consuming food at this restaurant showed bloody diarrhea or EHEC/HUS within 14 days (31 persons). All persons were questioned about which meals they ordered, using photos of the dishes as a reminder. Booking details and billing documents were utilized. Using these consumption data from the individual guests, the chef of the restaurant was interviewed about the detailed ingredients of each dish ordered by the guests. This included not only the major ingredients of each dish itself but also elements used for decoration of the dish or of the salad served separately. This approach provided reliable information about which food ingredients each guest had actually ordered and eaten. This interview technique and analysis had the major advantage that it was no longer necessary to depend on the memory of the guests to find out what they had eaten. Additional verification was obtained through photos taken at the table by a number of groups. These photos confirmed the details given for the nature of the ordered dish and its contents.
In univariate analysis the relative risk of disease was 14.2 times higher for persons eating sprouts compared to that of persons not eating sprouts (Buchholz et al., 2011). All 31 patients with EHEC/HUS had consumed sprouts. None of the guests who did not consume sprouts became ill. Based on these cohort studies, in a joint press conference of the RKI with the food safety authorities on June 10 the public announcement was made that sprouts were the vehicle of infection. The
earlier warning against the consumption of salad was now focused on a warning against consumption of the salad ingredient sprouts.
Origin of Bacterial Contamination of the Sprouts
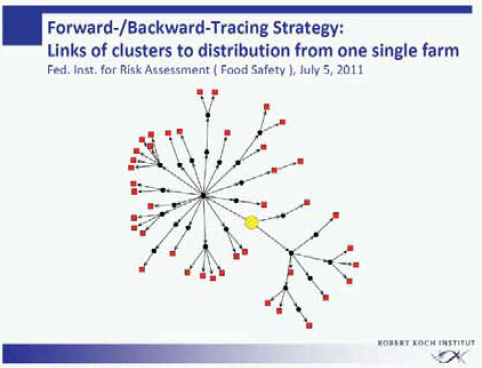
The more than 40 clusters within this outbreak were analyzed for a common denominator. The federal authorities responsible for food safety in Germany (the Federal Institute for Risk Assessment and the Federal Office of Consumer Protection and Food Safety) performed an intensive forward-backward tracing of the food supply chain of the various cluster locations (Figure A1-4). Through one or several distributors and intermediates, all clusters turned out to be connected to a specific food enterprise producing sprouts commercially. All infections within this outbreak in the state of Lower Saxony had in common that originally the supply of sprouts came from this single food enterprise.
Two clusters of infection independent of the outbreak in Lower Saxony provided information on the origin of the sprout contamination (Appel et al., 2011). Both clusters had definitely no connection to the sprout producer in Lower Saxony. One cluster consisted of so-called self-sprouters (i.e., consumers who grow their
SOURCE: Modified from Buchholz et al., NEJM, 365, 1763 (2011).
own sprouts at home from seeds provided by commercial suppliers). The second source of information was a small outbreak comprising 15 cases in the area of Bordeaux in France in mid-June. Detailed and labor-intensive tracing of the delivery channels revealed that the only common feature of the seeds used for growing sprouts in the food enterprises in Lower Saxony, in Bordeaux, and in the private households with the home-grown sprouts was a given lot of fenugreek seeds originating from Egypt. Fenugreek seeds (Trigonella foenum-graecum) are frequently used for the production of sprouts. The seeds are also used in many other food products (e.g., spices, cheese, and even tea) because of their very aromatic taste and intensive smell. The seeds are small (4-5 mm) and have a peanut-like colour.
Through a number of intermediates located in different countries this seed lot had been delivered to these three outbreak locations. No other common ingredient used for the production of sprouts was identified. This was clear evidence that contaminated seeds used for sprout growing were responsible for the outbreak (Appel et al., 2011). By nature, the epidemiological evidence is indirect or circumstantial but it explained the distribution of infections. The corresponding lot of fenugreek seeds was removed from the market. It is difficult to verify how complete this removal was.
“Stealth Food”
When the affected patients were interviewed initially during the first weeks of the outbreak, it became obvious that people do not remember in detail what they ate 1 or 2 weeks ago. Only in retrospect, after the second or third interview together with reports in the press, did they realize and remember that their dishes had in fact contained sprouts. Similar phenomena had been observed internationally in other outbreaks. In 2008, jalapeno chili peppers were contaminated with Salmonella Saintpaul in the United States. Chili peppers are used as an ingredient in tomato sauce-like salsa. The consumers were not aware that one of the spicy ingredients was chili peppers and, when interviewed, denied consumption of this food item, thereby delaying the identification of the vehicle. The identification of sprouts as a source in Germany within less than 3 weeks was quite rapid. The identification of the chili peppers took about 7 weeks. In another outbreak in 1996 with radish sprouts causing an outbreak of EHEC O157 in Japan, 7 weeks were required for the detection of the outbreak and 4 weeks to identify its source.
Microbiological Characterization of EHEC O104:H4
Once the outbreak had been recognized, EHEC O104:H4 was rapidly isolated from stool specimens of affected patients within a few days (Figure A1-5) (Askar et al., 2011, Bielaszewska et al., 2011). This is a rare serotype that had not been described previously in animals. As a rule, faecal contamination by ruminants is responsible for EHEC infections through vegetables or through
FIGURE A1-5 Electron micrograph of EHEC O104:H4.
SOURCE: Laue, Robert Koch Institute.
food products derived from animals (milk, meat). The usual EHEC strains (e.g., EHEC O157) are found in faeces of ruminants. EHEC O104:H4 has only rarely been identified previously in human beings (in a total of seven patients). A closely related EHEC strain, HUSEC041, was identified in 2001 by the laboratory of Karch at the University of Münster, Germany. Later, a few cases were identified in Korea in 2006, in Georgia in 2009, and in Finland in 2010.
A detailed microbiological characterization of EHEC O104:H4 was performed at the National Reference Centre for Gastrointestinal Bacteria at the RKI and the RKI-Consultant Laboratory of Karch in Münster (Bielaszewska et al., 2011; Brzuskiewicz et al., 2011). From the virulence markers, the outbreak strain was negative for Shiga toxin 1 and positive for Shiga toxin 2 (variant vtx2a of Shiga toxin 2). It was negative for Intimin (eae) and also negative for enterohaemolysin (hly). Macrorestriction analysis (pulsed-field gel electrophoresis) with a number of selected isolates obtained from various areas of Germany showed the same pattern, indicating early that the corresponding patients were all affected by one and the same outbreak event.
Surprisingly, the outbreak strain showed virulence characteristics of enteroaggregative E. coli (EAEC). It had the typical EAEC virulence plasmid with adhesion fimbriae type AAF/I. This virulence plasmid has not been described previously in EHEC isolates. All other previously identified EAEC or Shiga toxin—producing E. coli (STEC)/EAEC O104:H4 had AAF/III fimbriae. Subsequent sequencing revealed strong homology to an enteroaggregative E. coli (EAEC 55989). Obviously, the outbreak strain EHEC O104:H4 represents a virulence combination of two different pathogens. The origin of this outbreak strain with the characteristics of two different pathogens remains unclear for the time being. It is unclear whether the new EHEC O104:H4 pathotype had developed from two separate ancestors by horizontal gene transfer, leading to the observed acquisition of virulence factors (Figure A1-6) (Brzuskiewicz et al., 2011; Mellmann et al., 2011; Rasko et al., 2011). A number of mobile genetic elements can transfer traits in E. coli like the Stx-bacteriophage found in EHEC strains. Alternatively, an evolutionary model is discussed, postulating a common progenitor of EAEC
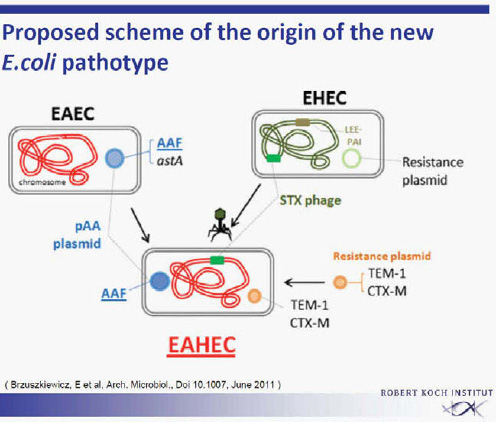
SOURCE: Brzuszkiewicz et al. (2011).
55989 and EHEC O104:H5 developing into two lines, each losing or acquiring virulence factors. The second explanation is favoured by the group from Karch, University of Münster.
The continuously updated EHEC datasheet on the RKI website summarized all known characteristics of the pathogen and suggested the proper microbiological diagnostic procedures.
ESBL Resistance Phenotype
The microbiological characterization revealed a resistance unusual for intestinal E. coli. The outbreak strain had an extended-spectrum β-lactamase (ESBL). This is an unusual property of intestinal E. coli. This resistance phenotype allowed efficient diagnostics of the outbreak strain. It permitted the use of the corresponding selective media for a targeted search in clinical samples, facilitating a rapid diagnosis. Colonies on an ESBL-agar plate were further characterized with multiplex polymerase chain reaction screening for genes of Shiga toxin 1 and 2 and Intimin.
Absence of Direct Microbiological Evidence for Contamination of Seeds with EHEC O104:H4
The identification of seeds as the source and sprouts as the vehicle of infection relied on sophisticated and elegant epidemiological analysis (i.e., indirect evidence). Direct microbiological evidence has not been obtained so far (Aurass et al., 2011). Intensive bacteriological screening of the fenugreek sprouts and seeds was performed. A large number of samples were also taken at the production site of the sprouts, including the water supply or waste water. All attempts to identify the outbreak strain on seeds or sprouts or in the samples obtained at the production site failed. Sampling sprouts in households with EHEC cases was successful in one or two cases. However, these results were more than questionable. One positive result was obtained from a single box of sprouts originating from the incriminated producer. However, it had already been opened in a household with EHEC cases and might simply have been contaminated by the handling. In another example the outbreak strain was identified in salad samples found in a trashcan days after disposal. Also here, the causal connection is unclear.
One reason for the failure to identify the outbreak strain through bacteriological screening may be the enormous size of the incriminated fenugreek seed lot. The lot size was around 15,000 kg. If only a minor part of this lot had indeed been contaminated, searching for contaminated seeds would resemble the search for a needle in a haystack. In addition, on the same day, the sprout-producing enterprise received another lot of seeds from the same seed distributor. The incriminated lot had been distributed to 70 different companies, 54 of them in Germany and 16 of them in 11 European countries (Appel et al., 2011). How-
ever, despite the two additional independent clusters (home-grown sprouts and the cluster in France; see above), no obvious other outbreaks were recognized. Despite all efforts to remove the incriminated lot from the supply chain, it is difficult to estimate how effective and complete this removal has been. Especially in private households, growing sprouts from small aliquots of seeds could lead to new infections. It is known that E. coli can survive on dried seeds for longer periods of time, potentially for years.
Incubation Time and Shedding Time
Detailed analysis revealed a median incubation time of 8 days. The maximum was 18 days. Seventy-five percent of the patients developed clinical symptoms after 10 days. Some of the patients showed a shedding of the pathogen for an extensive period. A few patients shed the pathogens for up to 8 months. It remains to be determined whether shedding might even be longer and whether a carrier status may develop. For enteroaggregative E. coli this extensive shedding period is not unexpected. It is known that aggregative bacteria adhere more strongly and remain in the gastrointestinal tract for longer periods of time. A close collaboration with the local health authorities proved to be important in the analysis of this outbreak (e.g., for these shedding studies) (Robert Koch Institute, 2011).
Secondary Infections
Even after the end of the outbreak had been announced, recommendations were made to enforce the standard hygiene rules, regarding both personal and hand hygiene and in particular kitchen and food hygiene. This included the recommendation to always clean kitchen utensils carefully when preparing food intended for raw consumption. A small number of secondary infections were observed, predominantly consisting of household members of patients. Therefore, stringent adherence to hygienic practices was strongly suggested in those households where EHEC patients or persons with diarrhea were present.
Single nosocomial infections occurred in hospitals (coloscopy). Transmission also occurred through the preparation or distribution of food. Also several laboratory infections were found. Therefore, raised awareness of the risk of infection was also emphasized in public announcements during the months after the official end of the outbreak.
Communication
The RKI made great efforts to inform the medical experts and the public health service and the professional societies (clinical and microbiological) about details of the outbreak in a very timely fashion. During the outbreak, at least daily updates were distributed by e-mail. The Internet proved to be the most important
tool for distribution of information. Usually visits to the RKI homepage result in 4 to 6 million page uses per month. During the outbreak months, May and June 2011, the numbers increased to 16.5 and 17.9 million, respectively. The information provided also included outbreak case definition, forms concerned with sample reporting, diagnostic procedures, information on hygienic measures, etc.
When a whole country is concerned about the safety of its food, the risk communication is important. It proved be helpful to clearly and reliably state the current knowledge and the known risks and their prevention. Also lack of knowledge or uncertainty should be stated clearly, as well as the point in time when new information might be expected. This is important in order to maintain public confidence in recommendations. Farmers requested information because a substantial number of farms suffered economically and were in danger of going out of business.
Conclusion
This outbreak of EHEC infections was the largest recorded outbreak of a bacterial infection observed in Germany in many decades. The enormous rate of HUS cases makes it the largest outbreak of HUS worldwide. It revealed how rapidly a food-borne pathogen can spread and cause serious illness and death. It demonstrates the importance of proper surveillance systems in order to detect an outbreak early and of a rapid reporting system in notifying the corresponding health authorities, in this case the RKI in Germany. According to the specifications of the German Infection Protection Act, a rapid report by the physician or the diagnostic laboratory to the local health authorities is required. In retrospect, between the onset of the disease, the visit to the doctor or hospital, diagnosis, and the report to the local health authority and subsequently to the state authorities and finally to the RKI, a substantial period of time passed, varying from a few days up to several weeks. Measures were taken to improve reporting and to prevent the notification delay. In the analysis of outbreak clusters a close cooperation of health authorities and food safety authorities and a rapid exchange of information is necessary.
The origin of the outbreak strain and how the seeds were contaminated remain unclear. It also remains to be determined whether EHEC O104:H4 will have a reservoir, in human beings, in animals, or in the environment. There is no evidence today that EHEC O104:H4 has become endemic anywhere in humans, animals, or in the environment in Germany. After the sprouts had been identified as the vehicle of this outbreak and after the sprout distribution ended, no further outbreak clusters were identified to be associated with the consumption of sprouts. It is unclear how frequently EHEC is present on sprouts, which are often consumed raw and represent a particularly vulnerable food for bacterial contamination. A rapid and sensitive EHEC diagnostic should also be available in routine diagnostic laboratories in order to identify outbreak events early and
reliably. Detailed subtyping should predominantly be performed in specialized laboratories, also in such an outbreak situation. It seems appropriate to observe these aspects or questions also in the future.
The outbreak had enormous consequences, not only for the patients affected but also economically because of strongly reduced trade in salads and salad ingredients. Spanish cucumbers had been discussed by a local health authority as a potential source of the pathogen. This assumption was not confirmed by laboratory analysis, and attempts to show a connection to the outbreak strain failed; however, it affected the sale and led to a major drop in the consumption and export of Spanish vegetables. Farmers in a number of vegetable-exporting countries were in turn compensated by the European Union in the amount of 220 million Euros for this loss in income.
In summary, the events in Germany during the summer of 2011 revealed the importance of functioning public health institutions, both at the county and state level and at the federal level.
A final detailed report of the EHEC O104:H4 outbreak in Germany is available through the RKI website (http://edoc.rki.de/documents/rki_ab/reQHS31jDrGxc/PDF/23NXL3JomOyAA.pdf) in an English version.
Declaration of Interest
The author declares no conflict of interest and has received no payment in preparation of this manuscript.
References
Altmann, M., A. Spode, D. Altmann, M. Wadl, J. Benzler, T. Eckmanns, G. Krause, and M. an der Heiden. 2011. Timeliness of surveillance during outbreak of Shiga toxin—producing Escherichia coli, Germany. Emerging Infectious Diseases 17:1906-1909.
Appel, B., G. F. Böl, M. Greiner, M. Lahrssen-Wiederholt, and A. Hensel A (Hrsg.). 2011. EHEC-Ausbruch 2011—Aufklärung des Ausbruchs entlang der Lebensmittelkette. Berlin: Bundes-institut für Risikobewertung (BfR-Wissenschaft 04/2011). http://www.bfr.bund.de/cm/350/ehec-ausbruch-2011-aufklaerung-des-ausbruchs-entlang-der-lebensmittelkette.pdf.
Askar, M., M. S. Faber, C. Frank, H. Bernard, A. Gilsdorf, A. Fruth, R. Prager, M. Höhle, T. Suess, M. Wadl, G. Krause, K. Stark, and D. Werber. 2011. Update on the ongoing outbreak of haemolytic uraemic syndrome due to Shiga toxin-producing Escherichia coli (STEC) serotype O104, Germany, May 2011. Eurosurveillance 16(22):pii=19883.
Aurass, P., R. Prager, and A. Flieger. 2011. EHEC/EAEC O104:H4 strain linked with the 2011 German outbreak of haemolytic uremic syndrome enters into the viable but non-culturable state in response to various stresses and resuscitates upon stress relief. Environmental Microbiology 13:3139-3148.
Bielaszewska, M., A. Mellmann, W. Zhang, R. Köck, A. Fruth, A. Bauwens, G. Peters, and H. Karch. 2011. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: A microbiological study. Lancet Infectious Diseases 11:671-676.
Brzuszkiewicz, E., A. Thürmer, J. Schuldes, A. Leimbach, H. Liesegang, F. D. Meyer, J. Boelter, H. Petersen, G. Gottschalk, and R. Daniel. 2011. Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero-aggregative-haemorrhagic Escherichia coli (EAHEC). Archives of Microbiology 193:883-891.
Buchholz, U., H. Bernard, D. Werber, M. M. Böhmer, C. Remschmidt, H. Wilking, Y. Deleré, M. an der Heiden, C. Adlhoch, J. Dreesman, J. Ehlers, S. Ethelberg, M. Faber, C. Frank, G. Fricke, M. Greiner, M. Höhle, S. Ivarsson, U. Jark, M. Kirchner, J. Koch, G. Krause, P. Luber, B. Rosner, K. Stark, M. Kühne, and RKI HUS investigation team (M. Abu Sin, K. Alpers, D. Altmann, M. Altmann, K. Arends, M. Askar, K. Atzpodien, S. Behnke, J. Benzler, A. Bergholz, J. Bielecke, B. Brodhun, R. Burger, W. Cai, H. Claus, C. Cyberski, M. Dehnert, S. Dudareva, T. Gunsenheimer-Bartmeyer, K. Haar, W. Haas, O. Hamouda, B. Hauer, W. Hellenbrand, J. Hermes, K. Köpke, K. Krügermann, G. Laude, M. H. Lee, I. Liss, M. Luchtenberg, M. Marx, D. Meyer, M. Mielke, A. Milde-Busch, K. Prahm, U. Preuß, S. Reiter, A. Reuß, U. Rexroth, M. Richter, T. Rieck, K. Rothe, A. Sailer, C. Santos-Hövener, L. Schaade, S. Schink, D. Schmidt, C. Schoene, I. Schöneberg, M. Schuster, F. Schwarz, B. Schweickert, P. Stöcker, T. Suess, A. Takla, E. Tietze, B. Ultsch, M. Ung-Zu Kang, E. Velasco, M. Wadl, D. Walter, B. Weiß, R. Zimmermann, W. Zhang, and J. Zunk). 2011. German outbreak of Escherichia coli O104:H4 associated with sprouts. New England Journal of Medicine 365:1763-1770.
Frank, C., M. S. Faber, M. Askar, H. Bernard, A. Fruth, A. Gilsdorf, M. Höhle, H. Karch, G. Krause, R. Prager, A. Spode, K. Stark, and D. Werber, on behalf of the HUS investigation team. 2011a. Large and ongoing outbreak of haemolytic uraemic syndrome, Germany, May 2011. Eurosurveillance 16(22):pii=19878.
Frank, C., D. Werber, J. P. Cramer, M. Askar, M. Faber, M. an der Heiden, H. Bernard, A. Fruth, R. Prager, A. Spode, M. Wadl, A. Zoufaly, S. Jordan, K. Stark, and G. Krause, for the HUS Investiation Team (M. Abu Sin, C. Adlhoch, K. Alpers, D. Altmann, M. Altmann, K. Erends, K. Atzpodien, S. Behnke, J. Benzler, A. Bergholz, J. Bielecke, M. Böhmer, B. Brodhun, U. Buchholz, R. Burger, W. Cai, H. Claus, M. Christner, C. Cyberski, M. Dehner, Y. Deleré, S. Dudareva, T. Eckmanns, W. Espelage, G. Falkenhorst, L. Fiebig, K. Fraedrich, A. Gilsdorf, B. Greutélaers, B. Gunsenheimer-Bartmeyer, K. Haar, W. Haas, O. Hamouda, B. Hauer, W. Hellenbrand, J. Hermes, M. Höhle, M.J. Kemper, J. Koch, K. Köpke, K. Krügermann, G. Laude, M.-H. Lee, I. Liss, A. W. Lohse, M. Luchtenberg, M. Marx, D. Meyer, M. Mielke, A. Milde-Busch, I. Mücke, L. Müller, M. Nachtnebel, J. Neifer, S. Nielsen, I. Noll, R. Offergeld, Y. Pfeifer, R. Pohland, K. Prahm, U. Preuß, S. Reiter, C. Remschmidt, A. Reuß, U. Rexroth, M. Richter, T. Rieck, H. Rohde, B. Rosner, A. Sailer, C. Santos-Hövener, L. Schaade, S. Schink, S. Schmiedel, D. Schmidt, C. Schoene, I. Schöneberg, M. Schuster, F. Schwarz, B. Schweickert, P. Stöcker, T. Süß, A. Takla, E. Tietze, B. Ultsch, M. U.-Z. Kang, E. Velasco, D. Walter, B. Weiß, H. Wilking, R. Zimmermann, W. Zhang, J. Zunk). 2011b. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. New England Journal of Medicine365:1771-1780.
Mellmann, A., D. Harmsen, C. A. Cummings, E. B. Zentz, S. R. Leopold, A. Rico, K Prior, R. Szczepanowski, Y. Ji, W. Zhang, S. R. McLaughlin, J. K. Henkhaus, B. Leopold, M. Bielaszewska, R. Prager, P. M. Brzoska, R. L. Moore, S. Guenther, J. M. Rothberg, and H. Karch. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS ONE 6:e22751, doi:10.1371/ journal.pone.0022751.
Rasko, D. A., D. R. Webster, J. W. Sahl, A. Bashir, N. Boisen, F. Scheutz, E. E. Paxinos, R. Sebra, C. S. Chin, D. Iliopoulos, A. Klammer, P. Peluso, L. Lee, A. O. Kislyuk, J. Bullard, A. Kasarskis, S. Wang, J. Eid, D. Rank, J. C. Redman, S. R. Steyert, J. Frimodt-Moller, C. Struve, A. Petersen, K. A. Krogfelt, J. P. Nataro, E. E. Schadt, and M. K. Waldor. 2011. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. New England Journal of Medicine 365:709-717.
Robert Koch Institute. 2011. Report: Final presentation and evaluation of epidemiological findings in the EHEC O104:H4 outbreak, Germany 2011. Berlin: Robert Koch Institute. http://www.rki.de/EN/Home/EHEC_final_report.pdf?__blob=publicationFile (accessed June 26, 2012).
Wadl, M., T. Rieck, M. Nachtnebel, B. Greutélaers, M. an der Heiden, D. Altmann, W. Hellenbrand, M. Faber, C. Frank, B. Schweickert, G. Krause, J. Benzler, and T. Eckmanns, on behalf of the HUS Surveillance and Laboratory Team. 2011. Enhanced surveillance during a large outbreak of bloody diarrhoea and haemolytic uraemic syndrome caused by Shiga toxin/verotoxin-producing Escherichia coli in Germany, May to June 2011. Eurosurveillance 16(22):pii=19893.
ONE HEALTH AND HOTSPOTS OF FOOD-BORNE EIDS
C. Zambrana-Torrelio, K. A. Murray, and P. Daszak3
Summary
In this section, we focus on a One Health approach to food-borne emerging infectious diseases (EIDs), their causes, global patterns, and the drivers of their emergence. First, we review two case studies that show the complexity of food-borne pathogen emergence across the One Health domain. Second, we examine the composition of food-borne diseases with respect to their causal agents (pathogen type), their association with pathogens of zoonotic origin, and their apparent disassociation with pathogens that show drug resistance. Third, we analyze the socioeconomic, environmental, and ecological drivers of food-borne EID events. Finally, we use published, spatially explicit information on the drivers of disease emergence to produce a preliminary “hotspot” map that reveals the epicentres, or hotspots, of food-borne EID events globally.
Introduction
One Health’s focus on the intersection of human, domestic animal, and environmental health is ideally suited to managing emerging zoonoses. However, the patterns of emergence are complex and poorly understood and for food-borne infections may involve multiple pathways. Food-borne infections can include directly transmitted or vector-borne diseases, for example, Rift Valley fever (Arzt et al., 2010). Single strains of drug-resistant microbes can infect livestock, wildlife, and humans (e.g., E. coli O157:H7) (Hughes et al., 2009; Nielsen et al., 2004; Rahn et al., 1997). Finally, viral pathogens that originate in wildlife may be driven to emerge by the intensification of livestock production (Pulliam et al., 2011) or by contamination of bush meat (Wolfe et al., 2005) or other food sources
_________________
3 EcoHealth Alliance, 460 West 34th Street, New York, NY 10001, USA.
(Khan et al., 2011). Our ability to predict the emergence of food-borne infections is hampered by this complexity. However, recent efforts to analyse disease emergence (Jones et al., 2008; Taylor et al., 2001) have provided a strategy that can be adapted to analyzing the origins of food-borne infections.
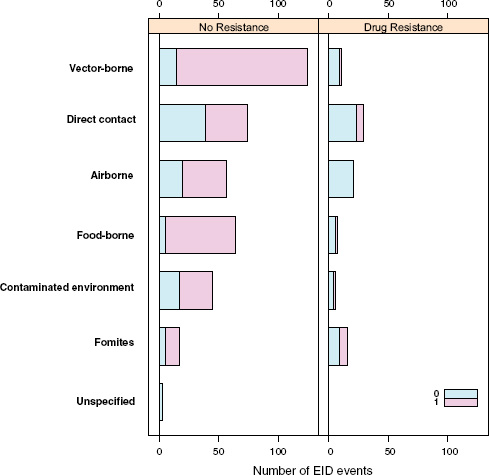
Following our first efforts to predict global patterns of disease emergence (Jones et al., 2008), we have continued to compile data on human EID events and their drivers under the aegis of the U.S. Agency for International Development—funded Emerging Pandemic Threats PREDICT project (Daszak, 2011). In the updated database, when the EID events are classified according to their disease transmission modes (Figure A2-1), we find that food-borne pathogens are responsible for 14.9 percent of known EIDs.
In this section, we focus on food-borne EIDs, their causes, global patterns, and the drivers of their emergence. First, we review two case studies that show
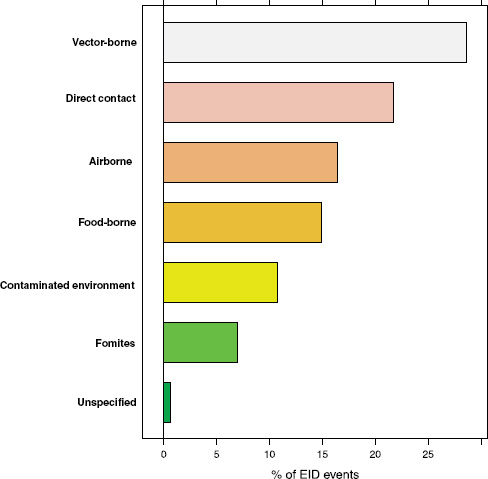
FIGURE A2-1 Proportion of EID events categorized by transmission mode.
the complexity of food-borne pathogen emergence across the One Health domain. Second, we examine the composition of food-borne diseases with respect to their causal agents (pathogen type), their association with pathogens of zoonotic origin, and their apparent disassociation with pathogens that show drug resistance. Third, we analyze the socioeconomic, environmental, and ecological drivers of food-borne EID events. Finally, we use published, spatially explicit information on the drivers of disease emergence to produce a preliminary “hotspot” map that reveals the epicentres, or hotspots, of food-borne EID events globally.
Food-Borne, Wildlife-Origin Pathogens: Two Case Studies
Nipah virus (NiV) is a paramyxovirus that first emerged in Malaysia in 1999, causing encephalitis with a 40 percent case fatality rate in humans (Chua et al., 2000). The virus originated in fruit bats of the genus Pteropus but was first transmitted to domestic pigs, which amplified the virus via a rapidly spreading respiratory infection. Subsequent transmission to people occurred via droplets or fomites contaminated with pig saliva. The initial spillover of NiV seems to have occurred when fruit bats fed on mango and other fruit trees planted next to pigsties at the index farm as a source of additional income and to increase shade. The question remained: Why did it suddenly emerge in this pig farm and not in pig farms 20 years earlier or 20 years later?
To answer this question we analyzed pig production and the age structure of NiV dynamics within the index farm population (Pulliam et al., 2011). We produced a mathematical model, parameterized with detailed data from the index farms and other similar farms still in existence in Malaysia today. This model allowed us to re-create the conditions of the farm when NiV first emerged and to test hypotheses on the drivers of its emergence. Our analyses suggest that repeated introduction of NiV from bats changed infection dynamics in pigs. Initial viral introduction produced an epizootic that drove itself to extinction within 1 to 2 months. Subsequent introduction into a now partially immune population, coupled with the gradual loss of maternal antibodies in pigs born to sows infected in the initial outbreak, led to ideal conditions for pathogen persistence and a prolonged window of spillover to people and regional spread as infected pigs were sold. The structured, compartmentalized nature of the index farm was critical to the emergence of NiV and was a product of agricultural intensification.
A similar scenario surrounds the emergence of highly pathogenic influenza A/H5N1. This virus is able to infect wild waterbirds, domestic poultry, and humans, and its emergence is linked to both intensive production of poultry and the patterns of rice farming within Southeast Asia. When rice is double-cropped, it attracts ducks throughout the year and allows greater potential for new strains of influenza to cross over into pigs and for subsequent crossover of those strains (Gilbert et al., 2008). Analysis of the patterns of double-cropping in Southeast Asia shows that it is possible to predict the risk of its presence throughout
the region based on the type of agricultural system (Gilbert et al., 2008). Poultry production in this region includes large intensive and small “backyard” farms, all connected via trade routes into markets and through the supply of breeding stock and their contact with wild birds. We have used a similar modeling approach for A/H5N1 to examine how farm size and connectivity matter as risk factors for the emergence of avian influenza. Our modeling shows that both factors interact to produce specific conditions conducive to outbreaks. When the vast majority of farms are of small size, outbreaks occur more frequently and last longer, but they involve few individual birds and therefore have a lower risk of infecting people. When farms are poorly connected these outbreaks die out because of stochastic factors. When large intensive farms predominate, outbreaks are very few in number, but their duration is relatively short because so many birds die in such a short space of time that the cause is rapidly recognized and the farm culled. The peak in duration and intensity of outbreaks occurs when there is a mixture of intensive and backyard farming. These are the conditions that occur most commonly in Southeast Asia because of the rapid growth of some economies and efforts to intensify poultry production.
Causes, Patterns, and Drivers of Food-Borne EIDs
How important are food-borne infections in the context of global disease emergence events? Going back to Figure A2-1, approximately 15 percent of human EID events are associated with food-borne transmission pathways. With 475 EID events in the updated database, this translates to 71 separate food-borne EIDs, at an average emergence rate of just under one completely new, previously unknown EID event per year reported globally.
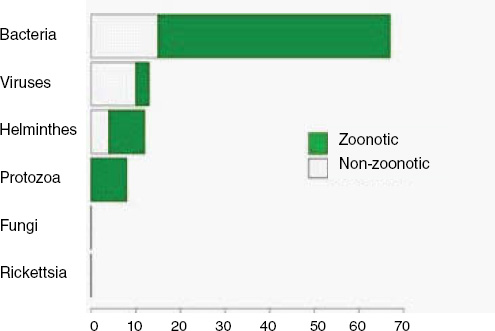
When broken down by causal pathogen type (Figure A2-2), food-borne EIDs are usually bacterial in origin, with smaller proportions of protozoan and helminth-driven diseases. While bacteria are also the major causes of EIDs associated with the contaminated environment and fomites, food-borne EIDs are generally more common and therefore account for the highest number of EIDs of bacterial origin (50) among all of the transmission modes. Hence, when bacteria are the causal agent implicated in EID events, they are more likely to be food-borne than of any other transmission mode. In contrast to the other transmission mode groups, food-borne EIDs are very rarely viral, accounting for only one (1.4 percent) food-borne EID (hepatitis A) compared to ~20 to 45 percent (average 30.9 percent) in the other groups. However, many viral pathogens (e.g., NiV and H5N1) are considered simply zoonotic because the role of food-borne transmission is either less well known or less well understood.
Our analyses suggest that the vast majority of food-borne EIDs are indeed zoonotic; in fact, an even higher proportion of food-borne EIDs are zoonotic (84.5 percent) than the background rate of all EIDs in the updated database (62.3 percent) and of any other transmission mode (Figure A2-3). Clearly, patho-
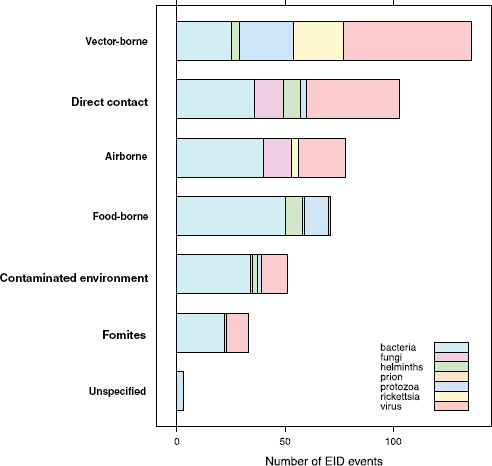
FIGURE A2-2 Number of EID events per transmission mode classified by pathogen type.
gens from animals entering the food-production chain are of significant concern for their potential to become EIDs.
One of our earlier findings (Jones et al., 2008) was that a majority (54.3 percent) of human EIDs were bacterial/rickettsial in origin, reflecting a large number (20.8 percent of all EIDs) of new drug-resistant pathogen strains. We show above that if an EID was identified as being caused by bacteria, it was most likely to be food-borne, and similarly if an EID was linked with food it was most likely to be bacterial than of another transmission mode. Given the propensity of bacteria to develop drug resistance, and the abundance of food-borne infections of bacterial origin, is there any evidence that food-borne pathogens are contributing to new drug-resistant diseases?
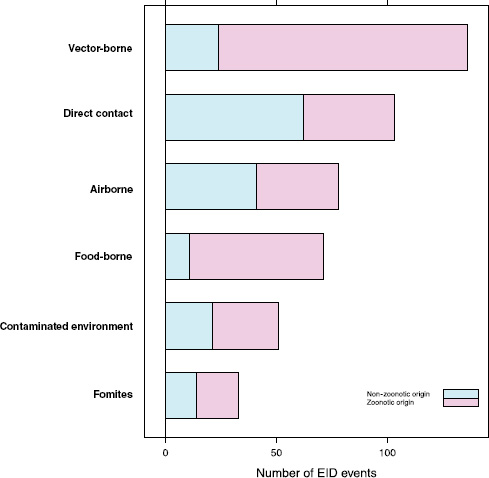
FIGURE A2-3 Number of EID events per transmission mode categorized by zoonotic origin.
Perhaps surprisingly, the answer is no: when EID events are split into categories reflecting the presence or absence of drug resistance (ignoring for a moment the secondary split on whether the pathogen was zoonotic or not), food-borne pathogens are very unlikely to be drug resistant (Figure A2-4). Although it is true that drug resistance is relatively infrequently observed across most transmission modes (the exception being fomite-associated EIDs), resistance is particularly infrequent in food-borne (as well as vector-borne) EIDs. Hence, even though bacteria are quite likely to cause food-borne EIDs and bacteria also cause the majority of new drug-resistant diseases, this is quite unlikely to occur together, resulting in very few drug-resistant food-borne EIDs. Why is drug resistance not more common in this group?
The answer may be related to whether the causal agent is zoonotic or not. Generally speaking, there is a low frequency of zoonotic EIDs that exhibit drug resistance (6.0 percent), regardless of the transmission mode (Figure A2-4). Non-zoonotic EIDs are far more likely to be associated with drug resistance (40.9 percent), again across all groups. This is consistent with the idea that new drug-resistant pathogens result from selection on our own circulating pathogens by the routine use of antimicrobial drugs, and not on the pathogens circulating in the food industry that originate in animals. In other words, even though most food-borne EIDs are caused by bacteria, which generally show high potential for becoming drug resistant, the fact that most food-borne EIDs are zoonotic means that the group is quite unlikely to experience strong selection pressure from
routine drug administration in human patients. Obviously there are limitations to this type of analysis, particularly in how extensive the data are, but it is clear that this issue is an important target for future research.
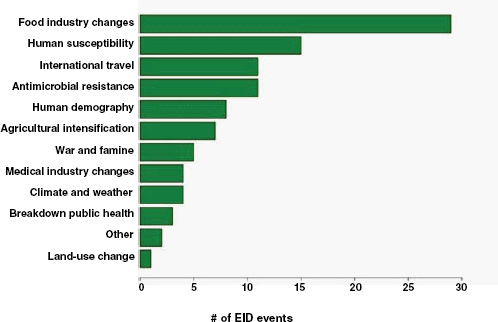
Finally, what are the drivers of food-borne pathogens, and are they an ongoing concern for their EID potential? As we have seen, food-borne EIDs are common, usually zoonotic, usually bacterial, and not likely to exhibit drug resistance. So what factors are driving them to emerge? What factors are allowing them to enter and circulate within the food-production system to subsequently cause disease in humans? In Figure A2-5, we analyse the underlying drivers listed in a previous Institute of Medicine report (IOM, 2003) for food-borne EIDs and find that the vast majority of food-borne EIDs are associated with “technology and industry,” and to a lesser extent with “international trade and commerce” and “human susceptibility to infection.” This is consistent with previous studies that have suggested that outbreaks of food-borne infections are likely to be associated with changes in livestock production and centralization of slaughtering
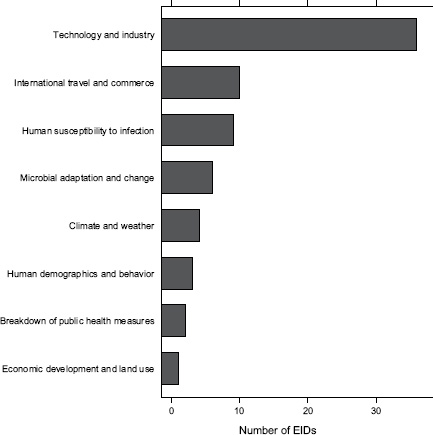
FIGURE A2-5 Association of food-borne EIDs with other drivers.
SOURCE: Following IOM (2003).
and processing (IOM, 2003; Tauxe, 1997). As a result of these analyses, we can hypothesize that the global distribution of food-borne EIDs is driven by a process of intensive production of livestock and food, not simply the number of livestock produced in a region.
Food-Borne EID Hotspots
Our previous approach to predicting the future geographic origins of new EIDs (Jones et al., 2008) can be adapted for food-borne EIDs. This approach involves identifying the geographic and temporal origins of previous disease emergence events and correcting them for surveillance biases. We then identify correlations between these and purported socioeconomic (demography, travel, trade), environmental (climate, land cover), and ecological drivers ( biodiversity, species interactions). Considering all EIDs together, these models suggest that surveillance should be directed toward regions of high biodiversity and dense human populations, which mainly occur in tropical and subtropical latitudes (Jones et al., 2008). When we adapt this approach to food-borne EID events and use the same drivers as in our earlier analysis, human population density and human population growth emerge as the most important in the emergence of novel food-borne outbreaks (Figure A2-6). This suggests that rapidly developing regions are the sites where most novel food-borne pathogens will emerge in future. This may appear to be in conflict with Figure A2-5; however, this is because the spatial analyses have so far been limited primarily by the availability of relevant spatial information. Human population density and growth are likely to be meaningful proxies for a range of other mechanistically more relevant drivers. One of our main goals more recently has thus been to improve our database of detailed drivers. We have, for
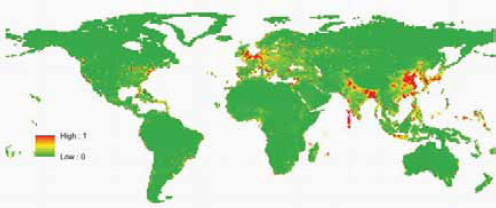
SOURCE: Reprinted by permission from Macmillan Publishers Ltd: Nature, (Jones et al., 2008).
example, begun to include spatial information on land-use change (cropping and pasture) and livestock density (including cattle, pigs, buffalo, goats, and sheep) into the predictive models. We are currently validating these new data sources for use in future models.
We conclude that food-borne EIDs are a common and important group within emerging diseases that emerge through complex pathways involving wildlife, livestock, and humans. They are therefore ideal candidates for a One Health approach but have rarely been considered in this way previously. Our analyses show that the majority of food-borne EIDs (1) are bacterial; (2) are, if bacterial, more likely to be food-borne than of any other transmission mode; (3) are zoonotic; (4) do not tend to be associated with drug resistance, perhaps because zoonotic pathogens in general show little tendency to become resistant; and (5) are driven by changes in human food-production systems, including intensification and centralization as human populations grow larger and more dense.
References
Arzt, J., W. R. White, B. V. Thomsen, and C. C. Brown. 2010. Agricultural diseases on the move early in the third millennium. Veterinary Pathology 47:15-27.
Chua, K. B., W. J. Bellini, P. A. Rota, B. H. Harcourt, A. Tamin, S. K. Lam, T. G. Ksiazek, P. E. Rollin, S. R. Zaki, W. Shieh, C. S. Goldsmith, D. J. Gubler, J. T. Roehrig, B. Eaton, A. R. Gould, J. Olson, H. Field, P. Daniels, A. E. Ling, C. J. Peters, L. J. Anderson, and B. W. Mahy. 2000. Nipah virus: A recently emergent deadly paramyxovirus. Science 288:1432-1435.
Daszak, P. 2011. Smart surveillance: Analyzing environmental drivers of emergence to predict and prevent pandemics. Ecohealth 7:S12-S13.
Gilbert, M., X. M. Xiao, D. U. Pfeiffer, M. Epprecht, S. Boles, C. Czarnecki, P. Chaitaweesub, W. Kalpravidh, P. Q. Minh, M. J. Otte, V. Martin, and J. Slingenbergh. 2008. Mapping H5N1 highly pathogenic avian influenza risk in Southeast Asia. Proceedings of the National Academy of Sciences of the United States of America 105:4769-4774.
Hughes, L. A., M. Bennett, P. Coffey, J. Elliott, T. R. Jones, R. C. Jones, A. Lahuerta-Marin, K. McNiffe, D. Norman, N. J. Williams, and J. Chantrey. 2009. Risk factors for the occurrence of Escherichia coli virulence genes eae, stx1 and stx2 in wild bird populations. Epidemiology and Infection 137:1574-1582.
IOM (Institute of Medicine). 2003. Microbial threats to health: Emergence, detection, and response. Washington, DC: The National Academies Press.
Jones, K. E., N. Patel, M. Levy, A. Storeygard, D. Balk, J. L. Gittleman, and P. Daszak. 2008. Global trends in emerging infectious diseases. Nature 451:990-994.
Khan, M. S. U., J. Hossain, E. S. Gurley, N. Nahar, R. Sultana, and S. P. Luby. 2011. Use of infrared camera to understand bats’ access to date palm sap: Implications for preventing Nipah virus transmission. Ecohealth 7:517-525.
Nielsen, E. M., M. N. Skov, J. J. Madsen, J. Lodal, J. B. Jespersen, and D. L. Baggesen. 2004. Verocytotoxin-producing Escherichia coli in wild birds and rodents in close proximity to farms. Applied and Environmental Microbiology 70:6944-6947.
Pulliam, J. R., J. H. Epstein, J. Dushoff, S. A. Rahman, M. Bunning, A. A. Jamaluddin, A. D. Hyatt, H. E. Field, A. P. Dobson, and P. Daszak. 2011. Agricultural intensification, priming for persistence and the emergence of Nipah virus: A lethal bat-borne zoonosis. Journal of the Royal Society Interface 9(66):89-101. doi: 10.1098/ rsif.2011.0223.
Rahn, K., S. A. Renwick, R. P. Johnson, J. B. Wilson, R. C. Clarke, D. Alves, S. McEwen, H. Lior, and J. Spika. 1997. Persistence of Escherichia coli O157:H7 in dairy cattle and the dairy farm environment. Epidemiology and Infection 119:251-259.
Tauxe, R. V. 1997. Emerging foodborne diseases: An evolving public health challenge. Emerging Infectious Diseases 3:425-434.
Taylor, L. H., S. M. Latham, and M. E. J. Woolhouse. 2001. Risk factors for human disease emergence. Philosophical Transactions of the Royal Society of London, B 356:983-989.
Wolfe, N. D., P. Daszak, A. M. Kilpatrick, and D. S. Burke. 2005. Bushmeat hunting, deforestation and prediction of zoonotic emergence. Emerging Infectious Diseases 11:1822-1827.
PLANT FOOD SAFETY ISSUES: LINKING PRODUCTION AGRICULTURE WITH ONE HEALTH
Marilyn C. Erickson and Michael P. Doyle4
During the past decade, fruits and vegetables have become leading vehicles of food-borne illnesses. Furthermore, many plant-based foods and ingredients, not previously considered a risk, have been associated with food-borne disease outbreaks. Most of the pathogens that have been identified as causative agents in these illnesses or outbreaks are enteric zoonotic pathogens that are typically associated with animal hosts. Transmission of zoonotic pathogens from animals to plant systems occurs by a variety of routes, but the initial contributing factor is the discharge of animal manure into the environment. Using a “One Health” approach that focuses on animal, human, and environmental health concurrently can provide practical and effective interventions for reducing the incidence of such outbreaks. This paper addresses this concept by providing recent food-borne disease outbreak data related to fruits and vegetables, delineating findings regarding the prevalence of pathogens in animal manures and describing the vehicles that transmit pathogens from manure to produce fields, and discussing the merits of reducing pathogen transmission through interventions that would not adversely affect the health of the environment or animals.
Outbreaks and Illnesses Associated with Fresh Fruits and Vegetables
Food-borne illnesses have been a persistent challenge to public health and are now being detected with greater frequency largely because of enhanced surveillance systems that have been implemented in many countries. These enhanced surveillance systems have during the past decade revealed that the proportion of total outbreaks attributed to produce is significant (Lynch et al., 2009) but varies with the country. For example, only 4 percent of all food-borne outbreaks
______________
4 Center for Food Safety, University of Georgia, 1109 Experiment Street, Griffin, GA 30223, USA.
reported in Australia from 2001 to 2005 were attributed to fresh produce (Kirk et al., 2008); similarly, in Canada, between 1976 and 2005, 3.7 percent of 5,745 outbreaks with a known vehicle of transmission were attributed to produce (Ravel et al., 2009). However, in contrast, data from the Centers for Disease Control and Prevention (CDC) identified produce as either the first or second leading vehicle in food-borne disease outbreaks attributed to a single commodity within the United States for the period 2006-2008 (Table A3-1). Furthermore, outbreak surveillance data of produce items compiled by the CDC during the period 2000-2009 revealed that leafy greens were the most common item associated with food-borne disease, followed by tomatoes and cantaloupes (Table A3-2). Moreover, attribution risk rankings of fresh produce—associated outbreaks in the United States identified enterohemorrhagic Escherichia coli in leafy greens as the leading pathogen-produce vehicle combination, followed by Salmonella spp. in tomatoes, and Salmonella spp. in leafy greens (Anderson et al., 2011). Further differentiation of vehicles of produce-associated outbreaks that occurred in the United States during the period of 1998-2008 revealed that fresh-cut produce accounted for 56 percent, 36 percent, and 17 percent of the outbreaks attributed to leafy greens, tomatoes, and melons, respectively (Sneed, 2010).
An evaluation of selected produce-associated outbreaks that occurred during the past 5 years revealed several common features (Table A3-3). These outbreaks often were multistate or multinational in nature and reflect the large areas to which the foods are distributed. With imports accounting for nearly 39 percent of fresh fruits and 14 percent of fresh vegetables in 2005 (Johnson, 2012), improved sampling and pathogen testing of produce at the borders of the United States offers one barrier for reducing the likelihood of contaminated produce
|
|
||||||
| Year | Rank | Food vehicle | Outbreaks (%) | |||
|
|
||||||
| 2006 | 1 | Produce | 23.5 | |||
| 2 | Meat | 19.3 | ||||
| 2 | Fish | 19.3 | ||||
| 4 | Poultry | 14.4 | ||||
| 2007 | 1 | Meat | 23.0 | |||
| 2 | Produce | 22.6 | ||||
| 3 | Fish | 17.4 | ||||
| 4 | Poultry | 17.0 | ||||
| 2008 | 1 | Produce | 27.5 | |||
| 2 | Meat | 23.4 | ||||
| 3 | Poultry | 14.7 | ||||
| 4 | Fish | 13.8 | ||||
|
|
||||||
SOURCE: CDC (2009a, 2010c, 2011e).
TABLE A3-2 Number of Outbreaks (illnesses) Reported Between 2000 and 2009 in the United States That Were Associated with Selected Fresh Produce Items as a Function of their Etiologya,b
| Bacterial agents | |||||
| Produce item |
Salmonella spp. |
Escherichia coli O157:H7c |
Shigella spp. |
Campylobacter jejuni |
Otherd |
| Cabbage | 1(8) | 1(41) | 2(68) | ||
| Lettuce | 10(456) | 14(364) | 1(4) | 2(16) | 3(114) |
| Spinach | 2(223) | 1(6) | |||
| Sprouts | 12(441) | 4(46) | 1(20) | ||
| Herbs | 3(70) | ||||
| Leafy green salads | 23 (997) | 15 (280) | 7(190) | 7(42) | 10(145) |
| Coleslaw | 1(26) | 4(22) | |||
| Peppers | 4(1,643) | 1(5) | 2(17) | ||
| Tomatoes | 25(1.867) | 1 (886) | 1(13) | 2(10) | |
| Cantaloupe/melons | 19(1,180) | 1(5) | 1(56) | 1(55) | |
a Data compiled from the CDC website on outbreak surveillance (http://www.cdc.gov/outbreaknet/surveillance_data.html).
b Outbreaks/illnesses attributed to each pathogen group includes both confirmed and suspected.
c Includes other Shiga toxin–producing Escherichia coli.
d Includes where multiple bacterial pathogens have been found and cases involving the agents of Bacillus cereus, Clostridium botulinum, C. perfringens, Listeria monocytogenes, and Staphylococcus aureus.
SOURCE: CDC.
from entering the retail sector. However, better implementation both domestically and abroad of best food safety practices for producing and processing fruits and vegetables would have even more impact on reducing pathogen contamination and the likelihood of produce-borne illnesses. This approach would address a significant contributing factor associated with several recent produce outbreaks, which is that contamination occurs on the farm where production and processing can occur. For example, in a multistate outbreak of listeriosis in 2011 that resulted in 34 deaths and was the most deadly food-borne outbreak in the United States since 1924, four outbreak-associated strains of Listeria monocytogenes were traced back to whole cantaloupes and packing equipment on Jensen Farms in Colorado (CDC, 2011c). In another 2011 outbreak, fenugreek seeds that were likely contaminated with fecal matter led to the largest outbreak in the number of cases of hemolytic uremic syndrome (22.3 percent of 4,075 total cases) ever reported in the world (WHO, 2011).
Surveillance of Pathogens in Retail Produce
A number of studies have been conducted to determine the prevalence of enteric pathogens on fruits and vegetables, and the results varied with respect
|
|
||||
| Viral agents | Other agents | |||
| Norovirus | Hepatitis A | Protozoan parasites | Unknown | Total |
|
|
||||
| 3 (78) | 1(16) | 3 (16) | 11 (227) | |
| 39 (999) | 1 (22) | 10 (60) | 80 (2,035) | |
| 3 (9) | 6 (238) | |||
| 1 (2) | 18 (509) | |||
| 1 (592) | 1 (20) | 5 (682) | ||
| 257 (8,520) | 3 (47) | 114 (1,419) | 436 (11,640) | |
| 20 (676) | 1 (8) | 1 (11) | 27 (743) | |
| 1 (2) | 8 (1,667) | |||
| 15 (399) | 1 (23) | 45 (3,198) | ||
| 12 (502) | 6 (79) | 40 (1,877 | ||
|
|
||||
to the country of origin and the target pathogen. For Salmonella, there was for most developed countries a very low prevalence in cabbage, lettuce, and mixed salads, whereas higher prevalences were observed for developing countries where agricultural production and hygienic practices were of a lower level of sanitation (Table A3-4). The presence of helminth and protozoan parasites in leafy greens (Table A3-5), however, likely reflects the ability of these pathogens to resist standard chlorine-based wastewater treatments (Erickson and Ortega, 2006; Graczyk et al., 2007). The relatively low occurrence of pathogen contamination on produce makes it inherently difficult to rank the degree of risk associated with the various sources of contamination by which enteric pathogens are transmitted from animals to plant production environments.
Pathogens in Manures from Domesticated Animal
A large number of zoonotic pathogens reside and grow in the gastrointestinal tract of domesticated animals (poultry, cattle, swine, sheep, and goats) and are shed in their feces asymptomatically, often in very large numbers. Those enteric pathogens associated with the largest number of food-borne disease outbreaks and illnesses include Campylobacter jejuni, Salmonella spp., Shiga toxin—producing
| Year | Pathogen | Number of cases | Country of origin |
| 2007 | Salmonella Wcltcvredcn | 45 | Italy, seed origin |
| 2009 | Salmonella Saimpaul | 228 | Domestic, seed company |
| 2010 | S. Newport | 44 | Domestic, processor |
| 2010 | Salmonella 1 4,[5],2:i:- | 112 | Domestic |
| 2011 | E. coli O104:114 | 3,911 | Egypt |
| 2007 | S. Senftenberg | 51 | Israel |
| 2007 | S. Senftenberg | 74 | Israel |
| 2006 | S. Saintpaul | 36 | Domestic |
| 2008 | S. Litchfield | 51 | Honduras |
| 2011 | S. Panama | 20 | Guatemala |
| 2011 | Listeria monocytogenes | 146 | Domestic |
| 2006 | Clostridium botulinum | 4 | Domestic |
| 2006 | Yersinia pseudotuberculosis | 427 | Domestic, traced to vegetable distributor |
| 2006 | Norovirus | 43 | China |
| 2006 | E. coli O157:117 | 71 | Not known |
| 2006 | E. coli O157:H7 | 81 | Domestic |
| 2011 | E. coli O157:117 | 60 | Domestic |
| 2008 | Salmonella Newport and Reading | 77 (Newport) 30 (Reading) | Domestic |
| 2010 | Norovirus and E. coli ETEC | 264 | Erance |
| 2010 | E. coli O145 | 33 | Domestic, processor |
| 2007 | E. coli O157:H-, PT8 | 50 | Netherlands, processing plant |
| 2008 | Cryptosporidium parvum | 21 | Italy |
| 2008 | S. Saintpaul | 1.442 | Mexico |
| 2007 | Shigella sonnei | 227 | Thailand |
| 2006 | E.coli O157:H7 | 204 | U.S. |
| 2006 | S. Typhimurium | 183 | Not known |
SOURCE: CDC.
enterohemorrhagic Escherichia coli (STEC), and Cryptosporidium parvum. Many studies have been conducted to determine the prevalence of these pathogens in the feces of domesticated animals. A selection of results of recent studies are shown in Tables A3-6 to A3-9 to illustrate the range of pathogen prevalences and cell numbers that may occur within animal wastes and between and within different groups of animals. For Cryptosporidium, not all species are pathogenic for humans. For example, currently there are at least 16 recognized species of Crypto-sporidium, of which two most affect humans, C. hominis and C. parvum (Jagai et al., 2010). Therefore, when results do not differentiate species of Cryptosporidium, the potential risk of those manures to human health may be overestimated.
| Affected regions | Implicated food | Reference |
| Norway, Denmark, Finland | Alfalfa sprouts | Emberland et al., 2007 |
| U.S., multistate | Alfalfa sprouts | CDC, 2009b |
| U.S., multistate | Alfalfa sprouts | CDC, 2010a |
| U.S., multistate | Alfalfa sprouts | CDC, 2011a |
| Multinational | Fenugreek sprouts | EFSA, 2011 |
| U.K., U.S., Denmark, Netherlands | Basil | Elviss et al., 2009 |
| U.K., Denmark, Netherlands, U.S. | Basil, fresh | Pezzoli et al., 2008 |
| Australia, multijurisdiction | Cantaloupe | Munnoch et al., 2009 |
| U.S., multistate | Cantaloupe | CDC, 2008a |
| U.S., multistate | Cantaloupe | CDC, 2011b |
| U.S., multistate | Cantaloupe | CDC, 2011c |
| U.S., Georgia | Carrot juice | CDC, 2006a |
| Finland | Carrots, grated | Rimhanen-Finne et al., 2009 |
| Sweden | Frozen raspberries | Hjertqvist et al., 2006 |
| U.S., multistate | Lettuce | FDA, 2006 |
| U.S., multistate | Lettuce | FDA, 2007 |
| U.S., multistate | Lettuce, romaine | CDC, 2011c |
| Finland | Lettuce | Lienemann et al., 2011 |
| Denmark, Norway | Lettuce, lollo biondo type | Ethelberg et al., 2010 |
| U.S., multistate | Lettuce, shredded romaine | CDC, 2010b |
| Netherlands, Iceland | Lettuce, shredded, prepacked | Friesema et al., 2007 |
| Sweden | Parsley | Insulander et al., 2008 |
| U.S., Canada | Peppers (jalapeño and Serrano), tomatoes | CDC, 2008b |
| Denmark, Australia | Raw baby corn | Lewis et al., 2009 |
| U.S., Canada | Spinach | Calvin, 2007 |
| U.S, multistate | Tomatoes | CDC, 2006b |
SOURCE: CDC.
Management of Wastes from Domesticated Animals
Globally, food animal production has increased more than fivefold in the past 50 years due in large part to the adoption of the industrialized concentrated animal production model. With multinational companies expanding their operations overseas, estimates indicate that concentrated animal feeding operations (CAFOs) provide 74 percent of poultry, 50 percent of pork, and 43 percent of beef produced worldwide (Halweil and Nierenberg, 2004). Accompanying this expansion in production has been the challenge of managing the massive quantities of animal wastes that are generated in one location. For example, in China, animal waste was estimated to be 3.2 billion tons, which was three times the amount of
| Produce item | Country | Sampling site | Number positive/ number sampled |
Prevalence ('*) | Reference |
| Cabbage | India | Fields | 4/33 | 12.1 | Rai and Tripathi, 2007 |
| India | Street vendors | 2/8 | 25.0 | Viswanathan and Kaur, 2001 | |
| Ireland | Supermarkets | 0/4 | 0 | McMahon and Wilson, 2001 | |
| Mexico | Supply station | 1/100 | 1.0 | Quiroz-Santiago et al. 2009 | |
| U.S. | Packing sheds, southern U.S. | 0/109 | 0 | Johnston et al., 2006 | |
| U.S. | Farms, organic, conventional, semiorganic | 0/291 | 0 | Mukhcrjee et al., 2004, 2006 | |
| Lettuce | Canada | Retail distribution centers/farmer's markets | 1/530 | 0.2 | Arthur eta)., 2007 |
| Ireland | Supermarkets | 0/8 | 0 | McMahon and Wilson, 2001 | |
| Italy | Producers | 2/62 | 3.2 | De Giusti et al., 2010 | |
| Korea | Department store, supermarket, restaurant | 1/30 | 3.3 | Seo et al., 2010 | |
| Mexico | Markets, supermarkets | 10/75 | 13 | Castancda-Ramirez et al., 2011 | |
| Nigeria | Fields | 0/55 | 0 | Okagoetal., 2003 | |
| Norway | Producers, organic | 0/179 | 0 | Loncarevic et al., 2005 | |
| Spain | Farms, organic, conventional | 0/72 | 0 | Oliveiraetal., 2010 | |
| Spain | Retail establishments | 1/29 | 3.4 | Abadias et al., 2008 | |
| U.S. | Farms, organic, conventional, semiorganic | 0/261 | 0 | Mukhcrjee et al., 2004, 2006 | |
| U.S. | Supermarkets, farmer's markets | 0/10 | 0 | Thunberg et al., 2002 | |
| U.S. | Markets and wholesale distribution centers | 2/5,453 | 0.04 | USDA, 2007, 2008, 2009 | |
| Mixed salads/vegetables | Brazil | Retailers | 1/21 | 4.8 | Fr6deretal.,2007 |
| Cyprus | Production sites, retail outlets | 6/294 | 2.0 | Fleftheriadou et al.. 2002 | |
| Korea | Department store, supermarket, restaurant | 1/129 | 0.8 | Seoetal., 2010 | |
| Malaysia | Wet markets | 40/112 | 35.7 | Salleh et al., 2003 | |
| U.K. | Catering, retail outlets | 5/10,002 | 0.05 | Sagoo et al., 2001, 2003a, 2003b | |
SOURCE: CDC.
industrial solid waste produced in that same year (Wang et al., 2005). Within the United States, it has also been reported that confined food animals produce approximately 335 million dry tons of waste per year, which is more than 40 times the amount of human biosolids waste generated from wastewater treatment plants (Graham and Nachman, 2010). The vast majority of this animal waste is applied to land without any required treatment for reduction of pathogens as is required for human biosolids (EPA, 2004).
There are two primary forms of animal wastes generated at CAFOs. In the case of broiler units, solid waste is generated either as single-use, partial reuse, or multiuse litter (Bolan et al., 2010). In confined swine and cattle operations, water is used to flush waste from the floors where the animals are housed, and the liquid slurry is channeled into large ponds for storage (Graham and Nachman, 2010). The application of animal wastes to land is largely based on agronomic requirements, geography, and commodity choices. For example, corn receives more than half of the land-applied manure, of which most of the manure is from dairy and hog stock because of the use of corn as a major feed crop for dairy and hog operations and the high growth nutrient requirement of corn for nitrogen-rich manure. Hay and grasses are the second largest of the crops fertilized by manure, which is mostly from hog, broiler, and dairy producers (MacDonald et al., 2009). Poultry litter, on the other hand, is frequently used as a fertilizer for cotton, peanuts, and fresh produce (Boyhan and Hill, 2008).
Direct Transmission of Enteric Pathogens from
Animal Wastes to Produce Fields
Animal manures applied to fields to be used for fruit and vegetable production have the potential to be a direct source of enteric pathogens if there has not been sufficient holding time between planting and harvest. The U.S. Department of Agriculture (USDA) National Organic Program permits the incorporation of raw manure into soil 120 days before harvest if the food crop has direct contact with the soil; however, only 90 days prior to harvest is required if crops have no contact with the soil (7 Code of Federal Regulations [CFR] 205.203). In contrast, more stringent requirements have been set by the Leafy Greens Marketing Agreement in which 1 year between application of raw manure and harvest of the crop is advocated (LGMA, 2012). As part of the Food Safety Modernization Act, it is anticipated that the Food and Drug Administration will include in its produce rule a required time interval between manure application to fields and either the planting or harvest of crops that would be consumed raw.
Transmission via Runoff of Enteric Pathogens from
Animal Waste—Applied Lands to Produce Fields
One of the routes by which enteric pathogens may be indirectly transferred to produce fields from domesticated animal waste deposited or stored on land
TABLE A3-5 Prevalence of Helminth and Protozoan Parasites in Leafy Greens from 2005-2010
| Ascaris spp. | Cryptosporidium spp. | |||||
| Produce item | Country | Sampling target | Number positive/number of samples | % | Number positive/number of samples | % |
| Cabbage | Ghana | Retail fruit, vegetable markets | 33/60 | 55.0 | ||
| Spain | Fields | 2/6 | 33.3 | |||
| Turkey | Wholesale markets | |||||
| Lettuce | Ghana | Retail fruit. vegetable markets | 36/60 | 60.0 | ||
| Libya | Wholesale, retail markets | 26/27 | 96.3 | |||
| Spain | Fields | 10/13 | 76.9 | |||
| Turkey | Field | 6/15 | 40.0 | |||
| Turkey | Wholesale markets | 2/35 | 5.7 | |||
SOURCE: CDC.
adjacent to produce fields is via storm runoff. Many studies have revealed that enteric pathogens can move both horizontally and vertically to contaminate land, surface waters, and ground waters adjacent to produce fields (Cooley et al., 2007; Forslund et al., 2011). In these situations, the risk of pathogen contamination of produce will be dependent on a number of factors, including the attachment strength of the pathogen to soil particles, the interval between the manure application and the precipitation events, the kinetic energy of the rainfall, the topographical slope that affects the direction and velocity of water flow, and the density of vegetation between the waste source and the destination site (Ferguson et al., 2007; Hodgon et al., 2009; Jamieson et al., 2002; Lewis et al., 2010; Mishra et al., 2008; Saini et al., 2003; Tyrrel and Quinton, 2003). In addition, the physical state of the waste will also affect the direction of movement of the pathogens with greater percolation occurring by a liquid slurry source and greater overland transport for a solid manure source (Forslund et al., 2011; Semenov et al., 2009).
Transmission of Enteric Pathogens from Waste-
Contaminated Water Sources to Produce Fields
Storm runoff carrying pathogens from animal wastes does not necessarily have to pass through agricultural produce fields to be a source of contamination. Collection in surface waters and subsequent use of that water to irrigate produce crops is another means to disseminate the pathogens. Surveys of environmental water sources for pathogen contamination have revealed significant contamina-
| Giardia spp. | Taenia spp. | Toxocara spp. | ||||
| Number positive/number of samples | % | Number positive/number of samples | % | Number positive/ number of samples | % | Reference |
| Amoah et al., 2006 | ||||||
| 2/6 | 33.3 | Amorós et al., 2010 | ||||
| 0/14 | 0 | Kozan et al., 2005 | ||||
| Amoah et al., 2006 | ||||||
| 1/27 | 3.7 | 9/27 | 33.3 | 23/27 | 85.2 | Abougrain et al., 2010 |
| 8/13 | 61.5 | Amorós et al., 2010 | ||||
| 3/15 | 20.0 | Erdoğrul and Şener, 2005 | ||||
| 2/35 | 5.7 | Kozan et al., 2005 | ||||
tion with Salmonella spp., STEC, and protozoan parasites (Table A3-10); however, contamination appears to be sporadic and is often associated with recent rain events and seasonality (Gaertner et al., 2009; Haley et al., 2009). Enhanced survival of pathogens in the sediment (Chandran et al., 2011; Garzio-Hadzick et al., 2010) and resuspension of the organisms into the water column may also perpetuate the risk. Contamination of surface waters, moreover, has been associated with the concentration of food animals raised in the area (Cooley et al., 2007; Johnson et al., 2003; Tserendorj et al., 2011; Wilkes et al., 2011). Salmonella and Cryptosporidium contamination of watersheds not impacted by human or domesticated animal production has been observed (Edge et al., 2012; Patchanee et al., 2010), which suggests that there is a level of natural occurrence of these pathogens from wildlife sources.
Several epidemiological studies lend support to the role that contaminated irrigation water serves as a transmission vehicle of enteric pathogens to fresh produce. In 2002 and 2005, two outbreaks of S. Newport infection in the United States were associated with eating tomatoes and the outbreak strain was isolated from the pond water used to irrigate the tomato fields (Greene et al., 2008). Irrigation of fields with contaminated irrigation waters was also indicated as a possible source of contamination of imported cantaloupe associated with an outbreak of S. Poona infection in the United States in consecutive years during 2000-2002 (CDC, 2002). Given the often sporadic nature of contamination of irrigation water, these documented cases linking irrigation water to an outbreak may represent only a small fraction of the contamination events that actually occur. World-
TABLE A3-6 Prevalence and Cell Numbers of Salmonella spp. in Manures from Domesticated Animals
| Pathogen | Source | Location | Prevalence (% of total samples positive) (average cell numbers in positive samples) |
Reference |
| Salmonella | Cattle feces | U.K., England and Wales | 7.7% of 810 samples were positive (4.6 log CFU/g for positive samples) | Hutchison et al., 2004 |
| Salmonella spp. | Cattle feces | Britain | 7.7-10.0% samples were positive; 3.3-3.4 log CFU/g in positive samples | Hutchison et al., 2005 |
| Salmonella | Cattle, beef and dairy farms, rectal fecal | U.S., TN. NC.AL, WA.CA | 0.2% of 480 beef cattle samples were positive; 0.4% of 480 dairy cattle samples were positive; 8.5% of 18 beef farms had at least one positive sample; 17.9% of 18 dairy farms had at least one positive sample |
Rodriguez et al., 2006 |
| Salmonella spp. | Cattle, beef, abattoir, feces | Ireland, northern | 3% of 200 samples were positive | Madden et al., 2007 |
| Salmonella | Cattle, dairy feces | New Zealand | 0% of 155 samples positive | Moriarty et al., 2008 |
| S. enterica | Cattle, feedlot feces | Australia | 6% of 32 samples were positive | Klein et al., 2010 |
| S. enterica | Cattle feces | California | 0.13% of 795 samples were positive | Gorski et al., 2011 |
| Salmonella | Chicken feces | U.K.. England and Wales | 17.9% of 67 samples were positive (3.7 log CFU/g for positive samples) |
Hutchison et al., 2004 |
| Salmonella spp. | Poultry feces | Britain | 11.5-17.9% samples were positive (2.3-3.6 log CFU/g in positive samples) |
Hutchison et al.. 2005 |
| Salmonella | Turkey feces and litter | U.S., NC | 70% of 48 composite fecal samples were positive; 79% of 48 composite litter samples positive (<1-5.3 log MPN/g in positive samples) |
Santos et al., 2005 |
| Salmonella | Chicken litter | Lebanon | 0% of 24 samples were positive | Omeira et al., 2006 |
| Salmonella | Chicken/turkey farms, rectal feces | U.S., TN. NC.AL, WA.CA | 0.2% of 480 samples were positive; 16.2% of 18 farms had at least one positive sample | Rodriguez et al., 2006 |
| Salmonella | Chickens, laying hens feces Chicken feces | Belgium | 0% of fecal samples collected on farm were positive | Van Hoorebeke et al., 2009 |
| Salmonella | Chicken feces | France | 8.6%' of 370 flocks had at least one positive sample; most prevalent serovar was S. hadar followed by S.anatum and S. mgandaka | LeBouquin et al., 2010 |
| Salmonella | Pig feces | U.K., Fngland and Wales | 7.9% of 126 samples were positive (4.0 log CFU/g for positive samples) |
Hutchison et al., 2004 |
| Salmonella spp. | Pig feces | Britain | 5.2-7.9% samples were positive (2.8 log CFU/g in positive samples) |
Hutchison et al., 2005 |
| Salmonella | Swine farms, rectal feces | U.S., TN, NC.AL, WA, CA | 6.0% of 480 samples were positive; 57.3% of 18 farms had at least one positive sample |
Rodriguez et al., 2006 |
| Salmonella | Swine, finishing pigs, feces | Italy, Piedmont | 9% of 75 fecal samples were positive | Lomonaco et al., 2009 |
| S. enlerica | Swine feces | Japan | 3.1% of 169 samples were positive 22.0% of farms had positive samples |
Kishima et al., 2008 |
| Salmonella | Sheep feces | U.K., Hngland and Wales | 8.3%' of 24 samples were positive (3.0 log CFU/g in positive samples) |
Hutchison et al., 2004 |
| Salmonella spp. | Sheep feces | Britain | 8.3-11.1% samples were positive (2.8-3.8 log CFU/g in positive samples) |
Hutchison et al., 2005 |
NOTE: CFU, colony forming unit; MPN, most probable number.
TABLE A3-7 Prevalence and Cell Numbers of Campylobacter spp. in Manures from Domesticated Animals
| Pathogen | Source | location | Prevalence (% of total samples positive)(average cell numbers in positive samples) | Reference |
| Campylobacter | Cattle feces | U.K., England and Wales | 12.8% of 810 samples were positive (3.9 log CFU/g in positive samples) |
Hutchison et al., 2004 |
| Campylobacter spp. | Cattle feces | Britain | 9.8-12.8% samples were positive (2.5-2.7 log CFU/g in positive samples) |
Hutchison et al., 2005 |
| Campylobacter | Cattle, dairy feces | New Zealand | 64% of 155 samples were positive (5.6 log CFU/g in positive samples) |
Moriarty et al., 2008 |
| Campylobacter spp. | Cattle, beef, abattoir, feces | Ireland, northern | 24.8% of 220 samples were positive | Madden et al., 2007 |
| C. jejuni | Cattle, feedlot feces | Australia | 94% of 32 samples were positive | Klein et al., 2010 |
| Campylobacter | Chicken feces | U.K., England and Wales | 19.4% of 67 samples were positive (3.6 log CFU/g for positive samples) |
Hutchison et al., 2004 |
| Campylobacter spp. | Poultry feces | Britain | 7.7-19.4% samples were positive: (2.4-2.8 log CFU/g in positive samples) |
Hutchison et al., 2005 |
| Campylobacter | Broilers ceca | Sweden | 47% of 540 ceca samples were positive: proportion of positive samples ranged from 10 to 100% within a flock. (1.7 (to 8.6 log CFU/g in positive samples) |
Hansson et al., 2010 |
| Campylobacter | Duck. Mallard, feces | U.K. | 93.3-100.0% of two groups of 60 farmed ducks tested at 28-56 days of age were positive | Colles et al., 2011 |
| Campylobacter | Pig feces | U.K., England and Wales | 13.5% of 126 samples were positive (3.3 log CFU/g for positive samples) |
Hutchison et al., 2004 |
| Campylobacter spp. | Pig feces | Britain | 10.3-13.5% samples were positive (2.5-3.2 log CFU/g in positive samples) |
Hutchison et al., 2005 |
| Campylobacter | Sheep feces | U.K., England and Wales | 20.8% of 24 samples positive (2.9 log CFU/g for positive samples) |
Hutchison et al.. 2004 |
| Campylobacter spp. | Sheep feces | Britain | 11.1-20.8% samples were positive (2.0-2.6 log CFU/g in positive samples) |
Hutchison et al., 2005 |
NOTE: CPU. colony forming unit.
| Pathogen | Source | Location | Prevalence (% of total samples positive) (average cell numbers in positive samples) | Reference |
| E.coli O157:H7 | Cattle, beef feedlot, feces | Canada, Alberta | 1.9% of 8,682 samples were positive | Berg et al., 2004 |
| E. coli O157 | Cattle, dairy beef, farms, rectal feces | Mexico, central | 1.2% of 240 samples were positive | Callaway et al., 2004 |
| E. coli O157:H7 | Cattle, beef and dairy, rectal feces | U.S., TN, NC, AL, WA, CA | 3.9% of 408 dairy cattle samples were positive 4.7% of 408 beef cattle samples positive | Doanc et al.. 2007 |
| E. coli O157:H7 | Cattle, cow and calf farms, rectal feces | U.S. | 2.5% of 408 samples were positive; 17.2% of 29 cow-calf farms positive | Dunn et al., 2004 |
| E. coli O157 | Cattle feces | U.K., England and Wales | 13.2% of 810 samples were positive (6.5 log CEU/g for positive samples) | Hutchison et al., 2004 |
| E. coli O157 | Cattle, beef and dairy farms, feces | Korea | 1.7% of 864 beef cattle samples were positive; 6.7% of 990 dairy cattle samples were positive | Jo et al., 2004 |
| E. coli O157:H7 | Cattle, feedlot | U.S., midwest | 10.2% of 10,622 cattle were positive; 52.0% of 711 pens had a positive animal; 95.9% of 73 feedlots sampled had a positive animal | Sargeant et al., 2004 |
| E. coli O157 | Cattle feces | Britain | 9.1-13.2% samples positive (2.4-3.1 log CEU/g in positive samples) | Hutchison et al., 2005 |
| E. coli STEC | Cattle, beef feces | Japan | 23% of 272 samples positive | Kijima-Tanaka et al., 2005 |
| E. coli O157:H7 | Cattle, dairy | Switzerland | 4.2% of 966 samples were positive | Kuhnert et al., 2005 |
| E. coli O157:H7 | Cattle feces | Norway | 7.0% of 156 samples were positive | Wasteson el al., 2005 |
| E. coli O157:117 | Cattle, feedlot feces | U.S., KS | 9.2%' of 891 samples were positive | Alam and Zurek, 2006 |
| E. coli STEC | Cattle, organic and conventional dairy feces | U.S., MN | 32.% of 2208 samples were positive; 71.4% of dairy farms had at least one positive sample | Cho et al., 2006 |
| Pathogen | Source | Location | Prevalence (% of total samples positive) (average cell numbers in positive samples) | Reference |
| E. coli O26 and O111 | Cattle, beef and dairy farms, feces | Korea | 6.7%, 4.6%, and 2.0% of 809 samples tested positive for O26, O111, and both O26 and O111, respectively. | Jeon et al., 2006 |
| E. coli O157 | Cattle, dairy, mature, rectal feces | U.S., Oil and Norway | 0.7% of 750 samples were positive in Ohio; 8% of 50 herds had at least one positive sample; 0% of 680 samples positive in Norway | LeJeune et al., 2006 |
| E. coli STEC | Cattle, dairy, mature, rectal fecal samples | U.S., Oil and Norway | 14% of 750 samples were positive in Ohio; 70% of 50 herds had at least one positive sample; 61% of 680 samples were positive in Norway; 100% of herds had at least one positive sample | LeJeune et al., 2006 |
| E. coli O157 | Cattle, feedlot and abattoir, fecal pats and rectal feces | U.S., CO, NE | 24.7% of 450 fecal pats were positive; 27.6% of 145 rectal fecal samples were positive | Wocmer et al., 2006 |
| E. coli O157:H7 | Cattle, rectal, feces | U.S., TX | 64.3% of 8 cattle were positive | Edrington et al., 2007 |
| E. coli O157 | Cattle, 12-30-month-old beef, feces | Scotland | 7.9% of 14,856 samples were positive; 22.8% of 952 farms had at least one positive sample | Gunn et al., 2007 |
| E. coli O157:H7 | Cattle feces | U.S., Central California | 33.8% of 77 samples tested were positive | Jay etal., 2007 |
| E. coli non-O157 | Cattle, feedlot, feces | Canada, Alberta | 0.7% of 2099 samples were positive; 57% of 21 fccdlots sampled had positive samples | Renter et al., 2007 |
| E. coli, non-O157 STEC | Cattle, beef, rectal fecal | Spain, northern | 46.0% of 124 samples were positive | Oporto et al., 2008 |
| E. coli, non-O157 STEC | Cattle, dairy, rectal fecal | Spain, northern | 20.7% of 82 samples were positive | Oporto et al., 2008 |
| E. coli O157:H7 | Cattle, beef, abattoir, feces | Ireland, northern | 0.9% of 220 samples were positive | Madden et al., 2007 |
| E. coli, STEC | Caule, dairy feces | New Zealand | 1.3% of 155 samples were positive | Moriarty et al., 2008 |
| E. coli O157:H7 | Caule, beef, rectal fecal | Spain, northern | 1.6% of 124 samples were positive; 6.7% of herds had positive samples | Oporto et al., 2008 |
| E. cO1i O157:H7 | Cattle, dairy, rectal fecal | Spain, northern | 7.0% of 82 samples were positive | Oporto et al., 2008 |
| E. cO1i O157:H7 | Cattle feces (perineal swab) | Canada | 7.2% of 2,125 cattle were identified as supershedders of E. cO1i O157:H7 (> 4 log CFU/g) in the spring/summer; 0.5% of 2,000 cattle were identified as supershedders of E. cO1i O157:H7 (> 4 log CFU/g) in the fall/winter | Stephens el al., 2009 |
| E. cO1i O157:H7 | Cattle, beef GI tract | U.S., KS | 20.3% of 815 samples were positive | Walker etal., 2010 |
| E. cO1i O157 | Chickens feces | Korea | 0% of 418 samples were positive | Jo el al., 2004 |
| E. cO1i STEC | Chicken broiler feces | Japan | 0% of 158 samples were positive | Kijima-Tanaka et al., 2005 |
| E. cO1i O157:H7 | Chicken feces | Norway | 13.6% of 22 samples were positive | Wasteson et al., 2005 |
| E. cO1i O157:H7 | Chicken and turkey, rectal fecal samples | U.S., TN, NC, AL, WA, CA | 2.7% of 444 samples were positive | Doane et al., 2007 |
| E. cO1i O157 | Swine, rectal feces | Mexico, central | 2.1% of 240 samples were positive | Callaway et al., 2004 |
| E. cO1i O157 | Pig feces | UK, England and Wales | 11.9% of 126 samples positive (4.8 log CFU/g for positive samples) | Hutchison et al., 2004 |
| E. cO1i O157 | Swine feces | Korea | 0.3% of 345 samples were positive | Jo el al., 2004 |
| E. cO1i O157 | Pig feces | Britain | 11.9-15.5% samples were positive (3.1-3.6 log CFU/g in positive samples) | Hutchison et al., 2005 |
| E. cO1i STEC | Swine feces | Japan | 14% of 179 samples were positive | Kijima-Tanaka et al., 2005 |
| E. cO1i O157:H7 | Swine, rectal fecal samples | U.S., TN, NC, AL, WA, CA | 8.8% of 426 samples were positive | Doane el al., 2007 |
| E. cO1i O157:H7 | Swine | Spain, northern | 0% of 17 samples were positive | Oporto et al., 2008 |
| Pathogen | Source | Location | Prevalence (% of total samples positive) (average cell numbers in positive samples) | Reference |
| E. coli, non-O157 STEC | Swine | Spain, Northern | 0% of 17 samples were positive | Oporto et al., 2008 |
| E. coli O157 | Sheep feces | UK, England and Wales | 20.8% of 24 samples positive (4.0 log CFU/g for positive samples) | Hutchison et al., 2004 |
| E. coli O157 | Sheep feces | Britain | 20.8-22.2% samples were positive (2.4-2.9 log CFU/g in positive samples) | Hutchison et al., 2005 |
| E. coli O157:H7 | Sheep feces | Norway | 17.1% of 117 samples positive | Wasteson et al., 2005 |
| E. coli O157 | Sheep feces | Turkey | 9.1% of 175 samples positive; 47% of 15 flocks had at least one positive sample | Turutoglu et al., 2007 |
| E. coli, non-0157 STEC | Sheep, dairy, rectal feces | Spain, northern | 50.7% of 122 samples were positive | Oporto et al., 2008 |
| E. coli O157:H7 | Sheep, dairy, rectal feces | Spain, northern | 8.7% of 122 samples were positive; 7.3% of herds had positive samples | Oporto et al., 2008 |
NOTE: CFU, colony forming unit; STEC, Shiga toxin-producing Escherichia coli.
TABLE A3-9 Prevalence and Cell Numbers of Cryptosporidium spp. in Manures from Domesticated Animals
| Pathogen | Source | Location | Prevalence (% of total samples positive) (average cell numbers in positive samples) | Reference |
| Pathogen | Source | Location | Prevalence (% of total samples positive) (average cell numbers in positive samples) |
Reference |
| C.parvum | Cattle feces | U.K.,England and Wales | 5.4% of 810 samples were positive (2.4 log CFU/g for positive samples) |
Hutchison et a)., 2004 |
| Cryptosporidium | Calf feces | Australia, Sydney watersheds |
57.1% of 7 samples were positive | Cox el al.,2005. |
| Cryptosporidium | Cattle, adult, feces |
Australia, Sydney watersheds |
22.2% of 9 samples were positive | Cox et al., 2005. |
| C.parvum | Cattle feces | Britain | 2.8-5.4% samples were positive (1.0-1.3 log CFU/g in positive samples) |
Hutchison et al., 2005 |
| Cryptosporidium | Cattle, dairy feces | New Zealand | 5.2% of 155 samples were positive | Moriarty et al., 2008 |
| Cryptosporidium spp. | Cattle, fccdlot feces | Australia | 13% of 32 samples were positive | Klein et al., 2010 |
| Cryptosporidium | Chicken feces | Australia, Sydney watersheds |
0% of 7 samples were positive | Cox el al., 2005 |
| C.parvum | Pig feces | U.K., England and Wales | 13.5% of 126 samples were positive (2.5 log CFU/g for positive samples) |
Hutchison et al., 2004 |
| Cryptosporidium | Pig feces | Australia, Sydney watersheds |
77.8% of 9 samples were positive | Cox el al., 2005 |
| C.parvum | Pig feces | Britain | 5.2-13.5% samples were positive (1.5-1.8 log CFU/g in positive samples) |
Hutchison et al., 2005 |
| Cryptosporidium | Pig slurry | Spain | 40% of 5 pig farms were positive | Bomay-Uinares et al., 2006 |
| C.parvum | Swine waste lagoons |
U.S., Southeast | 1.2% of 407 samples were positive | Jenkins et al., 2010 |
| Pathogen | Source | Location | Prevalence (% of total samples positive) (average cell numbers in positive samples) | Reference |
| Cryptosporidium | Swine feces | China, Shanghai | 34.4%. of 2,323 samples were positive; 82.6%. of positive samples were C. suis and 8.7% of positive samples were Cryptosporidium pig genotype II; 100% of 12 pig farms were infected with prevalence ranging from 14.1 to 90.6% |
Chen et al., 2011 |
| Cryptosporidium | Swine manure | Canada | 55.7% of 122 pooled samples from 10 farms were positive; 55.4% of positive samples were C. parvum and 37.5% of positive samples were Cryptosporidium sp. pig genotype II |
Farzan et al., 2011 |
| C.parvum | Sheep feces | U.K., England and Wales | 29.2% of 24 samples were positive (1.7 log CFU/g for positive samples) |
Hutchison et al., 2004 |
| Cryptosporidium | Sheep feces | Australia, Sydney watersheds |
66.6% of 9 samples were positive | Cox et al., 2005 |
| C.parvum | Sheep feces | Britain | 29.2% samples were positive (1.0 log CFU/g in positive samples) |
Hutchison et al., 2005 |
NOTE: CFU, colony forming unit.
TABLE A3-10 Prevalence of Salmonella spp., STEC, and Protozoan Parasites in Environmental Waters
| Pathogen | Source | Location | Prevalence (% positive of number of samples analyzed) | Reference |
| Salmonella spp. | Water, pond/creeks | Australia, Brisbane | 3% of 32 | Ahmed et al., 2009 |
| Water, river | Canada, Ontario | 62% of 32 | Droppo et al., 2009 | |
| Water, river | U.S., GA | 79.2% of 72 | Haley et al., 2009 | |
| Water, surface | Netherlands | 14.3% of 49 | Heuvelink et al., 2008 | |
| Watersheds, swine | U.S., NC | 41.7% of 12 | Patchanee et al., 2010 | |
| Watersheds, agriculture crops | U.S., NC | 50% of 12 | Patchanee et al., 2010 | |
| Watersheds, forestry | U.S., NC | 57.1% of 28 | Patchanee et al., 2010 | |
| Water, surface | Canada, Alberta | 6.2% of 1429 | Johnson el al., 2003 | |
| Water, irrigation | Nigeria | 8.2% of 196 | Okago et al., 2003 | |
| E. coli 0157 | Water, surface | Netherlands | 2.0% of 49 | Heuvelink et al., 2008 |
| E. coli 0157:H7 | Water, surface | U.S., Central CA | 3.83 of 79 | Jay et al., 2007 |
| Water, well | U.S., Central CA | 0% of 19 | Jay et al., 2007 | |
| Water, surface | Canada, Alberta | 0.9% of 1483 | Johnson el al., 2003 | |
| STEC | Water, pond and creeks | Australia, Brisbane | 9-15% of 32 | Ahmed et al., 2009 |
| Cryptosporium | Water, reclaimed | U.S. | 70% of 30 | Harwood et al., 2005 |
| Water, irrigation | Mexico | 18% of 11 | Thurston-Enriquez et al., 2002 | |
| Water, irrigation | U.S. | < l of 3 | Thurston-Enriquez et al.,2002 | |
| Giardia | Water, reclaimed | U.S. | 80% of 30 | Harwood et al., 2005 |
| Water, irrigation | Mexico | 64% of 11 | Thurston-Enriquez et al.,2002 | |
| Water, irrigation | U.S. | 67% of 3 | Thurston-Enriquez et al.,2002 | |
| Microsporidia | Water, irrigation | U.S. | 67% of 3 | Thurston-Enriquez et al.,2002 |
wide it is estimated that 17 percent of the world’s cropland (1.4 billion hectares) is irrigated and, of that, 20 million hectares are irrigated with untreated wastewater (Jimenez et al., 2010). In the United States and the United Kingdom, extensive irrigation of fresh produce crops occurs and, of the acreage irrigated, 48 percent and 78 percent, respectively, are derived from non-groundwater sources (Knox et al., 2011; USDA NASS, 2009), which are subject to intermittent inputs of pathogens from animal husbandry operations.
Contribution of Bioaerosols to Dissemination of Enteric Pathogens from Animal Production Operations to Produce Fields
Aerosolization of microbial pathogens is an inevitable consequence associated with animal production operations as well as the handling and disposition of animal manure. However, estimating the impact of bioaerosol dispersal on pathogen dissemination has been hampered by the notable absence of standardized and validated methods for enumeration of various types of microorganisms in outdoor bioaerosols. Hence, there has been a wide range of prevalence and cell number values reported across very diverse types of animal operations and landscapes (Millner, 2009).
Studies addressing bioaerosol levels in outdoor air generally address fecal indicator organisms because they are more abundant and easily identified in the aerosols, although it is acknowledged that they may behave differently than the pathogens. The general trend that has been observed is decreasing airborne microorganism concentrations as the distance from the source increases with relative humidity, temperature, and solar irradiance being major factors affecting viability (Dungan, 2010). Other pertinent observations made in studies addressing the levels of the indicator organism, E. coli, in aerosols of poultry houses are that the levels of airborne bacteria are intricately linked to the levels of those bacteria in the litter (Chinivasagam et al., 2009; Smith et al., 2012) and the type of ventilation system affects the distance that E. coli is disseminated, with E. coli traversing 11.1 and 7.5 m downwind from houses using tunnel and conventional fans, respectively (Smith et al., 2012).
Limited studies have been conducted addressing bioaerosol transport following land application of animal manures in contrast to those addressing the application of municipal wastes (Pillai and Ricke, 2002). Although there may be some similar behavior between these two sources, there could be differences given that they vary in their organic matter content that can provide differences in the degree of protection against ultraviolet radiation and drying (Dungan, 2010). In one of the few studies addressing land application of cattle and swine slurry and the method used to disperse the wastes, total bacterial counts in the air were greater at greater distances from spray guns that discharged the slurry upward into the air compared to tank spreading that sprayed the slurries closer to the ground (Boutin et al., 1988). In another study in which swine manure was ap-
plied through a center pivot irrigation system, coliform concentrations decreased to near background concentrations at 23 m downwind (Kim et al., 2008). Wind speed and topography, however, are likely to also factor into the distances traversed by pathogens and, hence, safe distances between produce fields and animal production activities will likely be site specific.
Wildlife as a Vehicle to Transmit Pathogens from
Domesticated Animal Waste to Produce Fields
The recent focus on wildlife as a potential source of pathogen contamination of produce fields was driven by the isolation of E. coli O157:H7 from feral swine that occupied areas near spinach fields and cattle farms in California following the 2006 spinach outbreak (Jay et al., 2007). More recently, Campylobacter jejuni was isolated both from Sandhill crane feces and raw peas and several of the isolates had pulsed-field gel electrophoresis (PFGE) patterns indistinguishable from clinical samples obtained during a C. jejuni gastroenteritis outbreak that occurred in Alaska in 2008 (Gardner et al., 2011). Attention was again focused on wildlife as a potential source of contamination when E. coli O157:H7 isolated from deer feces was determined to have an identical PFGE pattern as the isolates responsible for 15 people who were ill from eating contaminated fresh straw berries in Oregon in 2011 (IEH Laboratories & Consulting Group, 2011). Given that the same strain was also isolated from soil raises the question as to whether the deer were actually the source of the outbreak or were infected when they ate the contaminated strawberries. Most evidence indicating that wildlife is a potential source of food-borne contamination is from the isolation of clinically relevant pathogens from the animal’s feces. In one example, Renter et al. (2006) isolated from deer fecal samples four Salmonella serovars (Litchfield, Dessau, Infantis, and Enteritidis) known to be pathogenic to humans and animals. In another example, subtyping of STEC isolates from wildlife meat in Germany identified virulence genes associated with severe clinical outcome (stx2, stx2d, and eae) in 46 of the 140 STEC samples (Miko et al., 2009). More definitive proof that specific types of wildlife could be transmission vectors of pathogens from domesticated animal facilities was obtained with a study of European starlings (Williams et al., 2011). In that study, distinct molecular types of E. coli O157:H7 were similar in starlings and cattle on different farms, and these birds were capable of shedding the pathogen in their feces for more than 3 days (Kauffman and LeJeune, 2011). Hence, it is reasonable to assume that European starlings could serve as a vector of pathogens from cattle and dairy farms to produce fields.
In response to the limited studies linking wildlife to produce contamination, processors and buyers have become overreactive in many cases in requiring the absence of many types of wildlife from farms. To illustrate this trend, the percentage of growers that reported being told by their processors or buyers that feral pigs, deer, birds, rodents, and amphibians were a significant risk was 19, 28,
44, 47, and 28 percent, respectively (Lowell et al., 2010). Several studies, however, have revealed that some groups of animals have a very low prevalence of contamination with relevant human enteric pathogens (Table A3-11). It is likely that all animal groups have the potential to be contaminated with a food-borne pathogen, but whether they are significant harbingers of human enteric pathogens is likely dependent on their access to animal husbandry sites as well as on their social behavior (i.e., existence of a social group and its size). This would also be the case with insects. For example, filth flies collected in leafy green fields were believed to have originated from nearby rangelands that contained fresh cattle manure (Talley et al., 2009).
Persistence of Pathogens on Produce in Fields Requires a Systems Approach to Prevent and Monitor Pathogen Introduction
Many field studies have revealed the persistence of human enteric pathogens, albeit typically at low levels, in a number of different vegetables contaminated at various points during their cultivation (Erickson et al., 2010; Gutiérrez-Rodriguez et al., 2011; Islam et al., 2004a, 2004b, 2004c, 2005; Moyne et al., 2011). This is noteworthy because chemical disinfectants typically used during minimal processing of fresh produce are not fully effective in eliminating pathogen contamination (Doyle and Erickson, 2008). Hence, it is paramount to prevent the introduction of these pathogens into produce fields. The primary approach currently used to reduce the risk of pathogen contamination in fields is the application of good agricultural practices (GAPs). To prevent the introduction of pathogens through nontraditional vehicles (storm runoff, intrusions by pathogen-carrying wildlife) will require the development of novel approaches in addition to GAPs. Given that the environment surrounding the produce field would likely be impacted by these pathogen control practices, it is important to implement a systems approach and consider all ramifications to the adoption of any intervention practices. It is also important to be cognizant that storm runoff and fecal deposits from wildlife may only contaminate the plants at discrete locations within a field. The ability to detect this contamination by current sampling plans that rely on uniform contamination is therefore limited and efforts are needed to develop new monitoring systems that can detect contamination when such pathogen introductions occur.
Concluding Comments
Vegetables, fruits, and a variety of plant foods and ingredients are now recognized as major vehicles of food-borne disease outbreaks, and a primary source of pathogen contamination of this commodity group is animal manure. There are several routes by which pathogens can be transmitted from animal production sites to produce fields. The vehicles likely presenting the greatest risk are manure-contaminated soil amendments and irrigation water. Wildlife, insects,
TABLE A3-11 Prevalence of Enteric Food-Borne Pathogens in Wildlife and Insects
| Animal | Pathogen | Country | Prevalence (% positive of number of samples analyzed) | Reference |
| Wild boars/pigs | Ecoli O157:H7 | U.S. | 14.9% of 87 | Jay et al., 2007 |
| Ecoli O157:H7 | Spain | 3.3% of 212 | Sánchez et al., 2010 | |
| Non-O157 STEC | Spain | 5.2% of 212 | Sánchez et al., 2010 | |
| C.parvutn | Australia | 0% of 5 | Cox et al., 2005 | |
| Giardia | Australia | 0% of 5 | Cox et al., 2005 | |
| Coyotes | Salmonella | U.S. | 5% of 40 | Gorski et al., 2011 |
| Rabbits | E. coli (VTKC) | U.K. | 15.5% of 129 | Scaife et al., 2006 |
| Cryptosporidium | Australia | 50% of 2 | Cox et al., 2005 | |
| Giardia | Australia | 0% of 2 | Cox et al., 2005 | |
| Raccoons | Salmonella | U.S. | 0% of 2 | Gorski et al., 2011 |
| Skunks | Salmonella | U.S. | 30.7% of 13 | Gorski et al., 2011 |
| Deer | Ecoli O157:117 | U.S. | 0% of 4 | Jay et al., 2007 |
| Cryptosporidium | Australia | 100% of 1 | Cox et al., 2005 | |
| Giardia | Australia | 0% of 1 | Cox et al., 2005 | |
| Salmonella | U.S. | 1%. of 500 | Renter el al., 2006 | |
| Pigeons/sparrows | E. coli O157 | Czech Rep. | 0 %of 70 | Čižek et al., 1999 |
| Birds (cattle farm) | Ecoli O157:H7 | U.S. | 0.5% of 200 pooled | Hancock el al., 1998 |
| Geese, Canadian | E coli (EHEC) | U.S. | 6.0% of 151 | Kullas et al., 2002 |
| Multiple birds | Salmonella | Denmark | 1.5% of 1285 | Skov et al.. 2008 |
| (domestic animal farms) | ||||
| Multiple birds | Salmonella | U.S. | 6.6% of 105 | Gorskietal., 2011 |
| Reptiles | Salmonella | Spain | 41.5% of 94 | Briones et al., 2004 |
| Amphibians | Salmonella | Spain | 0% of 72 | Briones et al., 2004 |
| Rats | E. coli O157 | Czech Rep. | 40% of 10 | Čižek et al., 1999 |
| Mice, wood | E. coli O157 | Czech Rep. | 0% of 7 | Čižek et al., 1999 |
| Animal | Pathogen | Country | Prevalence (% positive of number of samples analyzed) | Reference |
| Rodents | E.coii O157:H7 | U.S. | 0% of 300 pooled | Hancock et al., 1998 |
| Rodents | Toxoplasma gondii | Netherlands | 9.1% of 77 | Kijlstra et al., 2008 |
| Rodents | Salmonella | Denmark | 2% of 135 | Skov el al., 2008 |
| Mice/rats | S. Enteritidis | U.S. | 16.2% of 715 | Henzler and Opiu. 1992 |
| Squirrels | S. enterica | U.S. | 0% of 28 | Gorskietal.,2011 |
| Mice/rats | C. parvutn | U.S. | 27.8% of 241 | Li et al., 2011 |
| Hies | E.coii O157:H7 | U.S. | 3.3% of 60 | Hancock et al., 1998 |
| House flies | E.coii O157:H7 | U.S. | 2.2% of 3,440 | Alam and Zurek. 2004 |
| Hies | Salmonella | Denmark | 22.6% of 31 | Skov et al., 2008 |
| llouseflies/dump Hies | Salmonella | U.S. | 18% of 22 | Olsen and Hammack, 2000 |
| House flies | S. Enteritidis | U.S. | ~50% of 120 | Holt et al., 2007 |
| Cockroaches | Salmonella | U.S. | 14.4% of 90 | Kopanic et al., 1994 |
| Slugs | E.coii O157:H7 | U.K. | 0.2% of 33 | Sproston et al., 2006 |
and vermin, however, may also serve as intermediate vectors of pathogens from animal wastes to plants in the field. The multifaceted routes by which pathogens may be transmitted to produce crops illuminates the value of a One Health approach to minimize pathogen contamination in the production environment while ensuring that adverse effects to the environment be minimized.
References
Abadias, M., J. Usall, M. Anguera, C. Solsona, and I. Viñas. 2008. Microbiological quality of fresh, minimally-processed fruit and vegetables, and sprouts from retail establishments. International Journal of Food Microbiology 123:121-129.
Abougrain, A. K., M. H. Nahaisi, N. S. Madi, M. M. Saied, and K. S. Ghenghesh. 2010. Parasitological contamination in salad vegetables in Tripoli-Libya. Food Control 21:760-762.
Ahmed, W., S. Sawant, F. Huygens, A. Goonetilleke, and T. Gardner. 2009. Prevalence and occurrence of zoonotic bacterial pathogens in surface waters determined by quantitative PCR. Water Research 43:4918-4928.
Alam, M. J., and L. Zurek. 2004. Association of Escherichia coli O157:H7 with houseflies on a cattle farm. Applied and Environmental Microbiology 70:7578-7580.
Alam, M. J., and L. Zurek. 2006. Seasonal prevalence of Escherichia coli O157:H7 in beef cattle feces. Journal of Food Protection 69:3018-3020.
Amoah, P., P. Drechsel, R. C. Abaidoo, and W. J. Ntow. 2006. Pesticide and pathogen contamination of vegetables in Ghana’s urban markets. Archives of Environmental Contamination and Toxicology 50:1-6.
Amorós, I., J. L. Alonso, and G. Cuesta. 2010. Cryptosporidium oocysts and Giardia cysts on salad products irrigated with contaminated water. Journal of Food Protection 73:1138-1140.
Anderson, M., L.-A. Jaykus, S. Beaulieu, and S. Dennis. 2011. Pathogen-produce pair attribution risk ranking tool to prioritize fresh produce commodity and pathogen combinations for further evaluation (P3ARRT). Food Control 22:1865-1872.
Arthur, L., S. Jones, M. Fabri, and J. Odumeru. 2007. Microbial survey of selected Ontario-grown fresh fruits and vegetables. Journal of Food Protection 70:2864-2867.
Berg, J., T. McAllister, S. Bach, R. Stilborn, D. Hancock, and J. LeJeune. 2004. Escherichia coli O157:H7 excretion by commercial feedlot cattle fed either barley- or corn-based finishing diets. Journal of Food Protection 67:666-671.
Bolan, N. S., A. A. Szogi, T. Chuasavathi, B. Seshadri, M. J. Rothrock, Jr., and P. Panneerselvam. 2010. Uses and management of poultry litter. World’s Poultry Science Journal 66:673-698.
Bornay-Llinares, F. J., L. Navarro-i-Martínez, F. García-Orenes, H. Araez, M. D. Pérez-Murcia, and R. Moral. 2006. Detection of intestinal parasites in pig slurry: A preliminary study from five farms in Spain. Livestock Science 102:237-242.
Boutin, P., M. Torre, R. Serceau, and P. J. Rideau. 1988. Atmospheric bacterial contamination from landspreading of animal wastes: Evaluation of the respiratory risk for people nearby. Journal of Agricultural Engineering Research 39:149-160.
Boyhan, G. E., and C. R. Hill. 2008. Organic fertility sources for the short-day organic onion transplants. HortTechnology 18:227-231.
Briones, V., S. Téllez, J. Goyache, C. Ballesteros, M. del Pilar Lanzarot, L. Domínguez, and J. F. Fernández-Garayzábal. 2004. Salmonella diversity associated with wild reptiles and amphibians in Spain. Environmental Microbiology 6:868-871.
Callaway, T. R., R. C. Anderson, G. Tellez, C. Rosario, G. M. Nava, C. Eslava, M. A. Blanco, M. A. Quiroz, A. Olguín, M. Herradora, T. S. Edrington, K. J. Genovese, R. B. Harvey, and D. J. Nisbet. 2004. Prevalence of Escherichia coli O157 in cattle and swine in Central Mexico. Journal of Food Protection 67:2274-2276.
Calvin, L. 2007. Outbreak linked to spinach forces reassessment of food safety practices. Amber Waves 5(3):24-31. http://www.ers.usda.gov/AmberWaves/June07/PDF/Spinach.pdf (accessed January 6, 2011).
Castañeda-Ramírez, C., V. Cortes-Rodríguez, N. de la Fuente-Salcido, D. K. Bideshi, M. Cristina, M. C. del Rincón-Castro, and J. E. Barboza-Corona. 2011. Isolation of Salmonella spp. from lettuce and evaluation of its susceptibility to novel bacteriocins of Bacillus thuringiensis and antibiotics. Journal of Food Protection 74:274-278.
CDC (Centers for Disease Control and Prevention). 2002. Multistate outbreaks of Salmonella serotype Poona infections associated with eating cantaloupe from Mexico—United States and Canada, 2000-2002. Morbidity and Mortality Weekly Report 51(46):1044-1047.
———. 2006a. Botulism associated with commercial carrot juice—Georgia and Florida, Septem ber 2006. Morbidity and Mortality Weekly Report 55:1098-1099. http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5540a5.htm (accessed January 8, 2011).
———. 2006b. Salmonellosis—outbreak investigation, October 2006. http://www.cdc.gov/ncidod/dbmd/diseaseinfo/salmonellosis_2006/outbreak_notice.htm (accessed January 6, 2011).
———. 2008a. Investigation of outbreak of infections caused by Salmonella Litchfield. http://www.cdc.gov/salmonella/litchfield/ (accessed January 6, 2011).
———. 2008b. Outbreak of Salmonella serotype Saintpaul infections associated with multiple raw produce items—United States, 2008. Morbidity and Mortality Weekly Report 57:929-934. http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5734a1.htm (accessed January 6, 2011).
———. 2009a. Surveillance for foodborne disease outbreaks—United States. 2006. Morbidity and Mortality Weekly Report 58:609-615, expanded Table 2. http://www.cdc.gov/outbreaknet/pdf/surveillance/2006_reported_outbreaks_illnesses.pdf (accessed March 14, 2012).
———. 2009b. Outbreak of Salmonella serotype Saintpaul infections associated with eating alfalfa sprouts—United States, 2009. Morbidity and Mortality Weekly Report 58:500-503.
———. 2010a. Investigation update: Multistate outbreak of human Salmonella Newport infections linked to raw alfalfa sprouts. http://www.cdc.gov/salmonella/newport/index.html (accessed January 6, 2011).
———. 2010b. Investigation update: Multistate outbreak of human E. coli O145 infections linked to shredded romaine lettuce from a single processing facility (final update). http://www.cdc.gov/ecoli/2010/ecoli_o145/index.html (accessed July 21, 2011).
———. 2010c. Surveillance for foodborne disease outbreaks—United States, 2007. Morbidity and Mortality Weekly Report 59:973-579, expanded Table 2. http://www.cdc.gov/outbreaknet/pdf/2007MMWRSurveillanceOutbreaks_ExpandedTable2_WEB.pdf (accessed March 14, 2012).
———. 2011a. Investigation of a multistate outbreak of human Salmonella I 4,[5],12:i:- infections linked to alfalfa sprouts (January 6, 2011 report). http://www.cdc.gov/salmonella/i4512i-/010611/index.html (accessed January 6, 2011).
———. 2011b. Investigation update: Multistate outbreak of Salmonella Panama infections linked to cantaloupe. http://www.cdc.gov/salmonella/panama0311/062311/index.html (accessed March 10, 2012).
———. 2011c. Multistate outbreak of listeriosis linked to whole cantaloupes from Jensen Farms, Colorado (final update). http://www.cdc.gov/listeria/outbreaks/cantaloupes-jensen-farms/120811/index.html (accessed March 10, 2012).
———. 2011d. Surveillance for foodborne disease outbreaks—United States, 2008. Morbidity and Mortality Weekly Report 60:1197-1202, expanded Table 2. http://www.cdc.gov/outbreaknet/pdf/2008MMWR-Table2.pdf (accessed March 14, 2012).
Chandran, A., S. Varghese, E. Kandeler, A. Thomas, M. Hatha, and A. Mazumder. 2011. An assessment of potential public health risk associated with the extended survival of indicator and pathogenic bacteria in freshwater lake sediments. International Journal of Hygiene and Environmental Health 214:258-264.
Chen, Z., R. Mi, H. Yu, Y. Shi, Y. Huang, Y. Chen, P. Zhou, Y. Cai, and J. Lin. 2011. Prevalence of Cryptosporidium spp. in pigs in Shanghai, China. Veterinary Parasitology 181:113-119.
Chinivasagam, H. N., T. Tran, L. Maddock, A. Gale, and P. J. Blackall. 2009. Mechically ventilated broiler sheds: A possible source of aerosolized Salmonella, Campylobacter, and Escherichia coli. Applied and Environmental Microbiology 75:7417-7425.
Cho, S., F. Diez-Gonzalez, C. P. Fossler, S. J. Wells, C. W. Hedberg, J. B. Kaneene, P. L. Ruegg, L. D. Warnick, and J. B. Bender. 2006. Prevalence of shiga toxin-encoding Escherichia coli isolates from dairy farms and county fairs. Veterinary Microbiology 118:289-298.
Čížek, A., P. Alexa, I. Literák, J. Hamrík, P. Novák, and J. Smola. 1999. Shiga toxin-producing Escherichia coli O157 in feedlot cattle and Norwegian rats from a large-scale farm. Letters in Applied Microbiology 28:435-439.
Colles, F. M., J. S. Ali, S. K. Sheppard, N. D. McCarthy, and M. C. J. Maiden. 2011. Campylobacter populations in wild and domesticated Mallard ducks (Anas platyrhynchos). Environmental Microbiology Reports 3:574-580.
Cooley, M., D. Carychao, L. Crawford-Miksza, M. T. Jay, C. Myers, C. Rose, C. Keys, J. Farrar, and R. E. Mandrell. 2007. Incidence and tracking of Escherichia coli O157:H7 in a major produce production region in California. PLoS ONE 11:E1159.
Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Applied and Environmental Microbiology 71:5929-5934.
De Giusti, M., C. Aurigemma, L. Marinelli, D. Tufi, D. De Medici, S. Di Pasquale, and C. De Vito. 2010. The evaluation of the microbial safety of fresh ready-to-eat vegetables produced by different technologies in Italy. Journal of Applied Microbiology 109:996-1006.
Doane, C. A., P. Pangloli, H. A. Richards, J. R. Mount, D. A. Golden, and F. A. Draughon. 2007. Occurrence of Escherichia coli O157:H7 in diverse farm environments. Journal of Food Protection 70:6-10.
Doyle, M. P., and M. C. Erickson. 2008. The problems with fresh produce: An overview. Journal of Applied Microbiology 105:313-330.
Droppo, I. G., S. N. Liss, D. Williams, T. Nelson, C. Jaskot, and B. Trapp. 2009. Dynamic existence of waterborne pathogens within river sediment compartments. Implications for water quality regulatory affairs. Environmental Science & Technology 43:1737-1743.
Dungan, R. S. 2010. Fate and transport of bioaerosols associated with livestock operations and manures. Journal of Animal Science 88:3693-3706.
Dunn, J. R., J. E. Keen, R. Del Vecchio, T. E. Wittum, and R. Alex Thompson. 2004. Escherichia coli O157:H7 in a cohort of weaned, preconditioned range beef calves. Journal of Food Protection 67:2391-2396.
Edge, T. A., A. El-Shaarawi, V. Gannon, C. Jokinen, R. Kent, I. U. H. Khan, W. Koning, D. Lapen, J. Miller, N. Neumann, R. Phillips, W. Robertson, H. Schreier, A. Scott, I. Shtepani, E. Topp, G. Wilkes, and E. van Bochove. 2012. Investigation of an Escherichia coli environmental benchmark for waterborne pathogens in agricultural watersheds in Canada. Journal of Environmental Quality 41:21-30.
Edrington, T. S., T. R. Callaway, D. M. Hallford, R. C. Anderson, and D. J. Nisbet. 2007. Influence of exogenous triiodothyronine (T3) on fecal shedding of Escherichia coli O157 in cattle. Microbial Ecology 53:664-663.
EFSA (European Food Safety Authority). 2011. Shiga toxin-producing E. coli (STEC) O104:H4 2011 outbreaks in Europe: Taking stock. EFSA Journal 9:2390.
Eleftheriadou, M., A. Varnava-Tello, M. Metta-Loizidou, A-S. Nikolaou, and D. Akkelidou. 2002. The microbiological profile of foods in the Republic of Cyprus: 1991-2000. Food Microbiology 19:463-471.
Elviss, N. C., C. L. Little, L. Hucklesby, S. Sagoo, S. Surman-Lee, E. de Pinna, and E. J. Threlfall. 2009. Microbiological study of fresh herbs from retail premises uncovers an international outbreak of salmonellosis. International Journal of Food Microbiology 134:83-88.
Emberland, K. E., S. Ethelberg, M. Kuusi, L. Vold, L. Jensvoll, B-A. Lindstedt, K. Nygård, C. Kjelsø, G. Sørensen, T. Jensen, S. Lukinmaa, T. Niskanen, and G. Kapperud. 2007. Outbreak of Salmonella Weltevreden infections in Norway, Denmark and Finland associated with alfalfa sprouts, July-October 2007. Eurosurveillance 12(48):Article 4. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=3321 (accessed January 6, 2011).
EPA (U.S. Environmental Protection Agency). 2004. Managing manure nutrients at concentrated animal feeding operations. Washington, DC. http://tammi.tamu.edu/final-manure-guidance.pdf (accessed March 11, 2012).
Erickson, M. C., and Y. R. Ortega. 2006. Inactivation of parasites in food, water, and environmental systems. Journal of Food Protection 69:2786-2808.
Erickson, M. C., C. C. Webb, J. C. Diaz-Perez, S. C. Phatak, J. J. Silvoy, L. E. Davey, A. S. Payton, J. Liao, L. Ma, and M. P. Doyle. 2010. Surface and internalized Escherichia coli O157:H7 on field-grown spinach and lettuce treated with spray-contaminated irrigation water. Journal of Food Protection 73:1023-1029.
Erdogrul, Ö., and H. Şener. 2005. The contamination of various fruits and vegetable with Enterobius vermicularis, Ascaris eggs, Entamoeba histolyca cysts and Giardia cysts. Food Control 16:559-562.
Ethelberg, S., M. Lisby, B. Böttiger, A. C. Schultz, A. Villif, T. Jensen, K. E. Olsen, F. Scheutz, C. Kjelsø, and L. Müller. 2010. Outbreaks of gastroenteritis linked to lettuce, Denmark, January 2010. Eurosurveillance 15(6):Article 1. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19484 (accessed January 6, 2011).
Farzan, A., L. Parrington, T. Coklin, A. Cook, K. Pintar, F. Pollari, R. Friendship, J. Farber, and B. Dixon. 2011. Detection and characterization of Giardia duodenalis and Cryptosporidium spp. on swine farms in Ontario, Canada. Foodborne Pathogens and Disease 8:1207-1213.
FDA (Food and Drug Administration). 2006. UPDATE: E. coli O157:H7 Outbreak at Taco Bell restaurants likely over. FDA traceback investigation continues. http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/2006/ucm108805.htm (accessed January 6, 2011).
———. 2007. FDA and states closer to identifying source of E. coli contamination associ ated with illnesses at Taco John’s restaurants. http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/2007/ucm108827.htm (accessed January 6, 2011).
Ferguson, C. M., C. M. Davies, C. Kaucner, M. Krogh, J. Rodehutskors, D. A. Deere, and N. J. Ashbolt. 2007. Field scale quantification of microbial transport from bovine feces under simulated rainfall events. Journal of Water and Health 5:83-95.
Forslund, A., B. Markussen, L. Toenner-Klank, T. B. Bech, O. S. Jacobsen, and A. Dalsgaard. 2011. Leaching of Cryptosporidium parvum oocysts, Escherichia coli, and a Salmonella enterica serovar Typhimurium bacteriophage through intact soil cores following surface application and injection of slurry. Applied and Environmental Microbiology 77:8129-8138.
Friesema, I., G. Sigmundsdottir, K. van der Zwaluw, A. Heuvelink, B. Schimmer, C. de Jager, B. Rump, H. Briem, H. Hardardottir, A. Atladottir, E. Gudmundsdottir, and W van Pelt. 2007. An international outbreak of Shiga toxin-producing Escherichia coli O157 infection due to lettuce, September-October 2007. Eurosurveillance 13:50. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19065 (accessed January 6, 2011).
Fröder, H., C. G. Martins, K. L. Oliveira de Souza, M. Landgraf, B. D. G. M. Franco, and M. T. Destro. 2007. Minimally processed vegetable salads: Microbial quality evaluation. Journal of Food Protection 70:1277-1280.
Gaertner, J. P., T. Garres, J. C. Becker, M. L. Jimenez, M. R. J. Forstner, and D. Hahn. 2009. Temporal analyses of Salmonellae in a headwater spring ecosystem reveals the effects of precipitation and runoff events. Journal of Water and Health 7:115-121.
Gardner, T. J., C. Fitzgerald, C. Xavier, R. Klein, J. Pruckler, S. Stroika, and J. B. McLaughlin. 2011. Outbreak of campylobacteriosis associated with consumption of raw peas. Clinical Infectious Diseases 53:16-21.
Garzio-Hadzick, A., D. R. Shelton, R. L. Hill, Y. A. Pachepsky, A. K. Guber, and R. Rowland. 2010. Survival of manure-borne E. coli in streambed sediment: Effects of temperature and sediment properties. Water Research 44:2753-2762.
Gorski, L., C. T. Parker, A. Liang, M. B. Cooley, T. M. Jay-Russell, A. G. Gordus, E. R. Atwill, and R. E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Applied and Environmental Microbiology 77:2734-2748.
Graczyk, T. K., F. E. Lucy, L. Tamang, and A. Miraflor. 2007. Human enteropathogen load in activated sewage sludge and corresponding sewage sludge end products. Applied and Environmental Microbiology 73:2013-2015.
Graham, J. P., and K. E. Nachman. 2010. Managing waste from confined animal feeding operations in the United States: The need for sanitary reform. Journal of Water and Health 8:646-670.
Greene, S. K., E. R. Daly, E. A. Talbot, L. J. Demma, S. Holzbauer, N. J. Patel, T. A. Hill, M. O. Walderhaug, R. M. Hoekstra, M. F. Lynch, and J. A. Painter. 2008. Recurrent multistate outbreak of Salmonella Newport associated with tomatoes from contaminated fields, 2005. Epidemiology & Infection 136:157-165.
Gunn, G. J., I. J. McKendrick, H. E. Ternent, F. Thomson-Carter, G. Foster, and B. A. Synge. 2007. An investigation of factors associated with the prevalence of verocytotoxin producing Escherichia coli O157 shedding in Scottish beef cattle. The Veterinary Journal 174:554-564.
Gutiérrez-Rodriguez, E., A. Gundersen, A. O. Sbodio, and T. V. Suslow. 2011. Variable agronomic practices, cultivar, strain source, and initial contamination dose differentially affect survival of Escherichia coli on spinach. Journal of Applied Microbiology 112:109-118.
Haley, B. J., D. Cole, and E. K. Lipp. 2009. Distribution, diversity and seasonality of water-borne Salmonella in a rural watershed. Applied and Environmental Microbiology 75:1248-1255.
Halweil, B., and D. Nierenberg. 2004. State of the world 2004: Special focus: The consumer society. In Watching what we eat. Washington, DC: The Worldwatch Institute.
Hancock, D. D., T. E. Besser, D. H. Rice, E. D. Ebel, D. E. Herriott, and L. V. Carpenter. 1998. Multiple sources of Escherichia coli O157 in feedlots and dairy farms in the northwestern USA. Preventive Veterinary Medicine 35:11-19.
Hansson, I., N. Pudas, B. Harborn, and E. O. Engvall. 2010. Within-flock variations of Campylobacter loads in caeca and on carcasses from broilers. International Journal of Food Microbiology 141:51-55.
Harwood, V. J., A. D. Levine, T. M. Scott, V. Chivukula, J. Lukasik, S. R. Farrah, and J. B. Rose. 2005. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Applied and Environmental Microbiology 71:3163-3170.
Henzler, D. J., and H. M. Opitz. 1992. The role of mice in the epizootiology of Salmonella enteritidis infection on chicken layer farms. Avian Diseases 36:625-631.
Heuvelink, A. E., J. T. M. Zwarkruis, C. Van Heerwaarden, B. Arends, V. Stortelder, and E. de Boer. 2008. Pathogenic bacteria and parasites in wildlife and surface water. Tijdschrift Voor Diergeneeskunde 133:330-335.
Hjertqvist, M., A. Johansson, N. Svensson, P. E. Aborn, C. Magnusson, M. Olsson, K. O. Hedlund, and Y. Andersson. 2006. Four outbreaks of norovirus gastroenteritis after consuming raspberries, Sweden, June—August 2006. Eurosurveillance 11(36). http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=3038 (accessed January 6, 2011).
Hodgon, C. J., N. Bulmer, D. R. Chadwick, D. M. Oliver, A. L. Heathwaite, R. D. Fish, and M. Winter. 2009. Establishing relative release kinetics of faecal indicator organisms from different fecal matrices. Letters in Applied Microbiology 49:124-130.
Holt, P. S., C. J. Geden, R. W. Moore, and R. K. Gast. 2007. Isolation of Salmonella enterica serovar Enteritidis from houseflies (Musca domestica) found in rooms containing Salmonella serovar Enteritidis-challenged hens. Applied and Environmental Microbiology 73:6030-6035.
Hutchison, M. L., L. D. Walters, S. M. Avery, B. A. Synge, and A. Moore. 2004. Levels of zoonotic agents in British livestock manures. Letters in Applied Microbiology 39:207-214.
Hutchison, M. L., L. D. Walters, S. M. Avery, F. Munro, and A. Moore. 2005. Analyses of livestock production, waste storage, and pathogen levels and prevalences in farm manures. Applied and Environmental Microbiology 71:1231-1236.
IEH Laboratories & Consulting Group. 2011. IEH isolates outbreak strain of E. coli O157:H7 from deer feces at implicated Oregon strawberry farm. http://www.iehinc.com/article/index.jek8_9_11 (accessed March 13, 2012).
Insulander, M., B. de Jong, B. Svenungsson. 2008. A food-borne outbreak of cryptosporidiosis among guests and staff at a hotel restaurant in Stockholm county, Sweden, September 2008. Eurosurveillance 13(51). http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19071 (accessed January 6, 2011).
Islam, M., M. P. Doyle, S. C. Phatak, P. Millner, and X. Jiang. 2004a. Persistence of entero-hemorrhagic Escherichia coli O157:H7 in soil and on leaf lettuce and parsley grown in fields treated with contaminated manure composts or irrigation water. Journal of Food Protection 67:1365-1370.
Islam, M., J. Morgan, M. P. Doyle, S. C. Phatak, P. Millner, and X. Jiang. 2004b. Persistence of Salmonella enterica serovar Typhimurium on lettuce and parsley and in soils on which they were grown in fields treated with contaminated manure composts or irrigation water. Foodborne Pathogens and Disease 1:27-35.
Islam, M., J. Morgan, M. P. Doyle, S. C. Phatak, P. Millner, and X. Jiang. 2004c. Fate of Salmonella enterica serovar Typhimurium on carrots and radishes grown in fields treated with contaminated manure composts or irrigation water. Applied and Environmental Microbiology 70:2497-2502.
Islam, M., M. P. Doyle, S. C. Phatak, P. Millner, and X. Jiang. 2005. Survival of Escherichia coli O157:H7 in soil and on carrots and onions grown in fields treated with contaminated manure composts or irrigation water. Food Microbiology 22:63-70.
Jagai, J. S., J. K. Griffiths, P. H. Kirshen, P. Webb, and E. N. Naumova. 2010. Patterns of protozoan infections: Spatiotemporal associations with cattle density. EcoHealth 7:33-46.
Jamieson, R. C., R. J. Gordon, K. E. Sharples, G. W. Stratton, and A. Madani. 2002. Movement and persistence of fecal bacteria in agricultural soils and subsurface drainage water: A review. Canadian Biosystems Engineering 44:1.1-1.9.
Jay, M. T., M. Cooley, D. Carychao, G. W. Wiscomb, R. A. Sweitzer, L. Crawford-Miksza, J. A. Farrar, D. K. Lau, J. O’Connell, A. Millington, R. V. Asmundson, E. R. Atwill, and R. E. Mandrell. 2007. Escherichia coli O157:H7 in feral swine near spinach fields and cattle, central California coast. Emerging Infectious Diseases 13:1908-1911.
Jenkins, M. B., J. L. Liotta, A. Lucio-Forster, and D. D. Bowman. 2010. Concentrations, viability, and distribution of Cryptosporidium genotypes in lagoons of swine facilities in the Southern Piedmont and in coastal plain watersheds of Georgia. Applied and Environmental Microbiology 76:5757-5763.
Jeon, B. -W., J. -M. Jeong, G. -Y. Won, H. Park, S. -K. Eo, H. -Y. Kang, J. Hur, and J. H. Lee. 2006. Prevalence and characteristics of Escherichia coli O26 and O111 from cattle in Korea. International Journal of Food Microbiology 110:123-126.
Jimenez, V., P. Drechsel, D. Kone, A. Bahri, L. Raschid-Sally, and M. Qadir. 2010. Wastewater, sludge and excreta use in developing countries: An overview. In Wastewater irrigation and health, edited by C. Drechsel, A. Scott, L. Raschid-Sally, M. Redwood, and A. Bahri. London: Earthscan.
Jo, M.-Y., J.-H. Kim, J.-H. Lim, M.-Y. Kang, H.-B. Koh, Y.-H. Park, D.-Y. Yoon, J.-S. Chae, S.-K. Eo, and J. H. Lee. 2004. Prevalence and characteristics of Escherichia coli O157 from major food animals in Korea. International Journal of Food Microbiology 95:41-49.
Johnson, J. Y. M., J. E. Thomas, T. A. Graham, I. Townshend, J. Byrne, L. B. Sellinger, and V. P. J. Gannon. 2003. Prevalence of Escherichia coli O157:H7 and Salmonella spp. in surface waters of southern Alberta and its relation to manure sources. Canadian Journal of Microbiology 49:326-335.
Johnson, R. 2012. The U.S. Trade Situation for Fruit and Vegetable Products. Congressional Research Service Report 7-5700. January 25, 2012. http://www.nationalaglawcenter.org/assets/crs/RL34468.pdf (accessed March 15, 2012). Johnston, L. M., L.-A. Jaykus, D. Moll, J. Anciso, B. Mora, and C. L. Moe. 2006. A field study of the microbiological quality of fresh produce of domestic and Mexican origin. International Journal of Food Microbiology 112:83-95.
Kauffman, M. D., and J. LeJeune. 2011. European starlings (Sturnus vulgaris) challenged with Escherichia coli O157 can carry and transmit the human pathogen to cattle. Letters in Applied Microbiology 53:596-601.
Kijima-Tanaka, M., K. Ishihara, A. Kojima, A. Morioka, R. Nagata, M. Kawanishi, M. Nakazawa, Y. Tamura, and T. Takahashi. 2005. A national surveillance of Shiga toxin-producing Escherichia coli in food-producing animals in Japan. Journal of Veterinary Medicine B 52:230-237.
Kijlstra, A., B. Meerburg, J. Cornelissen, S. De Craeye, P. Vereijken, and E. Jongert. 2008. The role of rodents and shrews in the transmission of Toxoplasma gondii to pigs. Veterinary Parasitology 156:183-190.
Kim, M., J. A. Thurston, and L. J. Hagen. 2008. Computational fluid dynamics (CFD) modeling to predict bioaerosol transport behavior during center pivot wastewater irrigation. Presented at the ASABE Annual International Meeting, Minneapolis, MN. Paper No. 074063.
Kirk, M. C., K. Fullerton, and J. Gregory. 2008. Fresh produce outbreaks in Australia 2001-2006. Board 21. In 2008 International Conference on Emerging Infectious Diseases Program and Abstracts Book. Atlanta, GA: CDC. Pp. 49-50.
Kishima, M., I. Uchida, T. Namimatsu, T. Osumi, S. Takahashi, K. Tanaka, H. Aoki, K. Matsuura, and K. Yamamoto. 2008. Nationwide surveillance of Salmonella in the faeces of pigs in Japan. Zoonoses Public Health 55:139-144.
Klein, M., L. Borwn, R. W. Tucker, N. J. Ashbolt, R. M. Stuetz, and D. J. Roser. 2010. Diversity and abundance of zoonotic pathogens and indicators in manures of feedlot cattle in Australia. Applied and Environmental Microbiology 76:6947-6950.
Knox, J. W., S. F. Tyrrel, A. Daccache, and E. K. Weatherhead. 2011. A geospatial approach to assessing microbiological water quality risks associated with irrigation abstraction. Water and Environment Journal 25:282-289.
Kopanic, R. J., Jr., B. W. Sheldon, and C. G. Wright. 1994. Cockroaches as vectors of Salmonella: Laboratory and field trials. Journal of Food Protection 57:125-132.
Kozan, E., B. Gonenc, O. Sarimehmetoglu, and H. Aycicek. 2005. Prevalence of helminth eggs on raw vegetables used for salads. Food Control 16:239-242.
Kuhnert, P., C. R. Dubosson, M. Roesch, E. Homfeld, M. G. Doherr, and J. W. Blum. 2005. Prevalence and risk-factor analysis of Shiga toxigenic Escherichia coli in faecal samples of organically and conventionally farmed dairy cattle. Veterinary Microbiology 109:37-45.
Kullas, H., M. Coles, J. Rhyan, and L. Clark. 2002. Prevalence of Escherichia coli serogroups and human virulence factors in feces of urban Canada geese (Branta canadensis). International Journal of Environmental Health Research 12:153-162.
Le Bouquin, S., V. Allain, S. Rouxel, I. Petetin, M. Picherot, V. Michel, and M. Chemaly. 2010. Prevalence and risk factors for Salmonella spp. contamination in French broiler-chicken flocks at the end of the rearing period. Preventive Veterinary Medicine 97:245-251.
LeJeune, J. T., D. Hancock, Y. Wasteson, E. Skjerve, and A. M. Urdahl. 2006. Comparison of E. coli O157 and Shiga toxin-encoding genes (stx) prevalence between Ohio, USA and Norwegian dairy cattle. International Journal of Food Microbiology 109:19-24.
Lewis, D. J., E. R. Atwill, M. S. Lennox, M. D. G. Pereira, W. A. Miller, P. A. Conrad, and K. W. Tate. 2010. Management of microbial contamination in storm runoff from California coastal dairy pastures. Journal of Environmental Quality 39:1782-1789.
Lewis, H. C., S. Ethelberg, K. E. P. Olsen, E. M. Nielsen, M. Lisby, S. B. Madsen, J. Boel, R. Stafford, M. Kirk, H. V. Smith, S. Tikumrum, A. Wisetrojana, A. Bangtrakulnonth, J. Vithayarungruangsri, P. Siriarayaporn, K. Ungchusak, J. Bishop, and K. Mølbak. 2009. Outbreaks of Shigella sonnei infections in Denmark and Australia linked to consumption of imported raw baby corn. Epidemiology & Infection 137:326-334.
LGMA (Leafy Green Marketing Agreement). 2012. Commodity specific food safety guidelines for the production and harvest of lettuce and leafy greens. January 20. http://www.caleafygreens.ca.gov/sites/default/files/01.20.12%20CALGMA%20GAPs%20-%20metrics.pdf (accessed March 14, 2012).
Li, X., E. Atwill, T. Vodovoz, E. Vivas, C. Xiao, C. Kilonzo, M. Jay-Russell, and T. Nguyen. 2011. Cryptosporidium spp. in wild rodent populations adjacent to produce production fields. IAFP Annual Meeting Abstracts, Madison, WI, P1-119.
Lienemann, T., T. Niskanen, S. Guedes, A. Siitonen, M. Kuusi, and R. Rimhanen-Finne. 2011. Iceberg lettuce as suggested source of a nationwide outbreak caused by two Salmonella serotypes, Newport and Reading, in Finland in 2008. Journal of Food Protection 74:1035-1040.
Lomonaco, S., L. Decastelli, D. M. Bianchi, D. Nucera, M. A. Grassi, V. Sperone, and T. Civera. 2009. Detection of Salmonella in finishing pigs on farm and at slaughter in Piedmont, Italy. Zoonoses and Public Health 56:137-144.
Loncarevic, S., G. S. Johannessen, and L. M. Rørvik. 2005. Bacteriological quality of organically grown leaf lettuce in Norway. Letters in Applied Microbiology 41:186-189.
Lowell, K., J. Langholz, and D. Stuart. 2010. Co-managing food safety and ecological health in California’s Central Coast region. http://caff.org/wp-content/uploads/2011/09/Safe_Sustainable1.pdf (accessed March 13, 2012).
Lynch, M. F., R. V. Tauxe, and C. W. Hedberg. 2009. The growing burden of foodborne outbreaks due to contaminated fresh produce: Risks and opportunities. Epidemiology & Infection 137:307-315.
MacDonald, J. M., M. O Ribaudo, M. J. Livingston, J. Beckman, and W. Huang. 2009. Manure use for fertilizer and for energy report to Congress. USDA Report AP-037. http://www.ers.usda.gov/Publications/AP/AP037/AP037.pdf (accessed March 11, 2012).
Madden, R. H., K. A. Murray, and A. Gilmour. 2007. Carriage of four bacterial pathogens by beef cattle in Northern Ireland at time of slaughter. Letters in Applied Microbiology 44:115-119.
McMahon, M. A. S., and I. G. Wilson. 2001. The occurrence of enteric pathogens and Aeromonas species in organic vegetables. International Journal of Food Microbiology 70:155-162.
Miko, A., K. Pries, S. Haby, K. Steege, N. Albrecht, G. Krause, and L. Beutin. 2009. Assessment of Shiga toxin-producing Escherichia coli isolates from wildlife meat as potential pathogens for humans. Applied and Environmental Microbiology 75:6462-6470.
Millner, P. D. 2009. Bioaerosols associated with animal production operations. Bioresource Technology 100:5379-5385.
Mishra, A., B. L. Benham, and S. Mostaghimi. 2008. Bacterial transport from agricultural lands fertilized with animal manure. Water, Air, & Soil Pollution 189:127-134.
Moriarty, E. M., L. W. Sinton, M. L. Mackenzie, N. Karki, and D. R. Wood. 2008. A survey of enteric bacteria and protozoans in fresh bovine feces on New Zealand dairy farms. Journal of Applied Microbiology 105:2015-2025.
Moyne, A.-L., M. R. Sudarshana, T. Blessington, S. T. Koike, M. D. Cahn, and L. J. Harris. 2011. Fate of Escherichia coli O157:H7 in field-inoculated lettuce. Food Microbiology 25:1417-1425.
Mukherjee, A., D. Speh, E. Dyck, and F. Diez-Gonzalez. 2004. Preharvest evaluation of coliforms, Escherichia coli O157:H7 in organic and conventional produce grown by Minnesota farmers. Journal of Food Protection 67:894-900.
Mukherjee, A., D. Speh, A. T. Jones, K. M. Buesing, and F. Diez-Gonzalez. 2006. Longitudinal microbiological survey of fresh produce grown by farmers in the Upper Midwest. Journal of Food Protection 69:1928-1936.
Munnoch, S. A., K. Ward, S. Sheridan, G. J. Fitzsimmons, C. T. Shadbolt, J. P. Piispanen, Q. Wang, T. J. Ward, T. L. M. Worgan, C. Oxenford, J. A. Musto, J. Mcanulty, and D. N. Durrheim. 2009. A multistate outbreak of Salmonella Saintpaul in Australia associated with cantaloupe consumption. Epidemiology & Infection 137:367-374.
Okago, C. N., V. J. Uoh, and M. Galadima. 2003. Occurrence of pathogens on vegetables harvested from soils irrigated with contaminated streams. Science of the Total Environment 311:49-56.
Oliveira, M., J. Usall, I. Viñas, M. Anguera, F. Gatius, and M. Abadias. 2010. Microbiological quality of fresh lettuce from organic and conventional production. Food Microbiology 27:679-684.
Olsen, A. R., and T. S. Hammack. 2000. Isolation of Salmonella spp. from the housefly Musca domestica L., and the dump fly, Hydrotaea aenescens (Wiedemann) (Diptera: Muscidae) at caged-layer houses. Journal of Food Protection 63:958-960.
Omeira, N., E. K. Barbour, P. A. Nehme, S. K. Hamadeh, R. Zuray, and I. Bashour. 2006. Microbiological and chemical properties of litter from different chicken types and production systems. Science of the Total Environment 367:156-162.
Oporto, B., J. I. Esteban, G. Aduriz, R. A. Juste, and A. Hurtado. 2008. Escherichia coli O157:H7 and non-O157 Shiga toxin-producing E. coli in healthy cattle, sheep, and swine herds in northern Spain. Zoonoses Public Health 55:73-81.
Patchanee, P., B. Molla, N. White, D. E. Line, and W. A. Gebreyes. 2010. Tracking Salmonella contamination in various watersheds and phenotypic and genotypic diversity. Foodborne Pathogens and Disease 7:1113-1120.
Pezzoli, L., R. Elson, C. L. Little, H. Yip, I. Fisher, R. Yishai, E. Anis, L. Valinsky, M. Biggerstaff, N. Patel, H. Mather, D. J. Brown, J. E. Coia, W. van Pelt, E. M. Nielsen, S. Ethelberg, E. de Pinna, M. D. Hampton, T. Peters, and J. Threlfall. 2008. Packed with Salmonella—investigation of an international outbreak of Salmonella Senftenberg infection linked to contamination of prepacked basil in 2007. Foodborne Pathogens and Disease 5:661-668.
Pillai, S. D., and S. C. Ricke. 2002. Bioaerosols from municipal and animal wastes: Background and contemporary issues. Canadian Journal of Microbiology 48:681-696.
Quiroz-Santiago, C., O. R. Rodas-Suárez, C. R. Vázquez, F. J. Fernández, E. I. Quiñones-Ramírez, and C. Vázquez-Salinas. 2009. Prevalence of Salmonella in vegetables from Mexico. Journal of Food Protection 72:1279-1282.
Rai, P. K., and B. D. Tripathi. 2007. Microbial contamination in vegetables due to irrigation with partially treated municipal wastewater in a tropical city. International Journal of Environmental Health Research 17:389-395.
Ravel, A., J. Grieg, C. Tinga, E. Todd, G. Campbell, M. Cassidy, B. Marshall, and E. Pollari. 2009. Exploring historical Canadian foodborne outbreak data sets for human illness attribution. Journal of Food Protection 72:1963-1976.
Renter, D. G., D. P. Gnad, J. M. Sargeant, and S. E. Hygnstrom. 2006. Prevalence and serovars of Salmonella in the feces of free-ranging white-tailed deer (Odocoileus virginianus) in Nebraska. Journal of Wildlife Diseases 42:699-703.
Renter, D. G., V. Bohaychuk, J. Van Donkersgoed, and R. King. 2007. Presence of non-O157 Shiga toxin-producing Escherichia coli in feces from feedlot cattle in Alberta and absence on corresponding beef carcasses. Canadian Journal of Veterinary Research 71:230-235.
Rimhanen-Finne, R., T. Niskanen, S. Hallanvuo, P. Makary, K. Haukka, S. Pajunen, A. Siitonen, R. Ristolainen, H. Pöyry, J. Ollgren, and M. Kuusi. 2009. Yersinia pseudotuberculosis causing a large outbreak associated with carrots in Finland, 2006. Epidemiology & Infection 137:342-347.
Rodriguez, A., P. Pangloli, H. A. Richards, J. R. Mount, and F. A. Draughon. 2006. Prevalence of Salmonella in diverse environmental farm samples. Journal of Food Protection 69:2576-2580.
Sagoo, S. K., C. L. Little, and R. T. Mitchell. 2001. The microbiological examination of ready-to-eat organic vegetables from retail establishments. Letters in Applied Microbiology 33:434-439.
Sagoo, S. K., C. L. Little, L. Ward, I. A. Gillespie, and R. T. Mitchell. 2003a. Microbiological study of ready-to-eat salad vegetables from retail establishments uncovers a national outbreak of Salmonellosis. Journal of Food Protection 66:403-409.
Sagoo, S. K., C. L. Little, and R. T. Mitchell. 2003b. Microbiological quality of open ready-to-eat salad vegetables: Effectiveness of food hygiene training of management. Journal of Food Protection 66:1581-1586.
Saini, R., L. J. Halverson, and J. C. Lorimar. 2003. Rainfall timing and frequency influence on leaching of Escherichia coli RS2G through soil following manure application. Journal of Environmental Quality 32:1865-1872.
Salleh, N. A., G. Rusul, Z. Hassan, A. Reezal, S. H. Isa, M. Nishibuchi, and S. Radu. 2003. Incidence of Salmonella spp. in raw vegetables in Selangor, Malaysia. Food Control 14:475-479.
Sánchez, S., R. Martínez, A. García, D. Vidal, J. Blanco, M. Blanco, J. E. Blanco, A. Mora, S. Herrera-León, A. Echeita, J. M. Alonso, and J. Rey. 2010. Detection and characterization of O157:H7 and non-O157 Shiga toxin-producing Escherichia coli in wild boars. Veterinary Microbiology 143:420-423.
Santos, F. B. O., X. Li, J. B. Payne, and B. W. Sheldon. 2005. Estimation of most probable number Salmonella populations on commercial North Carolina turkey farms. Journal of Applied Poultry Research 14:700-708.
Sargeant, J. M., M. W. Sanderson, R. A. Smith, and D. D. Griffin. 2004. Associations between management, climate, and Escherichia coli O157 in the faeces of feedlot cattle in the Midwestern USA. Preventive Veterinary Medicine 66:175-206.
Scaife, H. R., D. Cowan, J. Finney, S. F. Kinghorn-Perry, and B. Crook. 2006. Wild rabbits ( Oryctolagus cuniculus) as potential carriers of verocytotoxin-producing Escherichia coli. Veterinary Record 159:175-178.
Semenov, A. V., L. van Overbeek, and A. H. C. van Bruggen. 2009. Percolation and survival of Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in soil amended with contaminated dairy manure or slurry. Applied and Environmental Microbiology 75:3206-3215.
Seo, Y. H., J. H. Jang, and K. D. Moon. 2010. Microbial evaluation of minimally processed vegetables and sprouts produced in Seoul, Korea. Food Science and Biotechnology 19:1282-1288.
Skov, M. N., J. J. Madsen, C. Rahbek, J. Lodal, J. B. Jespersen, J. C. Jørgensen, H. H. Dietz, M. Chriél, and D. L. Baggesen. 2008. Transmission of Salmonella between wildlife and meat-production animals in Denmark. Journal of Applied Microbiology 105:1558-1568.
Smith, B. D., M. R. James, P. Millner, F. M. Hashem, C. P. Cotton, and L. E. Marsh. 2012. Pathogen aerosolization and deposition onto nearby leafy greens through various farm operations. Presented at Human Pathogens on Plants Workshop, College Park, MD, February 13-15. Abstract for Poster #14.
Sneed, J. 2010. Safety of fresh fruits and vegetables. Presentation for Association of State and Territorial Public Health Nutrition Directors, October 28. http://www.astphnd.org/resource_files/246/246_resource_file1.pdf (accessed March 8, 2012).
Sproston, E. L., M. Macrae, I. D. Ogden, M. J. Wilson, and N. J. C. Strachan. 2006. Slugs: Potential novel vectors of Escherichia coli O157. Applied and Environmental Microbiology 72:144-149.
Stephens, T. P., T. A. McAllister, and K. Stanford. 2009. Perineal swabs reveal effect of super shedders on the transmission of Escherichia coli O157:H7 in commercial feedlots. Journal of Animal Science 87:4151-4160.
Talley, J. L., A. C. Wayadande, L. P. Wasala, A. C. Gerry, J. Fletcher, U. DeSilva, and S. E. Gilliland. 2009. Association of Escherichia coli O157:H7 with filth flies captured in leafy green fields and experimental transmission of E. coli O157:H7 to spinach leaves by house flies. Journal of Food Protection 72:1547-1552.
Thunberg, R. L., T. T. Tran, R. W. Bennett, R. N. Matthews, and N. Belay. 2002. Microbial evaluation of selected fresh produce obtained at retail markets. Journal of Food Protection 65:677-682.
Thurston-Enriquez, J. A., P. Watt, S. E. Dowd, R. Enriquez, I. L. Pepper, and C. P. Gerba. 2002. Detection of protozoan parasites and microsporidia in irrigation waters used for crop production. Journal of Food Protection 65:378-382.
Tserendorj, A., A. J. Anceno, E. R. Houpt, C. R. Icenhour, O. Sethabutr, C. S. Mason, and O. V. Shipin. 2011. Molecular techniques in ecohealth research toolkit: Facilitating estimation of aggregate gastroenteritis burden in an irrigated periurban landscape. EcoHealth 8:349-364.
Turutoglu, H., D. Ozturk, L. Guler, and F. Pehlivanoglu. 2007. Presence and characteristics of sorbitol-negative Escherichia coli O157 in healthy sheep faeces. Veterinarni Medicina 52:301-307.
Tyrrel, S. F., and J. N. Quinton. 2003. Overland flow transport of pathogens from agricultural land receiving fecal wastes. Journal of Applied Microbiology 94:87S-93S.
USDA (U.S. Department of Agriculture). 2007. Microbiological Data Program progress update and 2009 data summary. http://www.ams.usda.gov/AMSv1.0/getfile?dDocName=STELPRDC5067866 (accessed January 25, 2011).
———. 2008. Microbiological Data Program progress update and 2009 data summary. http://www.ams.usda.gov/AMSv1.0/getfile?dDocName=STELPRDC5079908 (accessed January 25, 2011).
———. 2009. Microbiological Data Program progress update and 2009 data summary. http://www.ams.usda.gov/AMSv1.0/getfile?dDocName=STELPRDC5088761 (accessed January 25, 2011.
USDA NASS (National Agriculture Statistics Service). 2009. 2007 Census of Agriculture. Farm and Ranch Irrigation Survey. http://www.agcensus.usda.gov/Publications/2007/Online_Highlights/Farm_and_Ranch_Irrigation_Survey/index.php (accessed March 12, 2012).
Van Hoorebeke, S., F. Van Immerseel, J. De Vylder, R. Ducatelle, F. Haesebrouck, F. Pasmans, A. de Kruif, and J. Dewulf. 2009. Faecal sampling underestimates the actual prevalence of Salmonella in laying hen flocks. Zoonoses Public Health 56:471-476.
Viswanathan, P., and R. Kaur. 2001. Prevalence and growth of pathogens on salad vegetables, fruits, and sprouts. International Journal of Hygiene and Environmental Health 203:205-213.
Walker, C., X. Shi, M. Sanderson, J. Sargeant, and T. G. Nagaraja. 2010. Prevalence of Escherichia coli O157:H7 in gut contents of beef cattle at slaughter. Foodborne Pathogens and Disease 7:249-255.
Wang, F., Z. Dou, W. Ma, and F. Zhang. 2005. Challenges and opportunities of animal waste management in China. Proceedings of 2005 America Animal Waste Management Symposium. Pp.704-709.
Wasteson, Y., G. S. Johannessen, T. Bruheim, A. M. Urdahl, K. O’Sullivan, and L. M. Rørvik. 2005. Fluctuations in the occurrence of Escherichia coli O157:H7 on a Norwegian farm. Letters in Applied Microbiology 40:373-377.
Wilkes, G., T. A. Edge, V. P. J. Gannon, C. Jokinen, E. Lyautey, N. F. Neumann, N. Ruecker, A. Scott, M. Sunohara, E. Topp, and D. R. Lapen. 2011. Associations among pathogenic bacteria, parasites, and environmental and land use factors in multiple mixed-use watersheds. Water Research 45:5807-5825.
Williams, M. L., D. L. Pearl, and J. T. LeJeune. 2011. Multiple-locus variable-nucleotide tandem repeat subtype analysis implicates European starlings as biological vectors for Escherichia coli O157:H7 in Ohio, USA. Journal of Applied Microbiology 111:982-988.
Woerner, D. R., J. R. Ranson, J. N. Sofos, G. A. Dewell, G. C. Smith, M. D. Salman, and K. E. Belk. 2006. Determining the prevalence of Escherichia coli O157 in cattle and beef from the feedlot to the cooler. Journal of Food Protection 69:2824-2837.
WHO (World Health Organization). 2011. Outbreaks of E. coli O104:H4 infection: Update 30. July 22, 2011. http://www.euro.who.int/en/what-we-do/health-topics/emergencies/international-health-regulations/news/news/2011/07/outbreaks-of-e.-coli-o104h4-infection-update-30 (accessed March 10, 2012).
Jane Parmley,5Zee Leung,6David Léger,5Rita Finley,7Rebecca Irwin,5Katarina Pintar,5Frank Pollari,7Richard Reid-Smith,5,6David Waltner-Toews,6,8Mohamad Karmali,5and Rainer Engelhardt9
Introduction
This paper describes a holistic approach to the prevention and control of human food-borne illness from enteric pathogens, based on implementation of the “One Health” paradigm. Antimicrobial resistance (AMR) has been chosen as a particular illustrative theme for this overview to demonstrate the practical utility of a One Health approach.
The rapid emergence, global spread, morbidity, and mortality associated with emerging infections such as severe acute respiratory syndrome and avian and pandemic influenza is stimulating the global community to develop novel approaches for their prevention and control. Ongoing concerns about food-borne pathogens such as Escherichia coli O157:H7, Listeria monocytogenes, and various species of Salmonella, as well as the arrival and impact of new strains of food-borne pathogens such as E. coli O104 as observed during the 2011 outbreak in Germany, add to the need to take into account the complexity of infection from multiple dimensions. These include the following:
1. Burden of illness. The World Health Organization estimates that infectious and parasitic diseases are the second leading cause of death in the world (WHO, 2008). Enteric pathogens are the third leading cause of infectious disease worldwide and account for almost 2 million deaths every year ( Girard et al., 2006). As in many other countries, these pathogens also cause a significant disease burden in Canada, where there are an estimated 11 million food-borne enteric illnesses per year with an estimated cost of $3.7 billion dollars (Thomas et al., 2008). Although microbial enteric illness can be caused by bacteria, viruses, parasites, and protozoal organisms, bacteria play a major role in enteric disease (Girard et al., 2006)
_______________
5 Laboratory for Foodborne Zoonoses, Public Health Agency of Canada.
6 Department of Population Medicine, University of Guelph.
7 Centre for Foodborne, Environmental and Zoonotic Infectious Diseases, Public Health Agency of Canada.
8 Veterinarians without Borders/Vétérinaires sans Frontières, Canada.
9 Public Health Agency of Canada.
and are the major focus of enteric surveillance programs. Although most enteric bacterial infections result in subclinical or mild illness, their high rate of incidence in the population can be expected to have economic impact on a country simply through loss of short-term individual productivity. In addition, bacterial infections can cause severe disease, particularly in children, the elderly, and immunosuppressed individuals.
2. Zoonotic and environmental origins. More than 60 percent of new emerging and reemerging pathogens of humans, including those that are transmitted by food and water, arise from animals and the environment (Jones et al., 2008). The rate of emergence appears to be increasing, most likely related to factors such as human population growth, changing patterns of international trade, globalization, mass population migrations, climate change, and environmental degradation. With regard to food safety pathogens, it is anticipated that the increased industrialization of animal production, as is seen worldwide in both developed and developing countries, creates an environment for increased opportunity for entry of pathogens into the food chain.
3. Antimicrobial resistance. The severity of infections and our success in treating the associated clinical diseases are affected by the presence of antimicrobial resistance. Antimicrobial-resistant bacteria are those that are able to replicate in the presence of antimicrobials, here meaning antibiotics and their synthetic derivatives, at levels that normally suppress growth or kill the bacteria. Antimicrobial resistance is a growing concern that threatens animal and human health worldwide, driven mainly by antimicrobial use, both appropriate and inappropriate.
The One Health Paradigm
“One Health” has emerged as a strategic framework for reducing the risks of infectious diseases arising from the animal—human—ecosystems interface. Although a universal definition of One Health has not been achieved, and there are overlaps with integrative approaches used in international research and development, such as “ecosystem approaches to health” (Charron, 2011), there is consensus that One Health is an approach or method of practice that recognizes linkages among human, animal, ecosystem, and economic domains in the context of human health.
The One Health approach focuses on the dynamic interactions at the interfaces between multiple sectors that contribute to the expression of a public health risk. In that interactive context, the approach becomes a tool for disease prevention and control through more informed risk management, encompassing the separate elements of identification, assessment, avoidance, and mitigation of the public health risk. It is worth noting that One Health is bigger than the zoonotic infectious disease issues described below, and incorporates socioeconomic, cultural,
and community conditions (the social determinants of health), as well as individual lifestyle and hereditary health factors.
The economic relevance of early and comprehensive intervention is often overlooked, but can be significant. For instance, the direct economic impacts of individual zoonotic disease events that have occurred over the past 15 years can be in the billions (Figure A4-1).
Canada has been actively engaged in operationalizing the One Health concept through the development of a community of practice by participating and supporting international conferences encompassing the subject. The Public Health Agency of Canada, recognizing the emerging value of the One Health paradigm, hosted an Expert International Consultation on “One World One HealthTM: from Ideas to Action,” March 16-19, 2009, in Winnipeg, Manitoba (http://www.phac-aspc.gc.ca/publicat/2009/er-rc/index-eng.php). Many other major international meetings have helped define One Health, most recently, in November 2011, the High Level Technical Meeting on Health Risks at the Human—Animal—Ecosystems Interfaces in Mexico City. Upcoming is a meeting scheduled for February 2012 in Davos, Switzerland: Global Risk Forum One Health Summit 2012: One Health—One Planet—One Future.
One Health in relation to food safety has multiple dimensions, including science and research, optimizing animal health and ecosystem health, and food inspection and regulatory activities. In Canada work in this area is conducted by several federal government agencies, such as the Public Health Agency of Canada (surveillance, research, and epidemiology of food-borne illness), the Canadian
FIGURE A4-1 Economic impact examples.
SOURCE: Newcomb et al. (2011).
Food Inspection Agency (animal health and food inspection), Health Canada (food safety regulations and risk assessment), and Agriculture and AgriFood Canada (food animal production). Canada’s provincial and territorial jurisdictions have also started to embrace a One Health approach; for instance, Manitoba has a primer on One Health and food safety and has developed an animal health and food safety strategy for the future (“Protecting Animals, Food and People”), and Québec has an animal health and welfare strategy (“One Health, Health for All”). The Canadian academic sector is a critical contributor to the theme, particularly its five veterinary colleges.
Science and research activities include surveillance, detection, and public health risk assessment of nonhuman bacterial isolates, studies on the population and environmental determinants of food-borne zoonoses, systems modeling of the food chain to identify optimal points of intervention, development of intervention strategies such as vaccines and bacteriophage products, and knowledge translation for uptake by food production and processing workers. The activities also include characterization of impacts of particular practices, such as the use of antibiotics in commercial food animal production and its potential in giving rise to antimicrobial resistance in bacteria pathogenic to humans.
Mechanisms of Antimicrobial Resistance
Resistance can be intrinsic, conferred by naturally occurring characteristics of the bacteria, or acquired. Bacteria can acquire resistance through mutations of preexisting genes or through transfer of resistance determinants from other bacteria (horizontal gene transfer). Horizontal transfer occurs much more commonly than de novo development of resistance through mutation (White et al., 2008). It is through horizontal gene transfer that resistance genes, alone or in groups, can spread within bacterial populations and even to other bacterial species.
Resistance genes provide the molecular tools by which bacteria block or oppose the mechanism of action of antimicrobials. Some genes allow bacteria to physically modify their structure to evade drugs, while other genes express enzymes to directly degrade the antimicrobial agent. In addition, resistance mechanisms that are not specific to antimicrobial agents can also be present. For example, cell pumps that allow bacteria to excrete environmental toxins and prevent them from reaching harmful intracellular concentrations can also help bacteria to resist the harmful effects of antimicrobials.
Not all antimicrobial-resistant bacteria are harmful, and resistance genes can be found in nonpathogenic bacteria (Wright, 2007). However, these benign but resistant bacteria may also pose a threat through the transfer of resistance genes to pathogenic bacteria (Figure A4-2).
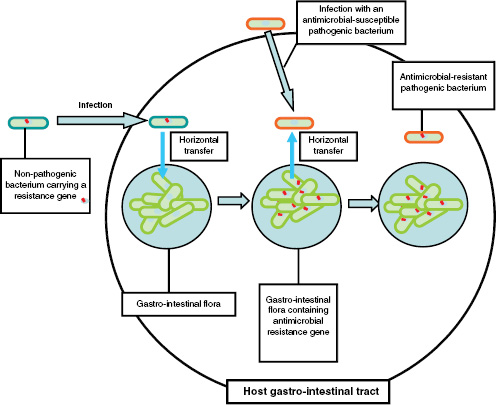
FIGURE A4-2 Transfer model for antimicrobial resistance genes.
Antimicrobial Usage and Resistance
Antimicrobial use (AMU) in animal and human populations is considered to be the major driver of AMR emergence and persistence. Use of antimicrobials exerts a powerful selective influence on bacteria, encouraging the survival and propagation of resistant strains and influencing how quickly AMR develops. Because different resistance genes are often clustered close together on the bacterial genome, especially on transmissible genetic elements such as plasmids and transposons, selection for resistance against one type of antimicrobial may also co-select for resistance against other unrelated antimicrobials. In addition, use of one antimicrobial can select for resistance to closely related antimicrobials (cross-resistance). For example, in Europe, use of avoparcin, an antimicrobial growth promoter used in food animals, has been linked with resistance to vancomycin, an antimicrobial “of last resort” in human medicine (Kruse et al., 1999).
Genetic mechanisms leading to the development and maintenance of AMR are complex. At one time, is was thought that AMR universally negatively im-
pacted the fitness of microorganisms and that, by removing the selective pressure imposed by antimicrobial usage, resistance genes would be selected against in future bacterial generations. However, Wright (2007) identified several genetic mechanisms that may be exceptions to this rule: resistance genes that increase fitness, resistance genes that do not have a fitness cost, and compensatory mutations that restore bacterial fitness. Finally, environmental factors may play a large role in the persistence of “unused” antimicrobial resistance genes. Selection of genes conferring protection against environmental stressors such as heavy metals and biocides may also co-select for resistance genes (Alonso et al., 2001).
The genetic regulation of AMR is complex and not fully understood. Despite our gaps in knowledge, prudent AMU and adherence to the principles of good anti microbial stewardship are recommended as key elements in a strategy directed at preserving the efficacy of antimicrobials, particularly those that are very important to human and veterinary medicine.
A Holistic Consideration of AMR and Enteric Disease
Figure A4-3 depicts the complex interactions between enteric organisms, animals, and humans, and the many determinants (socioeconomic, environmental, and geopolitical) that affect these relationships. Antimicrobial-resistant bacteria form the central component of our model. A number of different interactions can be described using this model, some of which require greater insight into their mechanisms and importance. For example, certain bacteria that cause disease in animal hosts may not cause disease in people but may exchange genetic material, including resistance genes, with human pathogens, causing community-acquired and nosocomial infections (Guardabassi et al., 2004).
Enteric infections in people generally occur through fecal—oral transmission, of which several risk factors can be identified: increased contact between humans and animals, extended hospitalization, poor hygiene, consumption of improperly handled and improperly cooked foods including meats, and ingestion of contaminated water. Prior treatment with antimicrobials can also increase an individual’s susceptibility to infection by pathogenic bacteria through disruption of the normal bacterial flora and by conferring a competitive advantage to resistant strains of pathogens such as Salmonella (Barza and Travers, 2002).
Previous infections with resistant bacteria can also predispose individuals to future resistant infections and disease. As seen in Figure A4-3, an individual may be infected with a commensal bacterium carrying resistance genes. Maintenance of resistance within the individual may occur through colonization of the gastrointestinal tract with this commensal bacterium or via horizontal transfer to gut flora as shown in the diagram. If this same individual is later infected with a pathogenic bacterium, then resistance may be transferred to this pathogen through horizontal transfer from the gut flora.
Implications on Global Health
A number of provincial and national reports, including the 2002 Walkerton Commission of Inquiry (Government of Ontario) and the 2004 Renewal of Public Health in Canada report (Government of Canada), have advocated for a holistic approach toward understanding enteric disease. This type of approach is especially useful given the complexity of enteric disease and its importance as a global health issue. AMR also has serious implications for global human and animal health.
AMR impairs our ability to treat infectious diseases and endangers the long-term efficacy of antimicrobial drugs available to human and veterinary medicine. Not only are infections caused by resistant bacteria more difficult and more expensive to treat, but also the longer duration of infection may increase disease shedding and spread. AMR thus has important effects on the pathogenicity and epidemiology of zoonotic bacterial agents.
Along with its global health implications, the emergence of resistant bacteria may have broad economic effects. Weakened public confidence over the safety of agricultural commodities, potential inclusion of AMR bacteria as a product adulterant leading to recalls, and changes to consumer buying patterns are major economic concerns to agricultural industries. At the patient level, AMR may reduce the efficacy of certain antimicrobials and thereby increase the cost of infection (e.g., longer hospital stays and changes in AMU for disease treatment and prevention) in people and animals. As discussed by Foster (2009), the economic burden of AMR may be most dramatic in developing nations because of the higher expense of second- or third-line drugs, and the lack of diagnostic capacity to detect resistance early, which may result in treatment failures and complications in antimicrobial selection.
Developing solutions to AMR and enteric disease requires synthesis of knowledge and analysis of data at the local, national, and global scales. Factors such as agricultural land-use patterns, attitudes toward antimicrobial usage, and the nature and extent of interactions between people and animals can have major effects on the development of AMR at the local and national levels. However, these local influences may also have global significance. Global interactions of people, animals, and animal products mean that AMU and the accompanying regulations in one country can affect the efficacy of a particular antimicrobial in another. Similarly, the global epidemiology of enteric pathogens is important in understanding the local burden of enteric disease. For example, it was estimated that 30 percent of all enteric disease cases at a sentinel site in Ontario, Canada, in 2008 were associated with international travel (Government of Canada, 2009).
Application of a Holistic Approach to
Zoonotic Bacterial Infections and AMR in Canada
The Public Health Agency of Canada supports two complementary surveillance programs that together provide a holistic approach to AMR and enteric
disease (Figure A4-3): (1) the Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) and (2) the National Integrated Enteric Pathogen Surveillance Program (C-EnterNet). Both were modeled after similar programs in other countries: NARMS (United States) and DANMAP (Denmark) for CIPARS and FoodNet (United States) and OzFoodNet (Australia) for C-EnterNet. The two Canadian programs generate and collect data that contribute to our understanding of the transmission of zoonotic bacteria, risk factors for infection, and the drivers of AMR and AMU. As surveillance systems, their ongoing and systematic designs allow for the identification of emerging trends and the ability to identify the impacts of prevention and control measures adopted at the national, provincial, and, occasionally, local levels in Canada.
Both programs also provide a research platform that aims to identify and understand how livestock husbandry and production methods, water-borne routes of exposure, wildlife, companion animals, exotic pets, and socioeconomic factors and high-risk human populations are affected by and contribute to zoonotic bacterial infections and AMR.
While CIPARS performs epidemiological surveillance on AMR and AMU through the generation and collection of nationwide data from farms, abattoirs, retail stores, and both human and animal diagnostic health laboratories, C-EnterNet performs epidemiological surveillance on enteric pathogens at intensively sampled local sentinel sites (currently one site in Ontario and one in British Columbia). Like CIPARS, C-EnterNet collects data at the level of the farm, retail store, and human community (via epidemiological and laboratory data on human cases in partnership with the local public health unit). C-EnterNet also performs environmental surveillance by collecting and testing untreated water samples. This parallel testing is critical to understanding the complex system of food and water-borne disease transmission. Results from both programs are publicly accessible through the Public Health Agency of Canada website as well as through annual reports and newsletters.
The epidemiological strength of CIPARS lies in its breadth of surveillance at major points along the farm-to-fork continuum. These data allow for temporal and spatial analyses of provincial and national trends in bacterial recovery and AMR. This is best demonstrated with the recent study of Salmonella Heidelberg and ceftiofur resistance (see the section titled Success Within CIPARS: A Case Example). While CIPARS is most effective at studying trends at broad scales, C-EnterNet’s value is in its ability to detect subtle epidemiological effects that may only be captured at the local level. In addition, it is one of the only systems that can delineate endemic versus travel-acquired human infections (see the section titled Success Within C-EnterNet: A Case Example). The sentinel-site surveillance approach provides rich data that would be cost-prohibitive to collect across all of Canada. But, by understanding sentinel populations, the information can be used to determine the predominant sources of enteric pathogens causing
infection and the risk factors (including individual behaviours) that contribute to the burden of enteric illness.
It is important to recognize the unique operational aspects of both CIPARS and C-EnterNet and their complementary nature. Having two different but linked surveillance models that encompass different scales is essential in providing a comprehensive look at the specific risk factors associated with AMR and enteric disease. When considered together, both programs provide a holistic picture of the complex relationships between enteric pathogens, the environment, and the health of humans and animals.
Success Within CIPARS: A Case Example
Recent analysis of CIPARS data identified a link between ceftiofur (an antimicrobial of high importance to human medicine) usage in poultry and ceftiofur-resistant Salmonella Heidelberg isolates obtained from people and chicken meat in Québec (Dutil et al., 2010), as shown in Figure A4-4. Because S. Heidelberg is a common serotype that infects and can cause disease in people, this finding had important human health implications.
Communication of this information led to a voluntary ban on the use of ceftiofur in 2005, and the ongoing collection of surveillance data provided the opportunity to follow trends in human and animal infection and in AMR. The findings from this work have provided strong evidence pointing toward changing patterns in AMU affecting clinical bacterial resistance in human and animal isolates. This study has been used to inform policy on the appropriate use of this antimicrobial and is helping to guide physicians and veterinarians in their selection of appropriate antimicrobials and how these drugs are dispensed.
Success Within C-EnterNet: A Case Example
The C-EnterNet program recently looked at 1,773 reported cases of disease caused by enteropathogens such as Salmonella, Campylobacter, and verotoxigenic Escherichia coli in Sentinel Site 1 (Region of Waterloo, Ontario) (Ravel et al., 2011). C-EnterNet and its local public health partners found that more than one in four reported cases of enteric infection were related to travel, including 9 percent involving new immigrants. The most popular destinations of the patients studied were the Caribbean, Latin America, and Asia.
The finding illustrates that travel-related cases of diseases caused by enteric pathogens represent a significant proportion of the burden of total diseases in Canada. These results will help to delineate domestically acquired infections from those acquired abroad. In the One Health framework, this will help target more effective prevention and control measures domestically, considering a broad suite of pathogens and the complex routes of transmission.
Conclusions and Key Policy Implications
The global, transdisciplinary, multiscalar, and multijurisdictional nature of AMR and enteric disease highlights the utility of the One Health approach in framing these health issues. One Health principles encourage public health practitioners to engage and collaborate with stakeholders and to consider the numerous socioeconomic, geopolitical, zoonotic, and environmental factors involved in health issues (Figure A4-2). Veterinarians and physicians as well as other human, animal, and ecosystem health professionals have important roles to play in preserving the efficacy of our antimicrobials through leadership roles in disease surveillance, AMU decision making, and health management decisions to prevent disease. Communication and collaboration with farms, industry, veterinarians, physicians, and other public health practitioners must be strengthened and is emphasized as key to the success of the approach to AMR and enteric disease.
C-EnterNet and CIPARS have successfully operated for 7 and 10 years, respectively. A large part of this success and the sustainability of these programs can be attributed to ongoing collaborations with multiple stakeholders and the flexibility of all the partners to adapt to changing needs and conditions. These programs serve as a model for how government agencies can address, in an integrated fashion, urgent problems and issues that cut across multiple departments and jurisdictions.
Acknowledgments
This article is based, in part, on one of 31 case studies included in One Health for One World: A Compendium of Case Studies, edited by David Waltner-Toews, Veterinarians without Borders/Vétérinaires sans Frontières—Canada, April 2010 (accessed on November 22, 2011, at http://www.vwb-vsf.ca/english/documents/OHOWCompendiumCaseStudies_001.pdf). This compendium was commissioned by the Public Health Agency of Canada (PHAC) and presented by PHAC and the United Nations System Influenza Coordinator at an Inter-ministerial Meeting of the International Partnership on Avian and Pandemic Influenza in Hanoi, Vietnam, in April 2010.
References
Alonso, A., P. Sánchez, and J. L. Martínez. 2001. Environmental selection of antibiotic resistance genes. Environmental Microbiology 3(1):1-9.
Barza, M., and K. Travers. 2002. Excess infections due to antimicrobial resistance: The “Attributable Fraction.” Clinical Infectious Diseases 34(Suppl. 3):S126-S130.
Charron, D. F. (editor). 2011. Ecohealth research in practice: Innovative applications of an ecosystem approach to health. New York: Springer/Ottawa, ON: International Development Research Centre.
Dutil, L., R. Irwin, R. Finley, L. K. Ng, B. Avery, P. Boerlin, A.-M. Bourgault, L. Cole, D. Daignault, A. Desruisseau, W. Demczuk, L. Hoang, G. B. Horsman, J. Ismail, F. Jamieson, A. Maki, A. Pacagnella, and D. R. Pillai. 2010. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerging Infectious Diseases 16(1):48-53.
Foster, S. D. 2009. The economic burden of resistance in the developing world. In Antimicrobial resistance in developing countries, edited by A. Sosa and D. K. Byarugaba. New York: Springer Science & Business Media.
Girard, M. P., D. Steele, C.-L. Chaignat, and M. P. Kieny. 2006. A review of vaccine research and development: Human enteric infections. Vaccine 24:2732-2750.
Government of Canada. 2004. Learning from SARS: Renewal of public health in Canada. Ottawa, ON: Health Canada.
Government of Canada. 2009. C-EnterNet Short Report 2008. Guelph, ON: Public Health Agency of Canada.
Government of Ontario. 2002. Report of the Walkerton Inquiry: A strategy for safe drinking water. Toronto: Ontario Ministry of the Attorney General.
Guardabassi, L., S. Schwarz, and D. H. Lloyd. 2004. Pet animals as reservoirs of antimicrobial-resistant bacteria. Journal of Antimicrobial Chemotherapy 54:321-332.
Jones, K. E., N. G. Patel, M. A. Levy, A. Storeygard, D. Balk, J. L. Gittleman, and P. Daszak. 2008. Global trends in emerging infectious diseases. Nature 451(7181):990-993.
Kruse, H., B. K. Johansen, L. M. Rørvik, and G. Schaller. 1999. The use of avoparcin as a growth promoter and the occurrence of vancomycin-resistant enterococcus species in Norwegian poultry and swine production. Microbial Drug Resistance 5(2):135-139.
Newcomb, J., T. Harrington, and S. Aldrich. 2011 (unpublished). The economic impact of selected infectious disease outbreaks.
Ravel, A., A. Nesbitt, B. Marshall, N. Sittler, and F. Pollari. 2011. Description and burden of travel-related cases caused by enteropathogens reported in a Canadian community. Journal of Travel Medicine 18(1):8-19.
Thomas, M. K., S. E. Majowicz, F. Pollari, and P. N. Sockett. 2008. Burden of acute gastrointestinal illness in Canada, 1999-2007: Interim summary of NSAGI activities. Canada Communicable Disease Report 34(5):8-13.
White, J. R., P. Escobar-Paramo, E. F. Mongodin, K. E. Nelson, and J. DiRuggiero. 2008. Extensive genome rearrangements and multiple horizontal gene transfers in a population of Pyrococcus isolates from Vulcano Island, Italy. Applied and Environmental Microbiology 74(20):6447-6451.
WHO (World Health Organization). 2008. The global burden of disease: 2004 update. http://www.who.int/healthinfo/global_burden_disease/GBD_report_2004update_part2.pdf (accessed June 26, 2012).
Wright, G. 2007. The antibiotic resistome: The nexus of chemical and genetic diversity. Nature 5:175-186.
OVERVIEW OF THE GLOBAL FOOD SYSTEM: CHANGES OVER TIME/SPACE AND LESSONS FOR FUTURE FOOD SAFETY
Will Hueston10,11and Anni McLeod10
Food systems emerged with the dawn of civilization when agriculture, including the domestication of animals, set the stage for permanent settlements. Inhabitants could grow more crops and raise more animals than necessary to feed those who tended them. This changed human culture; unlike earlier hunter-gatherers, agriculturalists did not need to be in constant motion to find new sources of food. Cultivating grain allowed for drying and storage of some of the harvest for later consumption. Different grain cultures emerged in each of the cradles of civilization: maize in Mexico, rice in China, and wheat and barley in the Middle East. The ability to produce a surplus of grain also set the stage for the development of art, religion, and government.
Since agriculture began, food systems have constantly evolved, each change bringing new advantages and challenges and ever-greater diversity and complexity. This paper looks backward to the drivers of change and forward to the challenges faced by producers, consumers, and policy makers of tomorrow.
Changes Over Time and Space
The emergence of city-states has been a major driver of food system changes, bringing together large populations within defined boundaries and requiring complex governance to deliver sufficient quantities and quality of food. Advances in food storage, with sealed containers and curing methods, the use of animal transport, sailing ships, and trains to move larger volume than can be carried by individuals; trade in ingredients like salt as well as live animals and agricultural products; and increasing political and military conflict for resources all have been developments of the city-state. Extensive trading routes have existed for salt, spices, tea, and pepper for thousands of years.
The Iron Age and the Roman Empire brought expanding empires and the beginning of global food systems, including regional specialization in products traded throughout empires. Food systems began to be organized on a grand scale to feed larger cities and fuel local economies. Trade networks for grain, nuts, oils, fruit, and wine developed using both road systems and sailing routes. Standardized weights and measures were established along with the expansion of money and accounting.
_______________
10 Global Initiative for Food Systems Leadership.
11 College of Veterinary Medicine and School of Public Health University of Minnesota.
The Middle Ages saw the emergence of the merchant class and banknotes. Prior to the Middle Ages, selling was considered a task for one of the lower classes of civilization, if not a sin. The equestrii in Roman times did the trading, not the citizens of Rome. The Middle Ages also saw banknotes replacing coinage, first with the Song dynasty in China and then later in Europe around 1661. As a wealthy class emerged, they became more sophisticated in their food preferences. The resulting demand of consumers began to affect trade in addition to supply.
Science and technology represent another major driver, changing the way that food is grown, processed, preserved, and transported. The Industrial Age brought a transition from manual labor and draft animal—based economies to machines. Further increases in agricultural productivity brought about by technology such as the seed drill, the iron plow, and the threshing machine freed up labor for the factories in the 1700s. The Industrial Revolution also created per-capita income growth. The emerging middle class had discretionary income to spend on its food preferences. Transportation breakthroughs were ushered in during the industrial age: canal systems, improved roadways, steam engines used for traction, railroads, and steamships. The Erie Canal, as an example, connected the Great Lakes and the northeastern United States with 363 miles of inland waterways by 1825.
Food preservation, important to both storage and transport of food, also changed over time. Drying was one of the early food preservation methods, certainly known in ancient times. Fermentation also was an early method of food preservation, with pasteurization applied to wine in China as early as 1117. Salting of food has been used for at least 500 years, beginning when the fishing fleets from Europe used drying and salting to store fish caught in Newfoundland and the Grand Banks in order to get them back to consumers in Europe.
Two preservation methods, canning and freezing, allowed food to be stored and transported in an almost-fresh state. Canning grew out of military research in 1810. Ice storage was developed in northern climates where ice could be cut from lakes in the winter for use later in the year. Commercial refrigeration followed in the 1800s. The first refrigerated ship, the SS Dunedin in 1882, revolutionized the meat and dairy industries in Australia and New Zealand. Refrigerated and frozen food products now could be traded globally.
The 20th century saw intensification of agricultural production with mechanization of planting and harvesting, selective breeding of animals and plants, and more attention to animal nutrition and feed input costs. Increased scale of production drove down the per-unit cost of products and fostered greater specialization in food systems. Advances in plant and animal disease control also helped, such as the movement of pigs and poultry indoors to decrease disease exposure and to enhance efficiency by controlling the environment.
Colonization and war have been important political influences on food systems, the first creating distributed ownership of food systems and the second highlighting a need for global agreements. Colonialism allowed for population growth of the industrialized countries when there were limited domestic opportunities to create employment or to grow food. Settler colonies captured market opportunities for the colonizing country’s exports and provided import sources for raw materials, including food and food ingredients.
Trade underwent dramatic changes in the 20th century as a result of the two world wars. The war-associated food shortages, economic crises, and disease spread set the stage for global trade agreements and organizations designed to address global public good issues. The 1947 General Agreement on Tariffs and Trade was created to reduce tariff-based trade barriers and to prevent the downward spiral of world trade seen in the Great Depression from 1929 to 1933. Monthly trade dropped from $3.0 billion in January 1929 to $0.9 billion in March 1933 as protectionist measures reduced trade worldwide (Personal communication, Christiane Wolff, World Trade Organization, March 2012).
Supply-driven to demand-driven Until the 20th century many countries had supply-driven economies, where policies favored increased agricultural production to ensure adequate domestic supplies of basic feedstuffs. Increasing the supply and reducing the costs of food were politically popular national priorities. Food self-sufficiency was a powerful motivation, especially for countries that had experienced food shortages in the past. Countries that exceeded domestic demand used export markets and food aid programs to deal with the excess.
Rising discretionary incomes in Europe and North America in the 20th century impacted food demand and global food trade. Rising consumer demand for chicken drove the development of the broiler industry, but, as marketing moved from whole birds to parts such as leg quarters or breasts, demand disequilibrium resulted. For example, many Americans prefer white meat and do not eat chicken feet, while in other parts of the world people prefer dark meat and consider chicken feet a delicacy. Global food trade provided an opportunity to sell the parts of animals for which there is little or no domestic demand. One reason that the developed world enjoys relatively inexpensive food is the ability to market commodities and specialized products worldwide.
Food systems are dynamic and ever changing in response to natural forces (e.g., weather), demographics (e.g., emergence of megacities), economics (e.g., currency values), technological advances in processing (e.g., high pressure pasteurization), entrepreneurism (e.g., development and marketing of new products), and consumer preferences (e.g., locavores). Every country in the world produces some of its own food and trades food. As a result of these constant changes, food systems are increasingly complex, as adding to the challenge of assuring global food safety.
The Complexity of Current Global Food Systems
and Implications for Food Safety
Today’s food systems are diverse and complex, involving everything from subsistence farming to multinational food companies. Everyone eats; therefore, everyone relies on food systems, local and global. The movement of food and food ingredients in food systems includes animals and animal products, plants and plant products, minerals, and vitamins. The classic cheeseburger provides an excellent example of the complexity of today’s supply chain. Researchers at the University of Minnesota mapped the global supply chain of the cheeseburger working with a large quick-service restaurant chain, Figures A5-1, A5-2, and A5-3 tell the story. Figure A5-1 demonstrates graphically the movement of different commodities from the farm through processing to the restaurant. Figure A5-2 lists all the ingredients found in this company’s cheeseburgers and Figure A5-3 provides an idea of the variety of companies supplying key ingredients like vinegar, garlic powder, tomatoes, beef, and wheat gluten. Each cheeseburger includes more than 50 ingredients sourced from countries in every continent of the world except the Arctic.

FIGURE A5-1 Global supply chain complexity. Movement of commodities.
SOURCE: Shaun Kennedy, Director, National Center for Food Protection and Defense, University of Minnesota.
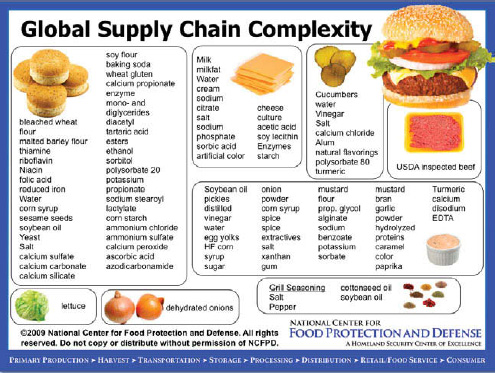
FIGURE A5-2 Global supply chain complexity. Ingredient list.
SOURCE: Shaun Kennedy, Director, National Center for Food Protection and Defense, University of Minnesota.
Food processing supplies also move globally and include processing equipment, packaging, and chemicals such as disinfectants and preservatives. Agricultural inputs move too, from feed to fertilizer, to vaccines and pharmaceuticals, to planting and harvesting equipment. As agricultural commodities are combined with other food ingredients to create processed foods, individual food items commonly include ingredients from multiple countries. The increasing consumer demand for “ready-to-eat” foods has fueled the growth of quick service restaurants and fully cooked, frozen dishes that only require reheating, further expanding supply chains. Government regulatory systems and private-sector initiatives are part of food systems, as are educational efforts and consumer actions.
Food systems are integrally related to food safety. Contamination can occur at any point in the food system, and prevention and control strategies can be implemented at any point. The scale and complexities of today’s food systems contribute to the likelihood and magnitude of food-borne illness (Ercsey-Ravasz et al., 2012). The more complex, the more opportunities for things to go wrong; the larger the scale, the more people are potentially affected.
Complex food systems each involve interconnected subsystems that, taken together, exhibit properties that are not predictable by the properties of the indi-

FIGURE A5-3 Globalizing the cheeseburger.
SOURCE: Shaun Kennedy, Director, National Center for Food Protection and Defense, University of Minnesota.
vidual subsystems or their parts. Food systems can be called complex adaptive systems. These have no boundaries; individual actions affect the food systems by what individuals produce and what they purchase. Complex adaptive systems have a memory. While food systems change over time, present behavior is affected by prior behavior. Food systems are nonlinear. A small perturbation in some part of the system may have a large effect, a proportional effect, or no effect. And the relationships of this system of systems have feedback loops. The adaptiveness and nonlinearity of food systems mean that food safety problems are also nonlinear; they can be anticipated but are hard to predict with accuracy or precision.
Feeding the world requires a multitude of systems. Each system is dynamic and the food systems are interdependent; there is no one best system that meets all needs. However, every success in improving the food system perturbs the whole system of systems and changes the nature of the food safety problems.
Lessons for the Future
Looking at existing global food systems and predicated demands for food, we can reasonably speculate the following over the next 10 to 20 years:
1. Food systems will continue to change, although with additional drivers. The drivers of urbanization, production and processing technology, transport technology, and political forces that have played a large part in shaping current food systems will continue to be relevant. Newer drivers playing an increasingly important part are a real prospect of a global population of 10 billion, aging populations changing the production and consumption base, climate change leading to constraints on water supplies, severe constraints on nonrenewable energy, and communication technology.
2. Food systems will continue to shift from being supply driven to being demand driven. The global quick service restaurant chains like McDonalds and big-box retailers like Walmart have had an enormous impact on food systems. Consumer groups demanding safety, fair trade, “green” production, and animal welfare-related changes in production practices put pressure on policy makers and retailers. The large processors are putting pressure on the primary producers of plants and animals for assurances on source, on identity preservation, on means of production, and on characteristics like animal welfare and labor standards.
3. Increasing prominence of private standards. Successful completion of the Uruguay Round of the multinational trade negotions under the framework of the General Agreement on Tariffs and Trade included approval of the Sanitary and Phytosanitary Agreement (SPS) in 1995 under a new organization, the World Trade Organization (WTO). The SPS established a framework for international standards for trade in animals, plants, and the products derived from them including food. More recently, coalitions of companies are forming to standardize specifications for food products, basically saying, “we can’t wait for the slow process of international standards organizations.” An example is the Global Food Safety Initiative, a nonprofit organization that benchmarks guidelines established by food processors, retail, and food service against the international standards recognized by WTO. Food safety standards used by the large companies who target premium market niches are often above and ahead of the minimum demanded by legislation.
4. Panarchy. The term “panarchy” is used in systems theory to describe systems interlinked in continual adaptive cycles of growth, restructuring, and renewal (Gunderson and Holling, 2001). The increased growth in connectedness and efficiency results in a lack of redundancy and at the same time makes individual food systems less resilient, more sensitive to stress, and therefore more susceptible to collapse. If subsystems within complex food systems collapse, the result is systems with greater resiliency that have fewer connections and less efficiency. And the cycle starts again.
Food systems have demonstrated adaptive cycles as they have evolved. Many current food systems have evolved to a point where they are both
complex and sensitive to stress, and the results of a collapse in a subsystem can be wide-reaching. For example, the concentration of production of an ingredient like a vitamin in a single company or country may be the most efficient approach, but if a production problem ensues or a disaster disrupts this supply chain, then all food processors using this vitamin as a food ingredient are affected. They must either remove the vitamin from their recipes or stop production because of lack of supply. Another example is the proliferation of “just-in-time” supply chains. Instead of stockpiling food supplies in warehouses, many large food retailers and food services have worked with food manufacturers to establish these supply chains. Real-time data on usage and inventories are provided directly to the supplier on a regular basis to allow for customized shipments of only those food products needed. If the supply chain is disrupted, there is very little food in reserve. Many cities have less than 2 days’ supply of perishable food like milk and eggs on the shelves at retail outlets. People in countries where systems regularly collapse have coping strategies: they store food, water, and alternative energy at home. Many of those in large modern cities do not. The urban poor have neither the finances nor the storage facilities to store reserves of food.
5. Culture clash. Disconnects exist between origination and destination countries because of differences in their cultures and differing levels of economic development. While developed countries have emphasized the importance of food safety and quality, less-developed countries may focus on the opportunity for exports to generate foreign currency reserves. The recent melamine incidents demonstrate economic adulteration in order to achieve greater profit in domestic and international markets.
What Do One Health Approaches Have to Offer Food
Safety in the Context of Food Systems?
Food safety is a “wicked problem.” We cannot completely understand the challenge; it is too complex. And yet food safety is compelling: people are getting sick and dying every day as a result of unsafe food and water. We must take action, and we recognize that every action we take perturbs the very food systems we are working to improve. The so-called wicked problem reflects the condition of a complex adaptive system.
If One Health is taken to imply holistic and multidisciplinary approaches to complex challenges (e.g., wicked problems), then a One Health approach offers the possibility of new perspectives on safety in food systems and new ways of working. It implies systems thinking, shared leadership, a holistic view, and a multifaceted approach.
Is this back to the future? The World Health Organization (WHO) definition of health in 1948 was quite broad: “Health is a state of complete physical,
social, and mental well-being, and not merely the absence of disease or infirmity.” However, the public health implementation of food safety focus often is limited to prevention and response to infectious diseases rather than a more holistic approach to food safety as an element of food security (availability, access, and nutrition as well as safety). More recently, the Food and Agriculture Organization and the WHO have developed a much broader definition of food safety: “All the conditions and measures necessary during production, processing, storage, distribution, and preparation of food to ensure that it is safe, sound, wholesome, and fit for human consumption.”
Successfully applying One Health approaches to food safety requires a sound understanding of the dynamics of food systems. Food safety must be addressed in a systemic manner rather than an ad hoc approach driven by reaction to crises. These One Health approaches have implications for what we record, measure, and analyze in food systems and how we share information about potential food safety problems as well as existing crises.
One Health approaches also require a new leadership model that is adaptive and shared, matching the adaptive nature of food systems and the many ways they are controlled and influenced. Five skill sets for adaptive leaders were identified by a small international working group at a session in Bellagio, Italy, sponsored by the Rockefeller Foundation: communications; getting things done and accomplishing change; working across boundaries, whether disciplinary, sectoral, or political; influence; and vision and strategy.
Applying these skills sets encourages a move from finger-pointing to shared leadership. It provides space to accept the fact that food-borne disease happens and will happen. Food safety programs are not always somebody’s fault. After all, “safe food” is an oxymoron. All food has risks and yet “safe” implies the absence of risk. Food systems can either contribute to the risks or be designed to help manage the risks. The very complexity of food systems also means that an infinite number of risk-management strategies are available, if we are only creative enough.
Incremental progress on complex food safety problems may also require a new model of partnership that engages producers and the food industry along with government. We do not have an ideal model for partnership or shared leadership, but several initiatives in fisheries and foods are trying to find or build models, and so are others outside of the food sector. A new partnership model would include a value proposition to engage industry (examples are beginning to emerge around agriculture and environment, where there is no alternative but for government and the private sector to work together) and a more flexible and realistic regulatory system. The idea of zero tolerance makes no scientific sense (zero risk is unachievable) and contributes to the very high levels of waste in U.S. food supply chains (e.g., supermarkets in the United Kingdom are moving to changes in the “use by” label to provide more flexibility in home-freezing, which is anticipated to reduce waste in kitchens with no reduction in food safety).
What Comes Next?
We have proposed a One Health approach that would match the complex, adaptive problems of food safety with shared, adaptive, and holistic problem solving that considers the entire food system. However, an approach is of little use while it remains on paper. The next challenge is to find a complex, subtle, pervasive, and wide-ranging food safety problem that will require adaptive leadership, partnerships, and a wide scope of action—the problem of mycotoxins is excellent example—and put the food systems community to work on it.
References
Ercsey-Ravasz, M., Z. Toroczkai, Z. Lakner, and J. Baranyi. 2012. Complexity of the International Agro-Food Trade Network and Its Impact on Food Safety. PLoSONE 7(5):e37810.
Gunderson, L., and C. S. Holling. 2001. Panarchy: Understanding transformations in systems of humans and nature. Washington, DC: Island Press.
THE AUSTRALIAN PERSPECTIVE, THE BIOSECURITY CONTINUUM FROM PREBORDER, TO BORDER AND POSTBORDER
Executive Summary
Biosecurity is of considerable importance to Australia and managing biosecurity risks through a One Health approach offers many attractive advantages. To date most of the international effort has been focused on adopting a One Health approach from the perspective of infectious diseases and the need to bring together multidisciplinary teams to most effectively understand and mitigate the risks. Central to understanding the skills and knowledge that are required is an appreciation that many recent outbreaks of infectious diseases arise in wildlife, create disease in livestock, and subsequently go on to cause infection in humans. While the drivers for this emergence are still not fully elucidated, a number of key factors play a part, including climate change.
While there are clear differences between the approaches to food safety versus infectious disease management, there is still the basic gain to be made by attacking the risks through reducing likelihood rather than addressing the consequences. This key concept underpins the approach undertaken in Australia,
_______________
12 Director, Australian Animal Health Laboratory, Geelong, Australia.
where biosecurity activities, preborder, at the border, and postborder, focus on early detection and rapid response. While the approach recognizes the continuum from pre- to postborder, resource allocation is currently being reviewed to ensure an appropriate balance for effective risk mitigation.
Underpinning the Australian biosecurity strategy is the recognition of the value of a One Health, multidisciplinary approach. There is a current awareness that much needs to be done to ensure that the maximum value is achieved from this approach and that a “business-as-usual” mentality does not prevail. Fortunately for Australia, the recent management of Hendra outbreaks in Queensland and New South Wales has provided an excellent example of the gains that can be made through a One Health approach. Similar examples need to be developed in the food safety arena.
Introduction
Biosecurity is the protection of the economy, the environment, social amenity, or human health from the negative impacts associated with the entry, establishment, or spread of animal or plant, pests and diseases, or invasive plant and animal species (Beale et al., 2008). Australia has an enviable biosecurity position having been free of many of the infectious diseases that infect livestock in most other parts of the world. Built on the “island status,” Australia has for many years maintained a stringent import policy around plants, livestock, and agricultural products to ensure the protection of this status. Australia has consistently adopted a precautionary policy, although international trade regulations (OIE, 2011) attempt to ensure that fair trading practices exist in the international agricultural marketplace. Notwithstanding this, the risks continue to increase and disease outbreaks are an unfortunate regular event. Recognizing this, the focus remains on early detection linked to a rapid and effective response. Eradication is the preferred option but not always achievable, particularly in the plant sector. Here a policy of containment is adopted that seeks to limit spread and reduce the impact on both productivity and the environment.
A number of frameworks have been developed to better enable the Australian biosecurity strategy. These include not-for-profit companies providing a framework for industry and government to work in partnership, such as Animal Health Australia (AHA, 2011a) and Plant Health Australia, agreed on plans for how to deal with outbreaks and agreed on processes for who will pay for what in the face of a major disease incursion (AHA, 2011b). Mostly developed for the livestock sector, this approach is now being applied to both the plant and environmental sectors.
There is a growing appreciation that the risks being addressed now encompass environment and human health as well as animals and plants. In order to effectively manage these risks, a One Health approach has much to offer.
The One Health Concept
One Health as a concept emerged some 10 years ago and has gained increasing acceptance as a process for addressing a range of issues involving environmental, animal, and human health (Leboeuf, 2011). Although there are many definitions of One Health, the current focus remains around emerging infectious disease (EID) and recognizes that 75 percent of EID in humans arise from animals, and in large part, from wildlife, often spilling over first into domestic livestock and then infecting humans (Woolhouse and Gowtage-Sequeria, 2005). This also includes the emergence of diseases affecting food safety and food security. A full understanding of these processes and the development of mitigating strategies to reduce the threats from EIDs will require input and engagements from people with a diverse set of skills and a range of disciplines (Vallet, 2009).
The emergence of disease requires an interaction between the pathogen, the host, and the environment. Understanding these interactions and developing effective mitigation strategies requires a complex of One Health disciplines. In the case of pathogen influences these involve such areas as quasispecies variation, genetic recombination, host/vector adaptation, tissue tropism, virulence determinants, and latency or persistence. For host influences it is necessary to understand reservoir host spillover, the range of intermediary hosts, various aspects of vector competence, the susceptible host range, the pathogenesis of the disease in different hosts, and the potential range of immune responses. In looking at the impact of anthropogenic influences it is important to appreciate the broader issues of globalization, urbanization, land-use changes, cultural changes, and regional and global conflicts. Finally in terms of geophysical influences, climate change and variability link to extreme weather events are critical (Cutler et al., 2010; Rushton, 2009; Wolfe et al., 2007).
The One Health approach strives to bring these many sciences and disciplines together to provide the best possible solution to health risk management. Presently much is being done at both the national and international levels to create effective One Health partnerships with the first One Health International Congress being held in 2011 in Melbourne, Australia (Ecohealth, 2011). Despite these efforts, few examples exist of real success, and it may require more drastic organizational changes to achieve the cultural changes needed to deliver the anticipated value and impact from a One Health approach.
Infectious Diseases Versus Food Safety
Ensuring the safety and quality of Australian foods within an integrated national biosecurity system is a current challenge for Australia. Although much has been done on characterization of food-borne hazards, on analysis of through-chain risks and the continual development of innovative risk management strategies, the approach is principally post-farm gate. In Australia these differences in infectious disease management versus food safety (Table A6-1) highlight areas
|
|
||
| Infectious Diseases | Food Safety | |
|
|
||
| Focus on effects of disease on host | Focus on impacts on humans | |
| Includes risk prevention as well as response (e.g., mainly on farm) | Focus on post-farm gate response (but clearly changing) | |
| Looks also at treatment in host (e.g., vaccination) | Looks at treatment of risk product | |
| Driven by government and primary product producers (e.g., farmers) | Driven by product processers and retailers | |
| Historically major outbreaks have driven change | Large events unusual and more about consumer impact | |
|
|
||
for a rethink and to consider how these two sectors can learn from each other. Central to this will be the application of the One Health principles.
Biosecurity Risk Management and the Biosecurity Continuum
The process of risk management for infectious diseases is concisely documented by the World Animal Health Organization (OIE) (Williberg, 2011) but can be simply considered in terms of the likelihood of a hazard occurring and the consequences if such an event did happen. In considering the likelihood, how the disease spreads and the survival of the pathogen are major components, but for the newer emerging infectious diseases understanding emergence and host switching are critical issues. Indeed, as an appreciation is gained of the emergence of pathogens from a wildlife reservoir into a livestock species and the subsequent potential to cause disease in humans, it becomes crucial to better understand those drivers that lead to a host switch.
On the consequence side of the risk profile, it is important to not only understand the process of disease in the affected host but also to appreciate this in terms of production and trade losses and the potential risks to humans and the environment. This expanded perception of the impact of disease lends even further credence to the concept of a multidisciplinary or One Health approach in managing effectively these consequences.
Australia has studied carefully the most effective approach to managing the risks from infectious disease and has come to the clear conclusion that the greatest return on investment lies in prevention and eradication rather than containment and allowing endemicity (see Figure A6-1).
It thus concludes that it is necessary to understand the risks for emergence and to tackle these directly to reduce or eliminate these risks. This approach, however, will never be 100 percent effective, and thus some resources will need
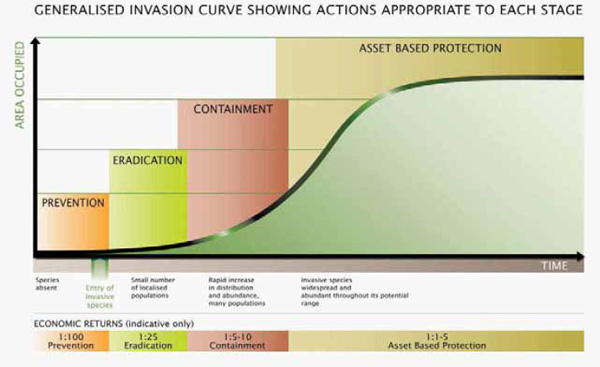
FIGURE A6-1 When to act: Generalized invasion curve showing actions appropriate to each stage.
SOURCE: © State of Victoria Department of Primary Industries, 2009, and reproduced with their kind permission.
to be allocated to consequence management but at a level that appreciates the lower risk if likelihood is significantly reduced.
In considering emerging risks, the movement of people to urbanized areas and the intensification of agriculture, often in close proximity to these urban areas, have clearly changed the risk profile in terms of both opportunities for the pathogen as well as the likely outcome of an infection. This risk is then exacerbated by the significant increased movement of people and products between these urbanized areas both nationally and internationally. There is little doubt that, overlying these issues, climate change has the potential to have a significant impact both directly through a change, for example, in available diseases carrying vectors and survival of the pathogen, but also in terms of change patterns of habitat and feeding by reservoir hosts (Rosenthal, 2009).
Australia has focused efforts for many years at the border and preborder areas in order to best manage the likelihood risks, to ensure detection as early as possible and thus a response that has the best chance to enable eradication. More recently and following a significant review of national biosecurity, an enhancement of postborder activities and the concept of the biosecurity continuum have emerged. To best manage this continuum it has been agreed between the Australian governments (Commonwealth, states, and territories), that the Common wealth government will focus and take responsibilities for preborder and border activities, with states and territories managing the majority of postborder activities. In recent times, the concept of a significant contribution from industry has emerged and, while government will retain primary responsibility for policy and standard setting, operational activities will in the future likely involve both government and industry working together in implementation of biosecurity activities.
Preborder Activities
It should be recognized from the outset, that although Australia is an island and this has been a significant advantage in maintaining a disease-free status, neighboring countries are in close proximity to Australia (via the Torres Strait) and the huge increase in international travel and trade considerably reduce the “safety factor” of being an island. It is therefore necessary to continually assess the threats from “abroad” and consider these threats in terms of market access and trade. There is a permanent pressure to broaden the trade in agricultural products, with an increase in demand to import from areas with a very different disease status to Australia. Managing these risks requires not only understanding the disease status of trading partners but also influencing the international regulations that govern such trade. Although continual risk analysis is a prerequisite for pre-border activities, threat reduction through a range of activities is also a major component. This starts with building trust and partnerships and has to grow into on-the-ground capacity-building support programs that
assist many countries, particularly in the Asian region, to better manage their own biosecurity programs.
A clear example of the need to reduce risk is that of the support by Australia to countries in the region to control foot and mouth disease (FMD). FMD represents the biggest risk to the Australian livestock industries, and for many years FMD was endemic in most countries in the region. Assistance to initially Indonesia and subsequently to the Southeast Asia FMD control program has considerably reduced this risk through both eradication and effective control in many countries in this region. Building an increased capacity in the region for countries to better manage their own biosecurity leads to a clear reduction of the likelihood risk of disease occurring in Australia. Various programs of the Australian Agency for International Development and the Australian Centre for International Agricultural Research focus in this area.
Border Activities
Australia’s Biosecurity Quarantine Operations Division manages those activities at the border that provide quarantine controls to minimize the risks of exotic pests and diseases entering the country. A further activity is the inspection of import and export certification to help retain Australia’s highly favorable health status and wide access to overseas markets. These inspections are targeted at activities involving aircraft, ships, and cargo and include the management of the National Australian Quarantine Strategy (NAQS).
Given the nature of the coastline of Australia, a large number of maritime programs are undertaken to ensure the effectiveness of these border operations. These include the management of unauthorized maritime arrivals, marine pollution, illegal activities in protected areas, issue around piracy, robbery or violence at sea, the illegal exploitation of natural marine resources, and maritime terrorism. Linked to this is the availability of a range of response assets including military naval vessels.
NAQS supports the government’s broader biosecurity objectives through conducting the monitoring and surveillance for exotic plants and animal disease across the north of Australia from Cairns to Broome and including the Torres Strait. These activities recognize the remote location of this region, the low human population, and the close proximity of neighboring countries and extend to collaborative surveillance and capacity building in Papua New Guinea, Indonesia, and Timor Leste, along with other neighboring countries. The overall strategy is clearly focused on early detection in a high-risk area, linked to an ability to mount an early response.
Postborder Activities
A wide range of activities are conducted postborder, principally through the governments of the states and territories. These aim for the early detection
of an emerging or exotic disease or disease-causing agent; the demonstration of freedom from a disease or disease-causing agent for trade purposes; the detection of changes in the distribution, prevalence, and incidence of a disease or disease-causing agent; and finally the detection of changes in factors or events that influence the risk of disease.
Increasingly a range of sophisticated geographical information systems and genetic-based tools have been used to better understand host and population structures with molecular epidemiology being used to understand the distribution of pathogens.
For the most part these activities have been targeted (or active) in nature with clear resource allocations and deliverables. Background (or passive) surveillance has been a lower priority for some time. The recent formation of a National Animal Health Surveillance System has recognized the critical component of passive surveillance in the overall approach, and increased activities in this area will be part of the future.
Conclusions and Discussions
• A One Health approach is essential to effectively managing the risks associated with both food safety and infectious diseases. Bringing together two necessary disciplines, skills and knowledge, is a real challenge given the current separation of management of environment, human, animal, and plant health. This may require real organizational change at the national level to achieve a genuine multidisciplinary and One Health approach.
• There are many similarities but some important differences between the management of food safety versus infectious disease. Having a whole systems approach (from farm to fork) has much to offer, and there are now real examples of success (e.g., control of salmonellosis in poultry in Denmark).
• Biosecurity (in the Australian concept) looks after both areas but remains somewhat fragmented. Currently biosecurity encompasses agricultural health (plants and animals) and environmental health; human health management remains outside of these activities except in exceptional cases (e.g., influenza management). A true One Health approach is perhaps some way off. The recent management of Hendra virus outbreaks, however, has clearly demonstrated the value of a One Health approach.
• Biosecurity is managed as a continuum from preborder to border and postborder activities.
• In managing biosecurity risks, investments in likelihood considerably out-weigh consequence management (but the latter cannot be ignored). Within this framework resource allocation across the preborder, border, and post-border need to be continually reassessed and currently in Australia there is an agreed-upon need to greater invest in postborder activities.
• Much still needs to be done to achieve a genuine One Health approach. Although progress is being made, real gain may require some fundamental changes in thinking and even reorganization at both the national and international levels with the creation of departments and divisions of One Health.
• Training and education in One Health is currently crucial in driving the longer-term cultures necessary to sustain a One Health approach. This needs to be linked to clear examples of success and the added value of a One Health approach.
Acknowledgments
Much of the material contained in this report was provided from the Australian Commonwealth Government’s Department of Agriculture, Forestry and Fisheries, State Departments of Primary Industries, and a number of agricultural institutes and universities in Australia. The author is indebted to the many staff of these government departments for these contributions.
References
AHA (Animal Health Australia). 2011a. Annual report 2010-2011. Canberra, Australia: Animal Health Australia.
———. 2011b. AUSVET Plans. http://www.animalhealthaustralia.com.au/programs/emergency-animal-disease-preperedness/ausvetplan/ (accessed January 11, 2012).
Beale, R., I. Fairbrother, R. Inglis, and W. Trebuk. 2008. Report to the Australian government.
Cutler, S. J., A. R. Fook, and W. H. van der Poel. 2010. Public health threat of new, reemerging, and neglected zoonoses in the industrialized world. Emerging Infectious Diseases 16(1):1-7.
Ecohealth. 2011. 7(Suppl. 1).
Leboeuf, A. 2011. Making sense of One Health: Cooperating at the human-animal-ecosystem health interface. Health and Environment Report No. 7. Paris: Institut Français des Relations Internationals.
OIE (World Organization for Animal Health). 2011. Terrestrial Animal Health Code I and II. Paris: World Organization for Animal Health.
Rosenthal, J. 2009. Climate change and the geographic distribution of infectious disease. Ecohealth 6(4):489-495.
Rushton, J. 2009. Economics of animal health and production. Wallingford, UK: CABI.
Vallet, B. 2009. Editorial. OIE Bulletin 2.
Willeberg, P. 2011. Models in the management of animal diseases. OIE Scientific and Technical Review 30(2).
Wolfe, N. D., C. Panosian Dunavan, and J. Diamond. 2007. Origins of major human infectious diseases. Nature 447:279-283.
Woolhouse, M. E., and S. Gowtage-Sequeria. 2005. Host range and emerging and reemerging pathogens. Emerging Infectious Diseases 11(12):1842-1847.
FOOD SAFETY: A VIEW FROM THE WILD SIDE
William B. Karesh, Elizabeth Loh, Catherine Machalaba13
Food-borne illnesses pose a serious threat to public health with growing economic and international trade ramifications. Past outbreaks of food-borne diseases have largely been viewed only through the lens of public health; yet food-borne illnesses are closely associated with the link between human and animal populations, and with the surrounding environment. For example, in 2006, approximately 200 people in 26 states were diagnosed with a particularly virulent strain of E.coli O157:H7 found in spinach. Viewed only from a human health perspective, our knowledge of this outbreak would have extended only to morbidity, mortality, outbreak investigation, laboratory diagnosis, and clinical treatment. However, once viewed through the lens of animal health and ecology, the E. coli O157:H7 isolates that caused the human deaths and serious illnesses related to spinach were also found in wild pig feces, the feces of several cows, and in a stream on one of the four spinach farms in the area (Warnert, 2007). Thus, a One Health perspective integrating our knowledge of the environment and ecology, in addition to human and animal health, was required to fully investigate and understand this outbreak and has great utility in the food-borne illness discussion.
Food-borne pathogens from wildlife span the taxonomic spectrum from helminthes to viruses (Figure A7-1). While the number of food-borne transmitted emerging disease events due to viruses is fewer than the number due to other groups of pathogens, some such as severe acute respiratory syndrome (SARS) have resulted in devastating consequences. As noted by Tauxe, the vast majority of food-borne illnesses in the United States due to known pathogens have emerged in the past two decades and an even larger percentage are due to yet-to-be-identified pathogens (Tauxe, 2002). Many of the known food-borne pathogens are zoonotic (Figure A7-2), and many may be linked to wildlife. As seen more generally with emerging infectious diseases (Jones et al., 2008), it is rational to assume that future outbreaks of new food-borne illnesses may be linked to wildlife.
From our review of the peer-reviewed literature, the main drivers of wildlife-related zoonotic disease emergence include land-use change and food industry changes. For food-borne disease, food industry changes, human susceptibility (reduced immune function), travel, and antimicrobial resistance were the primary drivers of past emergence events (Figure A7-3).
With more than 70 percent of food-borne EID events being zoonotic in nature (Figure A7-2), contact with or contamination of food or food ingredients
_______________
13 EcoHealth Alliance, 340 West 34th Street, New York, NY 10001.
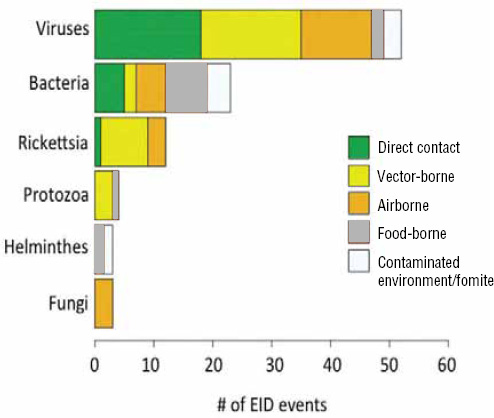
FIGURE A7-1 The number of infectious disease events that emerged from wildlife between 1940 and 2004 as published by Jones et al. (2008). These events (n = 96) are broken down by pathogen type, and each event is defined as the first emergence of a given pathogen. The colored areas within each bar depict the different transmission pathways as a percentage of the total number of emerging infectious disease (EID) events. For example, between 1940 and 2004, 52 zoonotic EIDs from wildlife were viruses. Of these events, 34.5 percent of these pathogens are transmitted by direct contact, 32.7 percent are vector-borne, 23.6 percent are air-borne, 5.5 percent are transmitted through contact with a contaminated environment or fomite, and 3.7 percent are food-borne.
by wild animals creates a serious potential for disease transmission. Commonly, both wild and domestic animals are implicated as sources of food contamination (Beuchat and Ryu, 1997; Cima, 2012; Doyle and Erickson, 2008; Gorski et al., 2011; Newell et al., 2010). Yet, definitive identification of a specific source animal or species is rare, particularly with wild species because they are typically no longer present at the time food-borne illness is detected in humans and investigations leading back to farms or processing plants are initiated. Despite the rarity of finding the “smoking gun,” we can operate on the fact that a large number of
food-borne illnesses are caused by pathogens frequently associated with wild species such as rodents, deer, feral pigs, reptiles, and birds (Figure A7-4).
Wild animals can provide the original source of pathogen contamination or serve to move pathogens from other infected sources. Similarly, insect reservoirs have been shown to introduce diseases into food processing systems (i.e., Chagas disease; Pereira et al., 2009) and have the potential for serving as mechanical vectors. Research by Wayadande et al. (2011) has identified filth flies as mechanical vectors that acquire and carry bacteria from their development stage environment (i.e., feces, carcasses, or decaying matter) including Salmonella typhii, Mycobacterium tuberculosis, and Rotavirus. The bacteria can also be carried in the excreta of blow flies, presenting risks of spread to external surfaces. Sela et al. (2005) reported similar findings for fruit flies.
In some cases, it may be feasible to limit contact or contamination of food during the production stage from larger wild mammals. Eliminating small-animal access to farm fields is impractical, although reducing exposure could be facilitated by a number of pest management techniques such as waste management and sanitation, eliminating hiding areas, etc. Postharvest contamination of food from small mammals and birds is more easily controlled by good pest management practices.
More directly, wildlife provides a substantial portion of our food globally, with nearly half of all seafood coming from wild sources. In some regions of the world, wild meat from terrestrial animals represents a primary source of protein on which populations are dependent. The volume of wild meat (“bush meat”) harvested from Central Africa alone totals more than 1 billion kg per year (Wilkie and Carpenter, 1999). This volume of meat, almost all of which is processed and distributed to consumers with few if any modern hygiene practices, provides a constant opportunity for human exposure to common food-borne pathogens (Karesh et al., 2005; Smith et al., 2012). Additionally, wild animals are sought after as delicacies based on cultural and consumer preferences. This demand for wildlife, which is both legally and illegally supplied, has global dimensions. Although there is a “not in my backyard” mentality that limits our concern for what diseases are circulating across the world, this is a problem in which the United States is deeply involved, as the United States is the main importer of wildlife (Asmüssen et al., 2011). What is present across the world can be in our backyard—and then on our plate—in a matter of days through our importation of tens of millions of legal and illegal animals (Jones et al., 2008; Smith et al., 2009). The United States also contributes to the potential spread of disease to other countries through its export of turtles destined for the food trade in Asia.
The emergence and transmission of food-borne zoonotic diseases from dietary habits and pressures are increasingly being documented. In simplest terms, we are seeing that the consumption of wild animals translates to “you get what you eat.” Hepatitis E (Vasickova et al., 2007), brucellosis (CDC, 2009), and trichinellosis (Roy et al., 2003) are examples of hunter-acquired food-borne illness. The origin
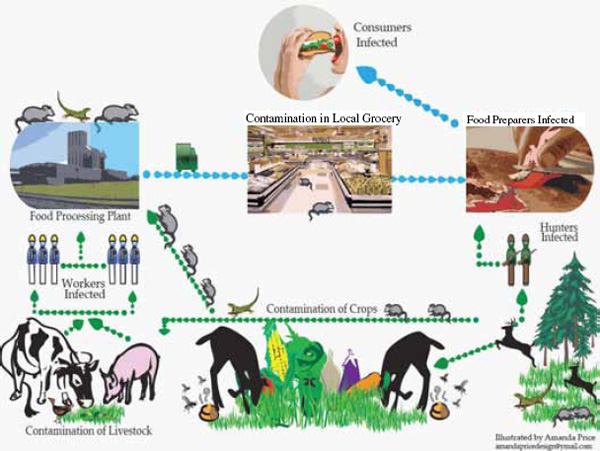
FIGURE A7-4 Routes of contamination resulting in food-borne illness linked to wildlife.
SOURCE: Illustrated by Amanda Price.
of HIV/AIDS through the transmission of nonhuman primate simian immunodeficiency viruses to humans via bush meat hunting represents a major example of how anthropogenic behaviors can lead to massive and pervasive public health threats, and newer evidence shows that the transmission of non human retroviruses to humans happens on a regular basis (Betsem et al., 2011; Calattini et al., 2011; Peeters et al., 2002). SARS is another well-known example of food-borne illness from animals, spreading to humans after the mixing of live reservoir hosts (e.g., bats) and intermediate hosts (e.g., civets) (Guan et al., 2003; Li et al., 2005).
Food-borne illnesses and the interactions that increase their presence in our food supply are not new. However, we are seeing increased detection of zoonotic food-borne illness as we engage more and more in the practices that drive disease emergence. There are challenges ahead, including climate change and its associated changes in animal migration, water supply demands, and possibly pathogen distribution and abundance. For the latter, Vezzulli et al. reported that, during the past half century, ubiquitous marine bacteria of the Vibrio genus, including V. cholerae, increased in occurrence within the plankton-associated bacterial community of the North Sea, where an unprecedented increase in bathing infections associated with these bacteria was recently reported. Among environmental variables, increased sea surface temperature explained 45 percent of the variance in Vibrio data, supporting the hypothesis that ocean warming is facilitating the spread of vibrios and may be the cause of the globally increasing trend in associated diseases (Vezzulli et al., 2012).
Opportunities for Food-Borne Disease Surveillance
Humans have commonly served as the sentinel species for food-borne illnesses, and as a result early detection and response systems such as PulseNet, FoodNet, the National Electronic Norovirus Outbreak Network (CalciNet), and the National Notifiable Diseases Surveillance System are based on human outbreak (or case) surveillance. There are new approaches in emerging disease surveillance that could possibly be adapted to food-borne disease surveillance to contribute to targeting surveillance efforts, early detection, control, and prevention. Some of these approaches are being developed and tested with the support of the U.S. Agency for International Development’s Emerging Pandemic Threats PREDICT program in an attempt to create more upstream focus for early detection of emerging diseases with pandemic potential.
PREDICT’s SMART (strategic, measurable adaptive, responsive, and targeted) surveillance method uses continuously refined predictive models, literature reviews and analyses, digital news surveillance, and input from front-line information by field personnel and the public. Predictive models can identify areas of greatest risk for outbreaks of food-borne illness (food-borne illness “hotspots”) by generating geospatial information on various human—animal interfaces, behaviors, activities, and presence of additional risk factors. These analyses
can be spatially explicit to account for host and pathogen niches and known distributions, as well as differing drivers of disease emergence in different regions of the world. For food-borne illnesses, these interfaces might include areas where food-production farms overlap with habitat used by wildlife (to include species and common pathogens), areas of hunting, wildlife—livestock conflict, natural resource extraction and land-use change, markets, and regions with high levels of global transportation.
Closely linked, and in some ways underpinning the predictive modeling, analyses of peer-reviewed publications can be conducted for food-borne diseases related to wildlife to determine species and human activities that present the highest risks. Databases on outbreaks or cases of wildlife-related food-borne illnesses could also be queried to identify products with high potential for food-borne diseases, drivers of disease emergence, and risky areas where surveillance and control efforts can be focused. Analysis of the individual drivers of disease emergence can help parse out the most likely routes of transmission in a given area, helping to set control measures in place. For the PREDICT program, these analyses are used to target surveillance to key taxonomic groups and key human activities and “interfaces” with wildlife. Similar approaches have been used in the food industry for years in determining the most effective “control points” or monitoring steps for known, common pathogens. Analyses of drivers might contribute to current approaches by revealing additional key points for surveillance and interventions preceding human infection with novel pathogens. Some of this information could be derived from the Foodborne Outbreak Online Database produced by the U.S. National Outbreak Reporting System, which reports annual outbreaks by year, month, state, etiology, location interface, total cases, hospitalizations, deaths, food vehicle, and contaminated ingredient. It currently contains data from 1998-2009. Wildlife-related food-borne illnesses present some additional challenges and opportunities that may require nonconventional approaches and include targeting surveillance outside of the normal farm-to-table production chain. For example, analyses may indicate that sampling by hunters or pest control operators could yield valuable surveillance data (both for pathogen and host species abundance) in a cost-effective manner.
Digital surveillance, the practice of seeking disease news and tracking disease trends via the Internet such as provided by HealthMap, could also help to deter mine where to target surveillance, control, and capacity-building efforts. Both HealthMap and ProMED-Mail are expanding their coverage of wildlife disease events with the support of the PREDICT program, providing easier access to that information for health workers and the public around the world. While historic, macro-level data can be useful for risk modeling and mapping, it is important to maintain on-the-ground approaches to continually update and target the specific and real-time events and factors driving food-borne disease risk.
Food-borne disease diagnostics. The increase in technological capabilities over the past decade has aided our ability to garner far more information from
surveillance efforts. These technologies can support rapid detection, diagnosis, and control of food-borne illness, as well as preventive measures around potential food-borne illness emergence. Expanded efforts in surveillance for pathogens in wildlife have created a need for simple, inexpensive broad-ranging tests that can be used locally for rapid screening, coupled with networks of labs that can follow up with more detailed, confirmatory testing. For food-borne illness, pathogen-specific and -sensitive tests will always play an important role, but as we expand our concern to wildlife-related and novel pathogens in food, there could be a growing need for some broad-level screening tests for family-, order-, or genus-level testing.
Intervention Strategies
Once a source or a transmission pathway has been established, the use of existing control mechanisms for food-borne illnesses will likely be effective. There is currently no evidence to suggest that wildlife-related food-borne illnesses are inherently different from other sources of contamination; in the case of consumption of wildlife itself, similar risk reduction practices for the safe handling and consumption of poultry, beef, or seafood should suffice. As with traditional food safety surveillance and interventions, stakeholder engagement is crucial for success. For diseases related to wildlife, this concept is the same, but the stakeholders may not be the traditional food safety partners that industry and public health agencies work with. Hunters, conservation organizations, and wildlife management authorities need to be engaged to most effectively develop and implement both surveillance systems and control strategies. As we are learning in most areas of public health, collaboration among multiple disciplines is key in the success of disease risk reduction interventions, and this is especially true as we try to reduce food-borne illnesses linked to wildlife in a variety of ways.
Agency Partnerships and Regulation
There is great potential to learn more about wildlife-related food-borne illnesses through collaboration with surveillance and regulatory agencies. Initial sampling efforts of confiscated wildlife through the Centers for Disease Control and Prevention (CDC) have begun to find pathogens (Smith et al., 2012), but there needs to be dedicated funding and sustained efforts to conduct sufficient inspections and pathogen testing across all agencies that regulate wildlife. This is especially relevant to the Food and Drug Administration, which has regulatory authority for food safety in meat or other animal-derived products from wildlife in interstate commerce not otherwise covered by U.S. Department of Agriculture authority. Greater understanding of wildlife trade risks can inform risk reduction regulations and practices. For example, short-term enforcement of traded animals
in Asian markets following the 2003 SARS outbreak shed light on the scale and composition of wildlife trade in the region (Karesh et al., 2005). In addition to protecting consumers, strong enforcement and surveillance of food-bound wildlife products can help the health, pet, and food industries proactively mitigate risks from potential food-borne threats.
The Convention on the International Trade of Endangered Species (CITES) regulates the trade of endangered wildlife in its 175 member countries. However, illegal wildlife trade is still widespread, with 87 percent of CITES member countries reporting illegal trade activity. Thus, trade regulations must be implemented and enforced at the individual country level as well. There are opportunities to use regulation to directly protect the public from wildlife diseases. The U.S. ban on small turtles resulted in a major decline in Salmonella transmission in children, leading to a 77 percent reduction of cases (Cohen et al., 1980). Food-borne disease surveillance can inform targeted regulations to reduce illness spread from high-risk sources.
Collaboration
Food-borne illness is a complex challenge that cannot merely be solved by an “expert solution” and demands diverse stakeholder participation. The public health sector and food industry should not be isolated in addressing food-borne illness concerns involving wildlife. There are ample opportunities for collaboration and mutual benefits among a wide array of stakeholder disciplines. The capture of endangered wildlife and detrimental impacts to wildlife habitats through land-use change—a major driver of emerging infectious diseases (Jones et al., 2008; Patz et al., 2004)—pose serious threats to the sustainability of biodiversity, bringing conservationists into the equation. Hunters and indigenous populations have a vested interest in both the food security and the health of their communities, and their direct involvement and ownership of disease reduction efforts is crucial for creating long-term success.
At the same time, although food-borne illness is a global concern, individual risks can be addressed at a microcosm level. Large-level suppliers can support the health of local communities where their products are sourced. This involves both reducing reliance on wildlife for food (e.g., in logging settlements) and taking measures to educate communities about safe and healthy practices (i.e., in hunting and animal butchering). These approaches have had marked success. For example, Northern Congo has not seen a human case of Ebola since 2005, despite its continuing presence in wildlife. The joint approach of education of village hunters on risk reduction (i.e., high-risk species, hand washing, and cooking techniques) and the assumption of responsibility taken on by hunters for protecting themselves and their families is credited with the prevention of new cases.
Going Forward
Without effective action, the world is slated for an increasing trend of negative health and economic consequences from food-borne illnesses. Fortunately, at present there are opportunities for intervention. Addressing food security issues will decrease reliance on high-risk food sources such as wildlife. Additionally, overall progress around disease surveillance, control, and prevention has allowed us to establish feasible disease monitoring systems and learn important lessons that can be applied to the risk reduction of food-borne illnesses. Integral to these lessons has been the necessity and value of One Health collaborations. The synergies formed by integrating environment, health, and wildlife sectors, in concert with local populations, can provide the perspectives and actions to reduce food-borne illnesses and provide appropriate intervention and prevention strategies before further outbreaks occur.
References
Asmüssen, M. V., J. R. Ferrer-Paris, et al. 2011 (accepted). Estimates and trends of illegal wildlife trade in the world. PLoS Biology.
Betsem, E., R. Rua, P. Tortevoye, A. Froment, and A. Gessain. 2011. Frequent and recent human acquisition of simian foamy viruses through apes’ bites in central Africa. PLoS Pathogens 7(10):e1002306.
Beuchat, L. R., and J. H. Ryu. 1997. Produce handling and processing practices. Emerging Infectious Diseases 3(4):459-465.
Calattini, S., E. Betsem, S. Bassot, S. A. Chevalier, P. Tortevoye, R. Njouom, R. Mahieux, A. Froment, and A. Gessain. 2011. Multiple retroviral infection by HTLV type 1, 2, 3 and simian foamy virus in a family of Pygmies from Cameroon. Virology 410(1):48-55.
CDC (Centers for Disease Control and Prevention). 2009. Brucella suis infection associated with feral swine hunting—three states, 2007-2008. Morbidity and Mortality Weekly Report 58(22):618-621.
Cima, G. 2012. Wildlife, trade, susceptibility amplify food risks. Journal of the American Veterinary Medical Association 240(4):352-355.
Cohen, M. L., M. Potter, R. Pollard, and R. A. Feldman. 1980. Turtle-associated salmonellosis in the United States. Effect of public health action, 1970 to 1976. Journal of the American Medical Association 243(12):1247-1249.
Doyle, M. P., and M. C. Erickson. 2008. Summer meeting 2007—the problems with fresh produce: An overview. Journal of Applied Microbiology 105(2):317-330.
Gorski, L., C. T. Parker, A. Liang, M. B. Cooley, M. T. Jay-Russell, A. G. Gordus, E. R. Atwill, and R. E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Applied and Environmental Microbiology 77(8):2734-2748.
Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302(5643):276-278.
Jones, K. E., N. G. Patel, M. A. Levy, A. Storeygard, D. Balk, J. L. Gittleman, and P. Daszak. 2008. Global trends in emerging infectious diseases. Nature 451:990-994.
Karesh, W. B., R. A. Cook, E. L. Bennett, and J. Newcomb. 2005. Wildlife trade and global disease emergence. Emerging Infectious Diseases 11(7):1000-1002.
Li, W., Z. Shi, M. Yu, W. Ren, C. Smith, J. H. Epstein, H. Wang, G. Crameri, Z. Hu, H. Zhang, J. Zhang, J. McEachern, H. Field, P. Daszak, B. T. Eaton, S. Zhang, and L. F. Wang. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310(5748):676-679.
Newell, D. G., M. Koopmans, L. Verhoef, E. Duizer, A. Aidara-Kane, H. Sprong, M. Opsteegh, M. Langelaar, J. Threfall, F. Scheutz, J. van der Giessen, and H. Kruse. 2010. Food-borne diseases—the challenges of 200 years ago still persist while new ones continue to emerge. International Journal of Food Microbiology 139(Suppl.):S3-S15.
Patz, J. A., P. Daszak, G. M. Tabor, A. A. Aguirre, M. Pearl, J. Epstein, N. D. Wolfe, A. M. Kilpatrick, J. Foufopoulos, D. Molyneux, and D. J. Bradley; Working Group on Land Use Change and Disease Emergence. 2004. Unhealthy landscapes: Policy recommendations on land use change and infectious disease emergence. Environmental Health Perspectives 112(10):1092-1098.
Peeters, M., V. Courgnaud, B. Abela, P. Auzel, X. Pourrut, F. Bibollet-Ruche, S. Loul, F. Liegeois, C. Butel, D. Koulagna, E. Mpoudi-Ngole, G. M. Shaw, B. H. Hahn, and E. Delaporte. 2002. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerging Infectious Diseases 8(5):451-457.
Pereira, K. S., F. L. Schmidt, A. M. Guaraldo, R. M. Franco, V. L. Dias, and L. A. Passos. 2009. Chagas’ disease as a foodborne illness. Journal of Food Protection 72(2):441-446.
Roy, S. L., A. S. Lopez, and P. M. Schantz. 2003. Trichinellosis surveillance—United States, 1997-2001. Morbidity and Mortality Weekly Report: Surveillance Summaries 52(6):1-8.
Sela, S., D. Nestel, R. Pinto, E. Nemny-Lavy, and M. Bar-Joseph. 2005. Mediterranean fruit fly as a p otential vector of bacterial pathogens. Applied and Environmental Microbiology 71(7):4052-4056.
Smith, K. F., M. Behrens, L. M. Schloegel, N. Marano, S. Burgiel, and P. Daszak. 2009. Ecology: Reducing the risks of the wildlife trade. Science 324(5927):594-595.
Smith, K. M., S. J. Anthony, W. M. Switzer, J. H. Epstein, T. Seimon, H. Jia, M. D. Sanchez, T. T. Huynh, G. G. Galland, S. E. Shapiro, J. M. Sleeman, D. McAloose, M. Stuchin, G. Amato, S. O. Kolokotronis, W. I. Lipkin, W. B. Karesh, P. Daszak, and N. Marano. 2012. Zoonotic viruses associated with illegally imported wildlife products. PloS One 7(1):e29505.
Tauxe, R. V. 2002. Emerging foodborne pathogens. International Journal of Food Microbiology 78(1-2):31-41.
Vasickova, P., I. Psikal, P. Chalupa, M. Holub, R. Svoboda, and I. Pavlik. 2007. Hepatitis E virus: A review. Veterinarni Medicina 52(9):365-384.
Vezzulli, L., I. Brettar, E. Pezzati, P. C. Reid, R. R. Colwell, M. G. Höfle, and C. Pruzzo. 2012. Long-term effects of ocean warming on the prokaryotic community: Evidence from the vibrios. ISME Journal 6(1):21-30.
Warnert, J. 2007. Expanded research to target E. coli outbreaks. California Agriculture 61(1):5.
Wayadande, A., J. Talley, A. Gerry, U. DeSilva, and J. Fletcher. 2011. Filth Flies as Disseminators of Human Pathogens. Chemical and Biological Defense Science and Technology Conference. Las Vegas; Presented November 15, 2011.
Wilkie, D. S., and J. F. Carpenter. 1999. Bushmeat hunting in the Congo Basin: An assessment of impacts and options for mitigation. Biodiversity and Conservation 8(7):927-955.
Introduction
The concept of One Health is not new but it has reemerged as an important concept to both understand and help address our contemporary challenges and threats to our health.
We live in a world that is rapidly changing, complex, and progressively more interconnected. The convergence of people, animals, and our environment has created a new dynamic—one in which the health of each group is now profoundly and inextricably linked and elaborately connected.
Inherent in this new dynamic is the changing interface between people and animals, including animal products. The human—animal interface is accelerating, expanding, and becoming increasingly more consequential. Over the past three decades, approximately 75 percent of new human infectious diseases have been zoonotic. The global population has now exceeded 7 billion people, and an estimated 30 billion food animals were produced to help feed this population and meet its growing demand for protein from animal sources. The result is a phenomenal global food system that is both a major agricultural and business accomplishment and an unparalleled challenge that is creating major societal issues that, to some extent, threaten human, animal, and environmental health (FAO, 2006).
As a further consequence, the safety of our food is being increasingly scrutinized and questioned by the public, and food-borne illnesses are significant, costly, and a global problem. There continue to be differences of opinion on how to improve food safety, and we lack an integrated and holistic strategy for implementation in the United States and much of the world. While we acknowledge some success in controlling and ameliorating food-borne illnesses and food contamination, these achievements are uneven, often transitory, and especially difficult. Ensuring a safe food supply will likely demand new levels of collaboration, understanding, and thinking. The application of a One Health model where potential solutions are viewed and delivered more holistically and with an emphasis on prevention is a compelling and timely strategy.
One Health Defined
One Health is the collaborative effort of multiple disciplines—working locally, nationally, and globally to attain optimal health for people, animals, and
_______________
14 The Ohio State University.
our environment (King et al., 2008). The scale and complexity of food safety issues demand that scientists, researchers, and others move beyond the confines of their own disciplines, professions, and mindsets and explore new organizational modes of team science, and the One Health concept embodies this declaration. The scope of One Health is impressive, broad, and growing. Much of the recent focus of One Health has been limited to emerging infectious diseases, yet the concept clearly embraces environmental and ecosystem health, social sciences, ecology, noninfectious diseases and chronic diseases, wildlife, land use, antimicrobial resistance, biodiversity, and much more. While these components are appreciated within our understanding of the broad dimensions of health, they also add to the complexity of One Health and the difficulty in implementing strategies, building effective coalitions, and mobilizing scientific communities who embrace One Health yet who have been trained and think in much narrower scope and scale. Although there may be disagreement on the exact definition of One Health there is broad consensus that a new framework for preventing food-borne diseases is essential rather than the alternative of constantly responding to them reactively.
“Wicked” Problems
We now live in a world that is complex, interconnected, and uncertain, with growing dilemmas and unprecedented societal problems. These problems have been referred to as “wicked problems” and are contrasted with “tame problems,” which can be solved with existing modes of inquiry, technological knowledge, and decision making. Wicked problems are complex, do not have yes-or-no answers, can generate unexpected consequences, may be symptomatic of other problems, and are unique in that past experiences and thinking are not helpful in addressing them. In addition, wicked problems and issues often crop up as organizations face constant change and unparalleled challenges, and they often occur in a social context with diverse opinions from numerous stakeholders who lack consensus in both identifying the total problem and how to resolve them (Brown et al., 2010).
Issues and problems connected with food safety, food security, sustainable production systems that ensure environmental protections, and the capacity to help feed more than 7 billion people collectively qualify as a societal and wicked dilemma. Ensuring safe, accessible, affordable, and nutritious food is increasingly difficult, especially in a global context. Central to this challenge is the development of a One Health strategy and a new level of thinking and acting.
The world population has a growth rate of 1.2 percent per year and the next century will represent a period of exponential growth. There is also a significant demographic fault line between the population growth in developed versus developing countries. Approximately 90 percent of the world’s population growth is occurring in the developing countries of the world. In addition almost 1 billion people live in peri-urban or slum settings in the developing world’s largest cities,
and these sites are where the most rapid growth in our human populations will continue (Smith and Kelly, 2008).
While there is also legitimate concern about the approximately 800 million people who are undernourished, we are concurrently observing a relative increase in wealth in the developing world and as per capita incomes rise; people eat more calories and consume different products, including a demand for meat and protein from animal sources. Today, 3 to 4 billion people consume very little meat but will consume more, should incomes increase. Thus, a new agricultural phenomenon is emerging: the Livestock Revolution. With relative increases in wealth and technological advances in livestock and poultry production, global increases in production and consumption of livestock products are unavoidable. The Food and Agriculture Organization (FAO) estimates that there will be a demand for a 50 percent increase for animal proteins in the next one to two decades. Thus, the entire global food system will adjust into a more intensive, specialized, and integrated system, and production systems will progressively shift to the developing world (Delgado et al., 1999).
As the Livestock Revolution ushers in a rapidly expanding animal agriculture production system in the developing world, there is real concern regarding the animal and public health infrastructures available to support this revolution. The United States now imports approximately 15 percent of its food, but it imports a much higher percentage of seasonable fruits, vegetables, and seafood (Acheson, 2010). The need for inspecting these products is growing much more quickly than the regulatory system now in place to implement such safeguards.
Concurrently, there is unprecedented immigration and movement of people worldwide. Unique diasporas have emerged, and there are large numbers of immunocompromised individuals dispersed throughout the United States and global populations who are especially susceptible to infections including food-and water-borne illnesses. In many countries, the population of seniors is one of the fastest growing cohorts.
There is also a disconnect between global commerce and the remarkable movement of food in trade channels and the commensurate emphasis and assurance of safe food. There is a significant gap between an emphasis on the rapidly growing commerce and business of global food companies and an equal emphasis and investment to address the potential health consequences generated by global food and animal commerce (Kimball, 2010). The 21st century has created a great mixing bowl of people, animals, and animal products and a group of wicked problems, including the protection and safety of our food, that demands a transformation of thought and actions to address these contemporary challenges, threats to our health and well-being, and threats to animal and environmental health that are under increasing pressure. A holistic and integrated approach considering these domains in a One Health strategy is both logical and essential to further success.
Food Safety: Trends and Concerns
The Centers for Disease Control and Prevention (CDC) now estimates that, in the United States, there are 48 million food-borne illnesses, 128,000 hospitalizations, and 3,000 deaths each year (Scallan et al., 2011). Thus, one out of six Americans will have at least one episode of a food-borne illness annually. Although we lack similar global data, a rough extrapolation would suggest that there could be at least 1 billion such illnesses worldwide each year. This would qualify as a global public health epidemic by any definition; however, there are few surveillance systems that can help us track and define the global burden of food-borne illnesses. With these estimates of the burden of illness, the global food system continues to grow increasingly more vulnerable and potentially riskier and progressively connects our global communities daily through our growing imports and exports of food.
Today, microbes can traverse the globe faster than their incubation period; our great convergence offers unique opportunities for them to cross species lines, become resistant to antimicrobial agents, adapt, change, and find new niches, and emerging and reemerging diseases result. Our current era of emerging infections and pace of emergence is accelerated with changing ecosystems, risky human behavior, poverty, travel, trade, globalization, population growth, and our interconnectiveness. Food as a potential vehicle for disease transmission is embedded in this complex system; food safety has taken on a growing importance and has become a critical public health imperative.
As we learn more about the burden of food-borne illness, we also appreciate and learn about new pathogens transmitted by food and the expansion of the types of food that can transmit potential food-borne pathogens. We are reminded that bacterial contamination of food is a critical issue; however, viruses, parasites, toxins, prions, chemicals, metals, and allergens may also be transmitted by food and water and result in an expanded burden of illness and growing spectrum of threats.
CDC studies have also demonstrated changing patterns of attribution. Plant-derived foods such as leafy greens, tomatoes, and sprouts have been implicated in more and more food-borne disease outbreaks. In the recent past, transmission has been linked to peanut butter, pizza, spinach, ice cream, cookie dough, pet food, melons, peppers, and carrot juice. We are also concerned about the concept of “stealth” vehicles in transmission. There are numerous food ingredients that are often mixed in with foods, such as spices, which can be vehicles for transmission. It is estimated that 75 percent of our food that has been processed has an ingredient from an international source (Doyle and Erickson, 2008).
In addition to the traditional food-borne pathogens such as Escherichia coli, Salmonella, Campylobacter, Listeria, and so on, new outbreaks often reveal new agents. The FoodNet system that analyzes outbreaks has revealed adenoviruses, sapoviruses, saffoldviruses, and picobirnaviruses as potential pathogens (Tauxe, 2008). To further complicate our understanding of the safety of our food, trans-
mission vehicles can change when microbes are given new opportunities. For example, the Nipah virus first found in a zoonotic disease outbreak in Malaysia that killed pigs and people associated with them has recently been found as a contaminant in date palm sap, a food source in Bangladesh. Pteropus fruit bats are the asymptomatic carriers. Trypanosoma cruzi is the parasite that causes Chagas disease and is usually transmitted to people via reduvid insects, yet it has recently been found in sugar cane juice in Brazil. There is a remarkable spectrum of foods and pathogens involved in food-borne illnesses, and this is an ever-changing dynamic. There is a growing importance of produce as a vehicle for food-borne pathogens, yet animal reservoirs are often the origin of these infections. One Health gives us the proper lens to view and better understand this linkage and, more importantly, to develop new insights for changing our interventions and prevention strategies. In many instances, ill people are the end point of a complicated epidemiological cycle and serve as indicator hosts; however, if we continue to focus exclusively on food-borne illness by responding to human outbreaks and just conducting retrospective analyses, then we will miss the true sites and origins of these diseases and we will forgo critical prevention strategies. To a certain extent, ill people serve as sentinels of a larger ecological problem and, as such, may not be the best focal point for our interventions. One Health is a mindset that is proactive and preventive and helps to shift our attention “upstream” to the ecological, animal, and environmental sources and influences responsible for these illnesses and helps us to identify the most effective points for the initiation of food safety actions.
According to Jared Diamond, in his book Guns, Germs, and Steel, diseases such as measles, smallpox, influenza, and tuberculosis likely evolved from animal diseases as the first group of zoonotic diseases (Diamond, 1999). The advent of agriculture and the domestication of animals approximately 8,000 to 10,000 years ago were drivers of a new human—animal interface and the first era of emerging zoonotic diseases. Although animal agriculture is much more sophisticated today, it is also growing more intensified and complex. Domestication has resulted in the development of new and more efficient food-animal species, and the human—animal interface has accelerated and multiplied through the globalization of our food system and has created the potential exposure of billions of people to potential pathogens. As our food-animal production and ecosystems continue to change to produce more and more, microbes are given further opportunities to adapt and find new niches. Transboundary diseases have again emerged at an alarming rate, suggesting that our new era of disease emergence has a troubling similarity to the past era that was created 8,000 to 10,000 years ago.
One Health Lens to View and Improve Food Safety
Dr. Gro Brundtland, former Director of the World Health Organization, stated, “In the modern world, bacteria and viruses travel almost as fast as money.
With globalization, a single microbial sea washes all of humankind. There are no health sanctuaries.” In actuality, that microbial sea washes not only over all humankind, but also across our animal and environmental domains. This dynamic exposes and connects the human, animal, and environmental domains in ways never previously experienced. Positive and negative actions and impacts in one domain now may significantly impact the others, and solutions to address threats in any single domain may have multiplier effects in the others. This is the essence of One Health, and the safety of our food must also be considered using this mindset.
Yet as our food systems grow ever more complex, larger, and more vulnerable, our scientific, medical, agricultural, environmental, and health systems and studies remain too isolated and entrenched. Perhaps our greatest challenge today may be our ability to reconcile the changes and challenges of our global convergence with our traditional thinking and habitual ways of working. For many zoonotic diseases, including certain food-borne illnesses, we focus on the risk to human health while the most effective control strategies are in animals, animal products, and the environment. There are divided constituents and responsibilities for animal and human health that must be integrated in order to make significant progress in the reduction of many food-borne illnesses. The microbes seldom distinguish among species as they just seek opportunities to survive and multiply. Our own bias and artificial separation between veterinary and animal health and public health is a critical barrier to the acceptance of One Health.
I have discussed the concept of wicked problems and the need to view many contemporary problems as interconnected issues that have created larger societal dilemmas. Patterns of thought of a previous era may not be useful to address current problems. Because wicked problems are part of the society that creates them, future solutions and actions must be based in that society. We can no longer focus on a single domain of health or any singular inquiry; we now must be open to new ways of thinking and be receptive to new ideas and directions that match our challenging times. The status quo in food safety must be replaced by a new transdisciplinarity and a new collective understanding of food safety characterized by a One Health mindset and approach. A One Health emergent community of practice now exists where new views, approaches, and knowledge can inform each other synergistically and more productively.
In Thomas Kuhn’s seminal book The Structure of Scientific Revolution (Kuhn, 1962), he discussed new paradigms and the conditions and factors that create them. A paradigm shift is often manifested because old models to solve problems do not work as well and new models have yet to be created or substantiated. Basic assumptions are questioned, and the evidence to change is not uniformly accepted. While we acknowledge that progress has been made in making our food safer, especially in the United States, breakthrough thinking is minimal. Kuhn suggested that changing mindsets can be difficult and protracted and that new paradigms are not necessarily led by a scientific community. In
the case of food safety, much of the force for change is being led by con sumers and more recently retailers. Also new paradigms often lead to new fields of study, inquiry, and work. One Health, although not new, is certainly a renewed field of inquiry and transdisciplinary thinking.
The convergence of people, animals, and our environment has created a new dynamic in which the health of each domain is inextricably interconnected. The challenges associated with this new reality are demanding, profound, and unprecedented. This remarkable convergence is a critical factor in disease emergence, and there is nothing on the horizon to suggest that this dynamic will be altered or abated. The safety of our food supply is a microcosm of this larger dynamic, and our food is increasingly vulnerable to both intentional and unintentional contaminations and changing microbial communities. Working successfully to address these threats will require new thinking, changing partnerships, and shifting our emphasis “upstream,” closer to the origin of pathogens in other domains. A One Health paradigm shift holds great promise but is also a new mindset that will be disruptive to the status quo; thus, old systems, habitual thinking, and working with old modes of inquiry that are sharply divided among diverse cultures and interests and that compete for resources and are part of strongly embedded belief systems remain as challenges.
Dr. Josh Lederberg, a Nobel Laureate and founder of this Forum, published an article in Science in 2000 titled “Infectious History.” He stated, “An axiomatic starting point for progress is the simple recognition that humans, animals, plants and microbes are cohabitants of this planet. That leads to refined questions that focus on the origin and dynamics of instabilities within this context of cohabitation. These instabilities rise from two main sources loosely definable as ecological and evolutionary” (Lederberg, 2000). I suggest that our dynamic, complex food system, and the challenge of its safety, is about controlling and preventing instabilities and using One Health as the construct to understand this ecological dilemma and as the foundation to devise new solutions and interventions. Dr. Lederberg further remarked that the future of humanity and microbes will be based on “our wits versus their genes.” This is a prophetic statement underpinning One Health’s application to food safety.
References
Acheson, D. 2010. Globalization of the food supply: Time for a change in approach. In Infectious disease movement in a borderless world. Washington, DC: The National Academies Press. P. 119.
Brown, V. A., J. A. Harris, and J. Y. Russell. 2010. Tackling wicked problems: Through the transdisciplinary imagination. Washington, DC: Earthscan.
Delgado, C., M. Rosegrant, H. Steinfeld, S. Ehui, and C. Courbois. 1999. Livestock to 2020: The next food revolution. ftp://ftp.fao.org/docrep/nonfao/lead/x6155e/x6155e00.pdf (accessed April 3, 2012).
Diamond, J. 1999. Guns, Germs, and Steel. New York: W. W. Norton.
Doyle, M. P., and M. C. Erickson. 2008. Imported foods: Microbiological issues and challenges. Washington, DC: ASM Press.
FAO (Food and Agriculture Organization). 2006. Agriculture and an animal feed industry. Business Meeting Presentation.
Kimball, A. M. 2010. Risky trade and emerging infections. In Infectious disease movement in a borderless world. Washington, DC: The National Academies Press. P. 117.
King, L. J., L. R. Anderson, C. G. Blackmore, M. J. Blackwell, E. A. Lautner, L. C. Marcus, T. E. Meyer, T. P. Monath, J. E. Nave, J. Ohle, M. Pappaioanou, J. Sobata, W. S. Stokes, R. M. Davis, J. H. Glasser, and R. K. Mahr. 2008. One Health Initiative Task Force Report. Journal of the American Veterinary Medical Association 233(2):259-261.
Kuhn, T. 1962. Structure of scientific revolutions. Chicago: University of Chicago Press.
Lederberg, J. 2000. Infections history. Science 288(5464):287-293.
Scallan, E., P. M. Griffin, F. J. Angulo, R. V. Tauxe, and R. M. Hoekstra. 2011. Foodborne illness acquired in the U.S. Emerging Infectious Diseases 17(1):7-22.
Smith, G., and A. M. Kelly. 2008. Food security in a global economy. Philadelphia: University of Pennsylvania Press.
Tauxe, R. 2008. Roots of foodborne illness. http://www.nyas.org/Publications/EBriefings/Detail.aspx?cid=d8f46e08-453f-463a-861d-f5df074b760b (accessed April 3, 2012).
FOOD-BORNE VIRUSES FROM A GLOBAL PERSPECTIVE
Abstract
Food-borne transmission has been described as one of the modes of transmission for many different viruses, associated with diseases ranging from mild diarrhea to severe neurological symptoms. The potential for such transmission can be studied by using common human pathogens as a model. By genomic epidemiology approaches, this has revealed significant food-related disease for noroviruses and hepatitis A viruses associated with food-handler transmission and sewage-contaminated foods. In the latter category, complex mixtures of viruses and other pathogens may be present in a single food item, creating potential for genetic recombination or reassortment and thus further expansion of the diversity of these pathogens. Therefore, bringing expertise and data together from veterinary, food, and clinical microbiology may help unravel these complexities and identify areas amenable to intervention and prevention.
Introduction
When it comes to food safety, most people would agree that food has become safer than ever. The potential for contamination with pathogenic bacteria, viruses, and parasites has been recognized and translated into control programs aimed at
_______________
15 Erasmus University.
reducing the burden of food-borne diseases in many parts of the world (Newell et al., 2010). Legislation exists to support countries in these control activities and to advise industries by developing guidelines targeting specific pathogens, commodities, or processes (Havelaar et al., 2004). Nevertheless, food-borne disease remains a significant cause of illness, of which the true burden is difficult to estimate (Scallan et al., 2011). The growing population density impacts upon the environment, for instance through sewage discharges, making it challenging to find clean waters for shellfish production in some parts of the world. Such environmental sources of contamination also may contain a mixture of human and animal pathogens, emphasizing the potential for introduction of animal pathogens into the food chain through routes that are not controlled (Figure A9-1). The increasing demand for seasonal produce year-round has globalised the food market, with the ensuing challenge to work with the same high hygienic standards across the world. While these production programs are largely successful, they also illustrate the vulnerability of the global food supply: if there is a flaw in the process, then contamination may occur with pathogens from across the globe, including those that have recently emerged (Newell et al., 2010). Therefore, thinking in terms of the future of food safety from a public health perspective does require a
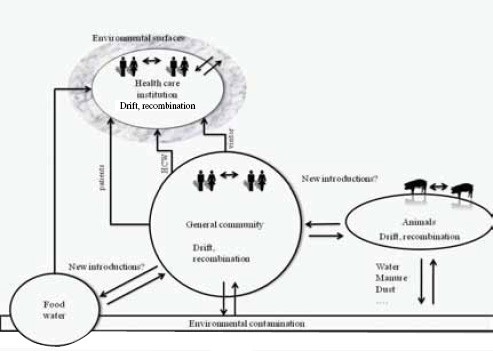
FIGURE A9-1 Epidemiology of food- and water-borne viruses, showing complexity of transmission and possible sources of infection. Which of the factors shown here apply may differ for different food-borne viruses.
holistic view, including the careful review of possible scenarios that may require our attention. Here, we focus on viral food-borne disease, reviewing the current state of knowledge with this forward-looking perspective. For detailed reviews of the state of the art, we refer to other recent publications (Baert et al., 2011; Iwamoto et al., 2010; Khuroo and Khuroo, 2008; Koopmans and Duizer, 2004; Strawn et al., 2011).
Most Common Viral Food Safety Concerns
Currently known viruses that can infect humans are grouped into 24 families. Food-borne transmission has been documented for viruses belonging to at least 10 of these, and the diseases associated with these infections range from mild diarrheal illness to severe encephalitis. The burden of food-borne illness is thought to be greatest for human viruses that are transmitted through poor hygienic practices, either by food handlers or during food production (Scallan et al., 2011). This applies to viruses that are transmitted by the faecal-oral route, hence infecting their host after ingestion, followed by invasion of cells in the epithelial lining of the gut, and subsequent replication in the same site or elsewhere in the body (Koopmans and Duizer, 2004).
Food-borne transmission can occur by contamination of food by infected food handlers, by contamination of food during the production process (e.g., in shellfish production), or more rarely by consumption of products of animal origin harboring a zoonotic virus. While intuitive, understanding these different potential sources is important because the disease ecology differs for these different sources of contamination. These differences are qualitative but, nevertheless, can help direct outbreak investigations.
Food Handler—Associated Illness
Food handler—associated food-borne illness results from the manual preparation of food by a food handler shedding viruses. The potential impact of such contamination events depends on the product type and preparation. There are numerous reports of food handler—associated viral outbreaks, usually resulting in limited outbreaks (Greig et al., 2007). Understandably, the most frequently identified viruses through this transmission route are highly prevalent. Priority concern in this category are noroviruses (NoVs) as the most common cause of gastroenteritis in all age groups, but outbreaks with several other enteric viruses are possible, particularly with hepatitis A (WHO, 2008). Contamination events are not limited to symptomatic persons, although there is no quantitative information about the relative contribution of symptomatic versus asymptomatic food handlers (Okabayashi et al., 2008; Todd et al., 2008). Food handling may occur throughout the food chain, but reported food handler—associated outbreaks often reflect contamination during the final food preparation or serving. This may be a
bias in surveillance, as end-of-the-chain food handler—associated outbreaks are easier to identify through regular outbreak investigations. Risk foods, therefore, are all foods that are handled manually and not further processed before consumption. Freezing is not sufficient to inactivate viral pathogens (Koopmans and Duizer, 2004).
Source Contamination
Food contamination at source occurs when food is contaminated during the primary production, as has been observed in particular in fresh produce such as berries and green onions, or bivalve filter-feeding shellfish. Here the nature of contamination may vary greatly, depending on location of the production area and nature of sewage contamination, but NoV and hepatitis A virus (HAV) were considered to be priority concerns in a coordinated expert meeting of the World Health Organization (WHO), the Food and Agriculture Organization (FAO), and the World Animal Health Organization (OIE) (WHO, 2008). In contrast with food handler—associated contamination, source contamination events may involve multiple pathogens that may be present in sewage, including animal viruses (Myrmel et al., 2006; Pommepuy et al., 2004; Costantini et al., 2006). This simultaneous exposure to mixtures of viruses theoretically increases the probability of recombination or reassortment of viral genomes when a person is simultaneously infected with multiple related viruses (Gallimore et al., 2005; Koopmans and Duizer, 2004; Le Guyader et al., 2006a; Symes et al., 2007). As with food handler—associated outbreaks, this mode of transmission typically involves the most common human viruses that are present in abundance in sewage (Iwai et al., 2009; Myrmel et al., 2006; Pommepuy et al., 2004; Shieh et al., 2003; Victoria et al., 2010; Wolf et al., 2010). However, treatment of sewage appears to selectively reduce levels of contamination with genogroup II NoV, possibly explaining the relatively high frequency of genogroup I viruses in sewage-related food-contamination events (van den Berg et al., 2005).
Zoonotic Food-Borne Viruses
Zoonotic food-borne infection occurs when meat, organs, or other products from an infected animal are consumed. For viruses, this is the least common mode of transmission, although the potential for such transmission is a cause for concern with every emerging disease outbreak. There is evidence that severe acute respiratory syndrome (SARS), monkeypox, and Nipah virus have been transmitted through food-related incidents (Leung et al., 2006; Luby et al., 2006; Rimoin et al., 2010; Wang et al., 2005a). However, more detailed review of these events suggests that it is more likely the process of food preparation (slaughter of the animal) that constitutes the greatest risk. For hepatitis E, there is documentation of food-borne infection through meat consumption.
In the WHO/FAO/OIE expert meeting, conclusions about priority food-commodity combinations of concern were based on available evidence from the literature, but it was also noted that large data gaps exist: trends in disease reporting are available in many parts of the world for hepatitis A, but not for the other viruses. Estimates of the proportion of illness caused by these pathogens that can be attributed to consumption of contaminated food are based on very few studies and would require the addition of systematic strain typing to routine surveillance, and more systematic studies to provide the data for burden estimates (Scallan et al., 2011; WHO, 2008). Finally, testing for viruses in commodities is difficult, and there is considerable debate over interpretation of findings from molecular assays, because these do not provide information on the viability of the pathogens detected (Baert et al., 2011). As a consequence, data from product monitoring are patchy at best.
Short Description of Common Food-borne Viruses
Norovirus
Virological aspects NoVs belong to the Family Caliciviridae, which is divided into genera. Norovirus and Sapovirus are the two out of five genera of the family Caliciviridae that contain viruses that cause infections in humans. NoVs have also been detected in pigs, cattle, mice, cats, dogs, and sheep, and sapoviruses in pigs (Han et al., 2004; Martella et al., 2007, 2008, 2011; Ntafis et al., 2010; Oliver et al., 2006; Smiley et al., 2003; Wang et al., 2005b, 2006; Wobus et al., 2006; Wolf et al., 2009). In humans, NoVs cause gastroenteritis, while the animal viruses can cause a range of different clinical syndromes, including oral lesions, systemic disease with hemorrhagic syndromes, upper respiratory tract infections, and others. Furthermore, one other potential genus comprising viruses detected in rhesus macaques has been described (Farkas et al., 2008). So far, the NoVs and sapoviruses are the only caliciviruses known to cause disease in humans, with the exception of anecdotal zoonotic infection with vesiviruses. NoVs can be divided into distinct genogroups, based on phylogenetic analyses of the capsid protein. To date, five norovirus genogroups (G) have been recognized (GI-GV) (Kroneman et al., 2011; Zheng et al., 2005). Viruses of GI, GII, and GIV are known to infect humans. GII viruses have additionally been detected in pigs, and GIV viruses have been detected in carnivores (a lion cub and a dog). GIII viruses infect cattle and sheep, and GV viruses infect mice. The host barrier is not absolute—a suggestion that there may be opportunity for genetic mixing if circumstances are favourable (Souza et al., 2007). Recombination between viruses from different genogroups is rare, suggesting that this constitutes a species level in taxonomy. Within each genogroup, viruses are further segregated into lineages, termed genotypes (Kroneman et al., 2011; Phan et al., 2007). Where known, these seem to have a global distribution, with little evidence for geographic clustering. Direct
comparison of data across countries is challenging because of differences in study design and laboratory diagnostics, resulting in poorly defined biases (Kroneman et al., 2008a, 2008b). This is particularly the case when trying to establish causes of food-borne illness. Here, the less common genotypes of norovirus are likely to play a bigger role, and it is these viruses that are less available for assay test validation studies (Duizer et al., 2007; Fisman et al., 2009; Gray et al., 2007). The development of quality assurance schemes for molecular diagnostics, therefore, is particularly important for detection of such highly diverse viruses.
Epidemiology The etiological importance of NoVs as causes of diarrheal illness has been documented worldwide, but few studies have been performed in a standardized way that allows international comparison and true burden of disease estimates (Hall et al., 2005; Scallan et al., 2011). Community studies have provided evidence for the abundance of NoVs and established that these viruses are the number one cause of community-acquired gastroenteritis, with one out of four or five persons infected per year (de Wit et al., 2001, 2003; Jansen et al., 2008; Kirkwood et al., 2005; Olesen et al., 2005; Patel et al., 2008; Tam et al., 2012; Tompkins et al., 1999; Wheeler et al., 1999). The burden of illness is highest in young children and the elderly (de Wit et al., 2001; Tompkins et al., 1999). The best described feature of NoVs is their propensity to cause outbreaks, resulting from some basic properties: the dose required for productive infection is very low (1-10 particles), and infected persons shed huge amounts of viruses (up to 1010 million per gram of stool) (Atmar et al., 2008; Teunis et al., 2008). In addition to this, the most common NoVs evolve through accumulation of mutations and selection of fitter variants that escape the receptor-blocking activities from antibodies triggered by prior infections (Lindesmith et al., 2008; Lochridge et al., 2005; Siebenga et al., 2007, 2010). In addition, the interaction of NoVs with histo-bloodgroup antigens determines the outcome of exposure, and strain-dependent differences in host susceptibility have been observed ( Donaldson et al., 2008; Marionneau et al., 2005; Rydell et al., 2011; Tan and Jiang, 2011). Although there is insufficient literature to substantiate this, the transmissibility is likely to differ between genotypes, and such differences may explain why relatively little diversity is seen in outbreak reporting, particularly when outbreaks notified include those in health care institutions: here, genogroup II.4 viruses are by far the most commonly identified outbreak strains (Kroneman et al., 2008; Sukhrie et al., 2011). In a study in hospitalized patients, the probability of secondary transmission of NoVs differed by age and genotype (Sukhrie et al., 2011). In recent years, the incidence of norovirus outbreaks has increased with the emergence of a particular variant (Lopman et al., 2004; Siebenga et al., 2010). More severe complications are seen in immunocompromised patients, and mortality in the elderly (Siebenga et al., 2008; van Asten et al., 2011; Westhoff et al., 2009).
Estimation of the burden of food-borne infection A challenging question, therefore, is how much disease caused by NoVs can be attributed to the different modes of transmission, in particular food-borne spread (Figure A9-1). One source of information comes from outbreak reporting, for instance the European Union (EU) Community Summary Report, the Centers for Disease Control and Prevention’s (CDC’s) FoodNet overviews, and the Australian FoodNet reports. These list NoVs as frequent causes of outbreaks (CDC, 2011; EFSA, 2010; Hall et al., 2005; OzFoodNet Working Group, 2009). In the EU, in 2008, crustaceans, shellfish, mollusks, and products thereof were the most frequently implicated food items in NoV and HAV outbreaks, but this may also reflect an ascertainment bias, because testing for the presence of viruses in shellfish is well established across Europe. The use of epidemiological criteria in the United States concluded that an estimated 28 percent of all reported outbreaks with unknown etiology were likely caused by NoVs (Turcios et al., 2006). An important caveat in using these data is that testing of patients with gastroenteritis for NoV is not yet an established routine, although this is rapidly changing (Tam et al., 2012). With that, numbers and proportion of reported viral outbreaks will most likely increase in the near future. In addition to the recognized food-borne outbreaks, the high rate of secondary infections in NoV outbreaks can rapidly mask an initial food-borne introduction. Therefore, a relevant question is what proportion of such outbreaks in fact were triggered by a food contamination event (Verhoef et al., 2010). What remains anecdotal is the geographic spread of most food-borne outbreaks, because this requires systematic incorporation of molecular typing into outbreak investigations and international data sharing to identify clusters (Koopmans et al., 2003). Therefore, the current reporting is likely to reflect the tip of the iceberg of true food-borne incidents. The available data also illustrates current challenges in using the notified outbreaks for action: only 5 percent of all reported NoV outbreaks are fully confirmed, reflecting the challenges of virus detection in or on food items (Kroneman et al., 2008a).
Given the paucity of evidence, few studies have attempted to quantify burden of food-borne illness attributable to viruses. In the Netherlands, approximately 12 to 15 percent of community cases of NoV gastroenteritis were attributed to food-borne transmission, based on risk factor analysis using questionnaire data. This makes NoV as common a cause of food-borne gastroenteritis as Campylobacter and more common than Salmonella (de Wit et al., 2003). A recent analysis of available data estimates that almost 60 percent of illness cases, 26 percent of hospitalizations, and 11 percent of deaths from food-borne illness are caused by NoV (Scallan et al., 2011). Similarly, an estimate based on data from Australia suggests that NoVs are important causes of food-borne illness (Hall et al., 2005).
In studies of outbreak reports, the term “food-borne” has been used loosely and has not been standardised. Also, the ultimate number of persons affected by a food-borne outbreak is rarely known, and reported outbreaks are likely to be biased (Kroneman et al., 2008b; Todd et al., 2008). The average size of reported
outbreaks is limited, but there are examples of widespread dissemination, for instance following consumption of wedding cake, sandwiches from an ill baker, deli meat during rafting trips down the Grand Canyon, frozen shellfish, or a manually prepared salad (de Wit et al., 2007; Friedman et al., 2005; Malek et al., 2009; Schmid et al., 2007; Webby et al., 2007). An interesting example was the simultaneous emergence of a new recombinant NoV in nine countries across Europe in 2001 (Ambert-Balay et al., 2005; Koopmans et al., 2003; Reuter et al., 2006). This variant was found in association with four different capsids until equilibrium was reached and the virus continued to circulate in combination with GII3 capsid. These viruses currently are the second most common cause of infection in children hospitalized with NoV (Beersma et al., 2009).
This example also raises the question of where to draw the line in terms of estimation burden of food-borne disease: could the widespread circulation of the GIIb strains have been prevented? Or is it only the first round of infections that should be attributed to food? While difficult to prove with certainty, these examples illustrate the contribution of food-borne introduction to the diversity of viruses circulating in the population, a situation that is not desirable from a virological standpoint: novel combinations of genes may have unpredictable effects on viral behavior and virulence and should be avoided when possible.
Hepatitis A (HAV)
Virology The hepatitis A virus belongs to the family Picornaviridae, genus Hepatovirus. Hepatoviruses have only been found in humans and primates, suggesting there is no risk of introduction from a reservoir. Based on genetic diversity, hepatitis A viruses are divided into six lineages or genotypes, of which genotypes I-III infect humans (Robertson et al., 1992). Genotypes I and II contain subgenotypes (Ia, Ib, IIa, and IIb). In regions with endemic HAV circulation, further segregation into geographically defined clusters is observed, a property that can be used to support source tracing activities in food-borne outbreaks (Costa-Mattioli et al., 2003; Robertson et al., 1992).
Epidemiology H AV is less transmissible than NoVs, and its incidence is greatly reduced in regions with proper sanitation and good hygienic conditions. As a consequence, great differences can be observed in the incidence of HAV in communities across the globe, related to socioeconomic status (Jacobsen and Wiersma, 2010; Mohd Hanafiah et al., 2011). These differences also affect the level of population immunity and, thus, the susceptibility to food-contamination events. In highly endemic regions, HAV is one of the childhood infections that, in the majority of cases, runs an asymptomatic course, while triggering a protective immune response that lasts long, possibly even lifelong (Hollinger and Emerson, 2007). In such regions, sustained circulation of HAV strains is found, resulting in geographically distinct genetic fingerprints (Barameechai et al., 2008;
Broman et al., 2010; Cao et al., 2011; Davidkin et al., 2007; Faber et al., 2009; Gharbi-Khelifi et al., 2006; Klevens et al., 2010; Kokkinos et al., 2010; Munné et al., 2007; Nejati et al., 2012; Pérez-Sautu et al., 2011; Sulbaran et al., 2010; Yun et al., 2008). Although this geographical diversity is not robustly defined, this information is used to support investigations into the possible source of an outbreak, or in defining where a patient most likely contracted the disease (Bialek et al., 2007; Petrignani et al., 2010; Shieh et al., 2007).
In regions with high socioeconomic status, HAV circulation is very limited and mostly restricted to risk groups such as men who have sex with men, to immigrant populations from regions with higher endemicity that may reintroduce viruses when infected during family visits in their country of origin, to travelers who contracted infection while visiting an endemic country and may transmit infection to nonimmune contacts, and to food- and water-borne infection. In such regions, population immunity builds up much slower, leading to an increase in the size of the susceptible population, and a right shift of first-time infections to higher age groups (Jacobsen and Wiersma, 2010). With increasing age, the probability of having symptomatic illness increases, and complications such as fulminant hepatitis are more common. This leads to the somewhat contrasting situation that food-contamination events may have a greater impact in regions with low endemicity of hepatitis A than in highly endemic regions (Greig et al., 2007; Koopmans and Duizer, 2004). This different epidemiological pattern also has consequences for the use of molecular typing in HAV source tracing; in low endemic regions, most people with HAV will have contracted the infection in a different region, and, as a consequence, a great diversity of HAV strains may be seen, reflecting the geographic fingerprints from the regions where they contracted the illness. This basic pattern can be greatly influenced by changing the population immune status through vaccination. Vaccination confers clinical protection that is thought to be long lasting (Van Damme et al., 2011). Whether vaccinated individuals contribute to shedding also is not well known.
Evidence for food-borne infection HAV is quite stable outside a host and, therefore, can persist on contaminated environments, food, and water. Food-and water-borne outbreaks have been documented, although again, as for NoVs, the most common mode of transmission occurs between persons (Bosch et al., 2001; Dentinger et al., 2001; Pinto et al., 2009; Sanchez et al., 2002). Because of the risk pattern described above, the biggest risk of food-borne HAV currently is introduction through food into regions where population immunity is relatively limited. Foods of primary importance, therefore, are those susceptible to contamination during the production phase, such as bivalve filter-feeding mol-lusks (oysters, clams, mussels) or produce that is irrigated with water that may be contaminated (e.g., lettuce, green onions, and soft fruits, such as raspberries and strawberries). An extreme example of the potential impact dates from 1988, when almost 300,000 cases were caused by consumption of clams harvested from
a sewage-polluted area (Halliday et al., 1991). A specific problem with shellfish is that the current microbiological quality control criteria are based on testing for bacterial contamination, which does not reliably predict the presence or absence of viruses. Also, mildly polluted products can be put on the market after “rinsing” the shellfish by storing them for a period of time in clean water in a process called depuration. Depurated shellfish have been associated with outbreaks of norovirus, hepatitis A, gastroenteritis, and other viral diseases (Ueki et al., 2007). For NoV, specific binding to histo-bloodgroup antigens in oyster tissues has been demonstrated, possibly further explaining the retention of viruses in these animals (LeGuyader et al., 2006b).
Estimation of the food-borne burden of illness In the CDC assessment of food-borne pathogens, hepatitis A is the second virus listed and is considered a significant cause of severe disease (Scallan et al., 2011). This may be related to the increased severity when HAV infection is first acquired during adulthood, although there also are differences in virulence between genotypes (Yoon et al., 2011).
Hepatitis E Virus (HEV)
Virology Hepatitis E viruses have been listed as genus Hepevirus in the family Hepeviridae in the database of the International Committee for Taxonomy of Viruses, along with the more distantly related avian hepatitis E viruses. The hepatitis E viruses can be grouped into four genotypes, with different geographical distribution and host range. Genotype 1 is endemic in Asia and Africa, and genotype 2 is endemic in Mexico and western Africa. Whereas these genotypes have been found exclusively in humans, genotypes 3 and 4 have also been detected in pigs and other animal species (e.g., wild boar and deer) (Lu et al., 2006; Teo, 2009). Genotype 3 is distributed worldwide, and genotype 4 is found commonly in Southeast Asia, although recent findings suggest these lineages also may be more widespread (Tessé et al., 2012). Nevertheless, current information suggests that the endemic strains found in pigs in Europe, Japan, and the United States are usually of genotype 3. In addition to the HEV genotypes 1 to 4, distinct HEV-like viruses with lower sequence identity to the strains found in humans have been detected in chicken, rats, and farmed rabbits in China (Huang et al., 2004; Johne et al., 2010; Zhao et al., 2009). In addition, serological data suggest the presence of HEV-related agents in cattle, horses, and some pet animals, but these remain to be confirmed by virological methods (Teo, 2009).
Epidemiology Historically, HEV has been considered to be endemic in developing countries, where genotype 1 and 2 HEV strains have been associated with large outbreaks of hepatitis, primarily in Asia and Africa. The most commonly
recognized mode of transmission in these outbreaks is water-borne, associated with poor-quality drinking water (Purcell and Emerson, 2001). Although HEV outbreaks are only observed in developing countries, antibodies have been found at lower prevalence levels globally, with estimates ranging from very low (around 1 percent) up to 33 percent. Some of these antibodies reflect exposures to genotypes 1 and 2 HEV in the recognized endemic regions through travel, but an increasing number of non-travel-related cases have been reported (Lewis et al., 2010). This follows the discovery of the presence of other lineages (genotypes 3 and 4) in farmed pigs across the world, with evidence for human infections with genotype 3 viruses in a wide geographic region 3 and for genotype 4 viruses in China, and recently in France (one case) (Liu et al., 2012; Tessé et al., 2012). The broader genetic diversity influences the use of existing commercial antibody tests that show large differences in baseline seroprevalence in populations where HEV genotype 3 strains are endemic in pigs, depending on the test used (Herremans et al., 2007). Therefore, type-specific validated methods are needed before robust conclusions can be drawn about the differences in population immunity across countries (Lewis et al., 2010). However, targeted studies suggest that HEV infections may be as common as HAV in some industrialized countries, although the risk profile of patients suggests that genotype 3 HEV is less virulent for humans because illness is mostly observed in persons with comorbidities (Borgen et al., 2008; Dalton et al., 2007; Fogeda et al., 2009; Wichmann et al., 2008). Men over 50 with comorbidities such as underlying chronic liver disease, liver cirrhosis, or a history of high alcohol consumption are at increased risk for symptomatic HEV. Chronic infections have been found in immunocompromised persons (Haagsma et al., 2008; Kamar et al., 2011).
Person-to-person transmission appears to be rare, but the exact mode of transmission of most HEV cases outside the previously recognized risk areas remains to be established. In addition to water-borne transmission, there is evidence for food-borne transmission, transmission by transfusion of blood products or organs, and maternofetal transmission (Aggarwal and Jameel, 2011).
Evidence for food-borne transmission As indicated above, the sources of most HEV infections remain unknown, but there is some evidence for food-borne transmission of genotype 3 HEV from undercooked wild boar and deer (Li et al., 2005; Tei et al., 2003). Epidemiological studies have provided evidence for consumption of undercooked or raw (wild) pork meat as risk factors for acquisition of HEV infection, but only very few systematic studies have been performed so far (Colson et al., 2010; Lewis et al., 2010; Wichmann et al., 2008).
Estimation of the food-borne burden of illness Currently, there is insufficient information to allow burden-of-illness estimates for food-borne HEV infection.
Detection of Food-borne Viral Disease: Specific Challenges
The detection of food-borne illness relies on a combination of laboratory diagnosis, epidemiological investigation, pathogen typing, and food traceback investigations. All of these activities need to be aligned for optimal detection, and the specific challenges differ for the different viruses discussed above (Figure A9-2).
Diagnosis and Genotyping of NoV, HAV, and HEV in Humans
For NoVs, the incidence in the community and the contribution of person-to-person spread dominate the picture (Figure A9-1). Testing of patients with diarrhea and vomiting for NoVs is not always routine because of the lack of low-cost rapid tests with adequate sensitivity and specificity, and in particular because it usually does not inform the decision making of the treating physician. For diarrheal disease outbreaks, norovirus testing is more common, and this has formed the basis of surveillance in most countries that have surveillance of food-borne viral disease in place. Again, however, the rapid secondary spread of NoVs leads to a bias for outbreaks with person-to-person transmission. More in-depth outbreak investigations that involve taking a detailed food consumption history are needed to identify those outbreaks related to food-borne introduction (Figure A9-2). Here, the use of genetic typing has shown to be informative: NoVs are a diverse genus, infecting humans and animals, and divided in lineages termed genotypes. Analysis of the aggregated data from outbreak reporting across Europe

SOURCE: Modified from http://www.cdc.gov/foodnet/surveillance_pages/burden_pyramid.htm.
has shown that the probability of a food- or water-borne source differs greatly between genotypes. Therefore, if outbreak investigations need to be triaged for lack of resources, genetic typing may be used to guide this decision making. Clearly, this is not ideal because food-related outbreaks also have been documented for the genotypes that spread most efficiently, hence dominating the reporting when outbreaks in health care settings are included.
For hepatitis A, diagnostic tests are part of the standard diagnostic repertoire; thus, underascertainment of the number of cases in vaccinated individuals is less of a problem than for hepatitis A. The challenge here, however, is the long incubation period, which may be between 15 and 50 days (CDC, 2008). Getting a reliable food consumption history this long after exposure is virtually impossible, unless an incident relates back to a specific event. Analysis of viral sequences may help identify the source of an outbreak (Bosch et al., 2001; Dentinger et al., 2001; Hutin et al., 1999; Sanchez et al., 2002; Shieh et al., 2007; Wheeler et al., 2005); systematic typing of outbreak strains has helped to identify clusters of patients related to food consumption that had not been recognized as such from the notifications, but this is done rarely (Petrignani et al., 2010).
For HEV, routine diagnostic evaluation of patients with acute hepatitis in regions with no known circulation of the human HEV genotypes (1 and 2) is rare, although the recent finding that genotype 3 HEV may cause chronic illness in immunocompromised individuals may change this practice. Therefore, HEV is likely to be largely underdiagnosed. Again, strain typing may be used to identify patient clusters, but this practice currently is limited to specific outbreak investigations and done in research settings.
Detection and Genotyping of NoV, HAV, and HEV in Food (Animals)
For all of the above viruses, there are great challenges in reliable detection in food products, a practice that is seen as an essential part of outbreak investigations (Gentry et al., 2009b; Le Guyader et al., 2008a, 2008b; Li et al., 2011; Rutjes et al., 2006). Recent publications have shown a high prevalence of viral genes on fresh produce, questioning the relevance of such findings as they do not reflect infectious articles (Baert et al., 2011; Stals et al., 2011). A practical problem is that there are no cell culture methods available for noroviruses (Duizer et al., 2004). An elegant study in Europe suggests a correlation between quantities of viral RNA in shellfish and illness in consumers, providing a possible basis for regulatory action (Lowther et al., 2010). Levels of virus contamination, however, vary greatly across production sites, typically reflecting population densities and the ensuing environmental impact from sewage contamination, particularly follow ing heavy rainfall (Boxman et al., 2006; Elamri et al., 2006; Gentry et al., 2009a; Groci et al., 2007; Le Guyader et al., 2008b; Lowther et al., 2010; Myrmel et al., 2004; 2006; Nishida et al., 2007; Nordgren et al., 2009; Pommepuy et al., 2004; Shieh et al., 2003; Suffredini et al., 2008).
Linking Epidemiological and Virological Data for Source Tracing and Attribution
In order to gain a better understanding of the trends in enteric viruses and the possible role of food-borne transmission, the Foodborne Viruses in Europe network was launched in 1999. Participating epidemiologists and virologists from academia, and clinical and public health laboratories, covering medical and food virology agreed to compile data related to outbreaks into a joint database. Since the launch of this network, data have been compiled for more than 8,000 outbreaks involving 13 countries, and some important importations were made. First of all, it became clear that the proportion of food-borne outbreaks reported differed greatly, reflecting differences in the surveillance setup of each county (Koopmans et al., 2003). This background also influenced the diversity of outbreak strains, with limited diversity and strong seasonal effect seen in healthcare—associated outbreaks and greater diversity with limited seasonality in outbreaks reported as food-related (Kroneman et al., 2008). For the common strains for which this was investigated, the strain diversity observed was very similar in different countries, showing that the epidemiology of these viruses is shaped by the global interlinked circulation of pathogens, with little evidence for geographic differences (Lopman et al., 2004; Siebenga et al., 2007, 2009, 2010; Verhoef et al., 2008). Food-borne outbreaks were rarely reported, but their number increased by almost 20-fold when genome sequencing was used to identify linked outbreaks (Verhoef et al., 2010, 2011). The analysis required the availability of both epidemiological and laboratory data, and it included approaches aiming to determine robustness of conclusions drawn, based on choice of target genes and fragment lengths. This was done because international standardization of molecular detection and genetic typing methods across clinical, public health, and food laboratories is very difficult because of the differences in focus and required levels of resolution at each level. In particular the virus detection in food requires such low detection limits that optimal target choice is a luxury that cannot be afforded. By using multiple genome targets to study food-borne NoV outbreaks, multiple recombinant genomes have been identified (Ambert-Balay et al., 2005; Bon et al., 2005; Reuter et al., 2006; Le Guyader et al., 2006). In food-related outbreaks where sewage contamination was the most likely cause, multiple viruses can be found within the same batch, thus favoring conditions for generation of recombinant genomes (Symes et al., 2007).
Emerging Viruses and Food-borne Transmission
Globalization and Risk of Introduction of New Diseases
With changing consumer behavior and the growing preference for consumption of fresh produce with year-round availability, food has become a commodity in the global market, dictated by availability and (low) cost. Seemingly unrelated events can lead to market shifts and, with that, to potential introduction of new
risks into the food chain. A recent example is the emergence of a highly lethal infection affecting a high proportion of oysters in European banks (Peeler et al., 2012). Although not documented, the lack of locally grown oysters may move the market to Southeast Asia, which has the fastest growth in the market of aquaculture products. Assuming that failures in the production system may occur, as evidenced from the NoV studies, such incidents would potentially lead to contamination of products with locally circulating strains, such as the distinct lineages of enterovirus 71 viruses causing large outbreaks of hand, foot, and mouth disease in that region only (van der Sanden et al., 2009). Even if this is not the prevailing way of spreading, dissemination of viruses via international food trade could disperse an otherwise localized outbreak. This concern has led to in-depth investigations during the emergence of SARS, highly pathogenic avian influenza, filoviruses in pigs in the Philippines, and Nipah virus outbreaks in Malaysia and Bangladesh (Leung et al., 2003; McKinney et al., 2006; Miranda and Miranda, 2011; Parashar et al., 2000). For all of these viruses, there is evidence of introduction of the viruses into the human population through the harvesting, preparation, and/or consumption of food. For all of these examples, the biggest concern is not widespread food-borne transmission, but the fact that this mode of transmission may favor cross-species infections that are not evident otherwise, with the potential for adaptation of these viruses to humans. A systematic review of emerging infectious disease outbreaks suggested that 76 percent of these resulted from zoonotic introductions, and the pressure on the environment from population growth is increasing the contact rates between humans and animals in biotopes that were previously untouched, attesting to the opportunity for cross-species transmissions (Jones et al., 2008). Consumption of virus-containing food, either through bush meat or food contaminated with excreta from animals, is one of the potential routes (Costantini et al., 2006).
Food Safety and the Era of Virus Discovery
The classical toolbox for virology was greatly expanded when sequencing-based technologies entered the playing field, and with this it also became clear that viruses are among the most prevalent entities in the world. Unbiased sequencing has established that a large proportion of ocean waters contain viral sequences, many of them unknown (Breitbart et al., 2004; Rosario et al., 2009). Based on these studies, an estimated 104 genotypes per kilogram of sediment have been identified, and the current view is that viral communities are powerful manipulators of microbial diversity, biochemistry, and evolution in the marine environment. Similarly, samples collected from humans when subjected to unbiased analysis of the gene content contain high quantities of viral information, with a dominance of plant and bacterial viruses, but also typically multiple human viruses (Breitbart et al., 2003). These findings are opening an entirely new field of research in host/microbiome and pathogen interaction that is likely to fundamen-
tally change how we view infectious diseases. Sequence-based virus discovery programs identify new viruses in humans and (wild) animals with high frequency (Allander et al., 2005). While most of these newly discovered viruses likely have been present for a long time, these observations do underscore the notion that there is ample potential for new human pathogens. There is consensus among virologists that the probability of the emergence of new viruses or the evolution of old viruses into new forms is inevitable, given the demographic, economical, and sociological changes that we are now facing. Therefore, having mechanisms in place to rapidly address the probability and possible consequences of food-borne transmission of a new infectious disease when it emerges should be a priority.
Another consequence is a revision of how we view the detection of viruses in food or clinical samples (Nakamura et al., 2009; Svraka et al., 2010). As the methods develop further, more diversity of viruses (and microbes) are found in any of the samples that have been tested, calling for the challenging task to answer what these findings signify. This is no different in clinical virology, where applications of multiplex polymerase chain reaction—based methods or deep sequencing increasingly find complex mixtures of potential pathogens in patients that are tested. This makes it difficult to decide which one or which combination of these was the cause of the symptoms. Methods will be needed to filter the data for relevance for the question addressed.
Conclusion
Food-borne transmission is common but largely underdiagnosed. While viruses from at least 10 families have been associated with food-borne transmission, NoV and HAV have been listed as priority concerns. By genomic epidemiology approaches, significant food-related disease associated with food handler transmission and sewage-contaminated foods has been identified for these viruses. In the latter category, complex mixtures of human and animal viruses and other pathogens may be present in a single food item, creating the potential for genetic recombination or reassortment and, thus, further expansion of the diversity of these pathogens. Bringing expertise together from veterinary, food, and clinical microbiology may help unravel these complexities and identify areas amenable to intervention and prevention.
References
Aggarwal, R., and S. Jameel. 2011. Hepatitis E. Hepatology 54(6):2218-2226, doi: 10.1002/hep.24674.
Allander, T., M. T. Tammi, M. Eriksson, A. Bjerkner, A. Tiveljung-Lindell, and B. Andersson. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proceedings of the National Academy of Sciences of the United States of America 102:12891-12896.
Ambert-Balay, K., F. Bon, F. Le Guyader, P. Pothier, and E. Kohli. 2005. Characterization of new recombinant noroviruses. Journal of Clinical Microbiology 43(10):5179-5186.
Atmar, R. L., A. R. Opekun, M. A. Gilger, M. K. Estes, S. E. Crawford, F. H. Neill, and D. Y. Graham. 2008. Norwalk virus shedding after experimental human infection. Emerging Infectious Diseases 14(10):1553-1557.
Baert, L., K. Mattison, F. Loisy-Hamon, J. Harlow, A. Martyres, B. Lebeau, A. Stals, E. Van Coillie, L. Herman, and M. Uyttendaele. 2011. Review: Norovirus prevalence in Belgian, Canadian and French fresh produce: A threat to human health? International Journal of Food Microbiology 151(3):261-269.
Barameechai, K., P. Sa-Nguanmoo, K. Suwannakarn, C. Thongmee, S. Payungporn, V. Chongsrisawat, A. Theamboonlers, and Y. Poovorawan. 2008. Molecular characterisation of the hepatitis A virus circulating in the 2001-2005 outbreaks in Thailand. Annals of Tropical Medicine and Parasitology 102(3):247-257.
Beersma, M. F., M. Schutten, H. Vennema, N. G. Hartwig, T. H. Mes, A. D. Osterhaus, G. J. van Doornum, and M. Koopmans. 2009. Norovirus in a Dutch tertiary care hospital (2002-2007): Frequent nosocomial transmission and dominance of GIIb strains in young children. Journal of Hospital Infection 71(3):199-205.
Bialek, S. R., P. A. George, G. L. Xia, M. B. Glatzer, M. L. Motes, J. E. Veazey, R. M. Hammond, T. Jones, Y. C. Shieh, J. Wamnes, G. Vaughan, Y. Khudyakov, and A. E. Fiore. 2007. Use of molecular epidemiology to confirm a multistate outbreak of hepatitis A caused by consumption of oysters. Clinical Infectious Diseases 44(6):838-840.
Bon, F., K. Ambert-Balay, H. Giraudon, J. Kaplon, S. Le Guyader, M. Pommepuy, A. Gallay, V. Vaillant, H. de Valk, R. Chikhi-Brache, A. Flahaut, P. Pothier, and E. Kohli. 2005. Molecular epidemiology of caliciviruses detected in sporadic and outbreak casesof gastroenteritis in France from December 1998 to February 2004. Journal of Clinical Microbiology 43(9):4659-4664.
Borgen, K., T. Herremans, E. Duizer, H. Vennema, S. Rutjes, A. Bosman, A. M. de Roda Husman, and M. Koopmans. 2008. Non-travel related Hepatitis E virus genotype 3 infections in the Netherlands: A case series 2004-2006. BMC Infectious Diseases 8:61.
Bosch, A., G. Sánchez, F. Le Guyader, H. Vanaclocha, L. Haugarreau, and R. M. Pintó. 2001. Human enteric viruses in Coquina clams associated with a large hepatitis A outbreak. Water Science and Technology 43:61-65.
Boxman, I. L., J. J. Tilburg, N. A. Te Loeke, H. Vennema, K. Jonker, E. de Boer, and M. Koopmans. 2006. Detection of noroviruses in shellfish in the Netherlands. International Journal of Food Microbiology 108(3):391-396.
Breitbart, M., I. Hewson, B. Felts, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2003. Metagenomic analyses of an uncultured viral community from human feces. Journal of Bacteriology 185(20):6220-6223.
Breitbart, M., B. Felts, S. Kelley, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2004. Diversity and population structure of a near-shore marine-sediment viral community. Proceedings of the Royal Society B: Biological Sciences 271(1539):565-574.
Broman, M., S. Jokinen, M. Kuusi, M. Lappalainen, M. Roivainen, K. Liitsola, and I. Davidkin. 2010. Epidemiology of hepatitis A in Finland in 1990-2007. Journal of Medical Virology 82(6):934-941.
Cao, J., S. Bi, Q. Meng, L. Shen, H. Zheng, and Y. Zhang. 2011. Genotyping of acute hepatitis A virus isolates from China, 2003-2008. Journal of Medical Virology 83(7):1134-1141, doi: 10.1002/jmv.22086.
CDC (Centers for Disease Control and Prevention). 2008. Hepatitis A FAQs for health professionals. http://www.cdc.gov/hepatitis/HAV/HAVfaq.htm (accessed April 2, 2012).
———. 2011. Surveillance for foodborne disease outbreaks—United States, 2008. Morbidity and Mortality Weekly Report 60(35):1197-1202.
Colson, P., P. Borentain, B. Queyriaux, M. Kaba, V. Moal, P. Gallian, L. Heyries, D. Raoult, and R. Gerolami. 2010. Pig liver sausage as a source of hepatitis E virus transmission to humans. Journal of Infectious Diseases 202(6):825-834.
Costa-Mattioli, M., A. Di Napoli, V. Ferré, S. Billaudel, R. Perez-Bercoff, and J. Cristina. 2003. Genetic variability of hepatitis A virus. Journal of General Virology 84(Pt. 12):3191-3201.
Costantini, V., F. Loisy, L. Joens, F. S. Le Guyader, and L. J. Saif. 2006. Human and animal enteric caliciviruses in oysters from different coastal regions of the United States. Applied and Environmental Microbiology 72(3):1800-1809.
Dalton, H. R., P. H. Thurairajah, H. J. Fellows, H. S. Hussaini, J. Mitchel, R. Bendall, M. Banks, S. Ijaz, C. G. Teo, and D. F. Levine. 2007. Autochthonous hepatitis E in southwest England. Journal of Viral Hepatitis 14(5):304-309.
Davidkin, I., N. Zheleznova, S. Jokinen, O. Gorchakova, M. Broman, and S. Mukomolov. 2007. Molecular epidemiology of hepatitis A in St. Petersburg, Russia, 1997-2003. Journal of Medical Virology 79(6):657-662.
de Wit, M. A., M. P. Koopmans, L. M. Kortbeek, W. J. Wannet, J. Vinjé, F. van Leusden, A. I. Bartelds, and Y. T. van Duynhoven. 2001. Sensor, a population-based cohort study on gastroenteritis in the Netherlands: Incidence and etiology. American Journal of Epidemiology 154(7):666-674.
de Wit, M. A., M. P. Koopmans, and Y. T. van Duynhoven. 2003. Risk factors for norovirus, Sapporolike virus, and group A rotavirus gastroenteritis. Emerging Infectious Diseases 9(12):1563-1570.
de Wit, M. A., M. A. Widdowson, H. Vennema, E. de Bruin, T. Fernandes, and M. Koopmans. 2007. Large outbreak of norovirus: The baker who should have known better. Journal of Infection 55(2):188-193.
Dentinger, C. M., W. A. Bower, O. V. Nainan, S. M. Cotter, G. Myers, L. M. Dubusky, S. Fowler, E. D. Salehi, and B. P. Bell. 2001. An outbreak of hepatitis A associated with green onions. Journal of Infectious Diseases 183:1273-1276.
Donaldson, E. F., L. C. Lindesmith, A. D. Lobue, and R. S. Baric. 2008. Norovirus pathogenesis: Mechanisms of persistence and immune evasion in human populations. Immunological Reviews 225:190-211.
Duizer, E., K. J. Schwab, F. H. Neill, R. L. Atmar, M. P. Koopmans, and M. K. Estes. 2004. Laboratory efforts to cultivate noroviruses. Journal of General Virology 85(Pt. 1):79-87.
Duizer, E., A. Pielaat, H. Vennema, A. Kroneman, and M. Koopmans. 2007. Probabilities in norovirus outbreak diagnosis. Journal of Clinical Virology 40(1):38-42.
EFSA (European Food Safety Authority). 2010. The Community Summary Report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in the European Union in 2008. EFSA Journal 1496.
Elamri, D. E., M. Aouni, S. Parnaudeau, and F. S. Le Guyader. 2006. Detection of human enteric viruses in shellfish collected in Tunisia. Letters in Applied Microbiology 43(4):399-404.
Faber, M. S., K. Stark, S. C. Behnke, E. Schreier, and C. Frank. 2009. Epidemiology of hepatitis A virus infections, Germany, 2007-2008. Emerging Infectious Diseases 15(11):1760-1768.
Farkas, T., K. Sestak, C. Wei, and X. Jiang. 2008. Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. Journal of Virology 82(11):5408-5416.
Fisman, D. N., A. L. Greer, G. Brouhanski, and S. J. Drews. 2009. Of gastro and the gold standard: Evaluation and policy implications of norovirus test performance for outbreak detection. Journal of Translational Medicine 7:23.
Fogeda, M., A. Avellón, C. G. Cilla, and J. M. Echevarría. 2009. Imported and autochthonous hepatitis E virus strains in Spain. Journal of Medical Virology 81(10):1743-1749.
Friedman, D. S., D. Heisey-Grove, F. Argyros, E. Berl, J. Nsubuga, T. Stiles, J. Fontana, R. S. Beard, S. Monroe, M. E. McGrath, H. Sutherby, R. C. Dicker, A. DeMaria, and B. T. Matyas. 2005. An outbreak of norovirus gastroenteritis associated with wedding cakes. Epidemiology & Infection 133(6):1057-1063.
Gallimore, C. I., J. S. Cheesbrough, K. Lamden, C. Bingham, and J. J. Gray. 2005. Multiple norovirus genotypes characterised from an oyster-associated outbreak of gastroenteritis. International Journal of Food Microbiology 103(3):323-330.
Gentry, J., J. Vinjé, D. Guadagnoli, and E. K. Lipp. 2009a. Norovirus distribution within an estuarine environment. Applied and Environmental Microbiology 75(17):5474-5480.
Gentry, J., J. Vinjé, and E. K. Lipp. 2009b. A rapid and efficient method for quantitation of genogroups I and II norovirus from oysters and application in other complex environmental samples. Journal of Virological Methods 156(1-2):59-65.
Gharbi-Khelifi, H., V. Ferre, K. Sdiri, M. Berthome, L. Fki, R. Harrath, S. Billaudel, and M. Aouni. 2006. Hepatitis A in Tunisia: Phylogenetic analysis of hepatitis A virus from 2001 to 2004. Journal of Virological Methods 138(1-2):109-116.
Gray, J. J., E. Kohli, F. M. Ruggeri, H. Vennema, A. Sánchez-Fauquier, E, Schreier, C. I. Gallimore, M. Iturriza-Gomara, H. Giraudon, P. Pothier, I. Di Bartolo, N. Inglese, E. de Bruin, B. van der Veer, S. Moreno, V. Montero, M. C. de Llano, M. Höhne, and S. M. Diedrich. 2007. European multicenter evaluation of commercial enzyme immunoassays for detecting norovirus antigen in fecal samples. Clinical and Vaccine Immunology 14(10):1349-1355.
Greig, J. D., E. C. Todd, C. A. Bartleson, and B. S. Michaels. 2007. Outbreaks where food workers have been implicated in the spread of foodborne disease. Part 1. Description of the problem, methods, and agents involved. Journal of Food Protection 70(7):1752-1761.
Groci, L., M. N. Losio, E. Suffredini, E. Pavoni, S. Di Pasquale, F. Fallacara, and G. Arcangeli. 2007. Assessment of human enteric viruses in shellfish from the northern Adriatic sea. International Journal of Food Microbiology 114(2):252-257.
Haagsma, E. B., A. P. van den Berg, R. J. Porte, C. A. Benne, H. Vennema, J. H. Reimerink, and M. P. Koopmans. 2008. Chronic hepatitis E virus infection in liver transplant recipients. Liver Transplantation 14(4):547-553.
Hall, G., M. D. Kirk, N. Becker, J. E. Gregory, L. Unicomb, G. Millard, R. Stafford, and K. Lalor; OzFoodNet Working Group. 2005. Estimating foodborne gastroenteritis, Australia. Emerging Infectious Diseases 11(8):1257-1264.
Halliday, M. L., L.-Y. Kang, T.-Z. Zhou, M.-D. Hu, Q.-C. Pan, T.-Y. Fu, Y.-S. Huang, and S.-L Hu. 1991. An epidemic of hepatitis A attributable to the ingestion of raw clams in Shanghai, China. Journal of Infectious Diseases 164:852-859.
Han, M. G., J. R. Smiley, C. Thomas, and L. J. Saif. 2004. Genetic recombination between two genotypes of genogroup III bovine noroviruses (BoNVs) and capsid sequence diversity among BoNVs and Nebraska-like bovine enteric caliciviruses. Journal of Clinical Microbiology 42(11):5214-5224.
Havelaar, A. H., M. J. Nauta, and J. T. Jansen. 2004. Fine-tuning food safety objectives and risk assessment. International Journal of Food Microbiology 93(1):11-29.
Herremans, M., J. Bakker, E. Duizer, H. Vennema, and M. P. Koopmans. 2007. Use of serologi-cal assays for diagnosis of hepatitis E virus genotype 1 and 3 infections in a setting of low endemicity. Clinical and Vaccine Immunology 14(5):562-568.
Hollinger, F. B., and S. U. Emerson. 2007. Hepatitis A virus. In Fields virology, edited by D. M. Knipe and P. M. Howley. Philadelphia: Lippincott Williams and Wilkins. Pp. 911-947.
Huang, F. F., Z. F. Sun, S. U. Emerson, R. H. Purcell, H. L. Shivaprasad, F. W. Pierson, T. E. Toth, and X. J. Meng. 2004. Determination and analysis of the complete genomic sequence of avian hepatitis E virus (avian HEV) and attempts to infect rhesus monkeys with avian HEV. Journal of General Virology 85(Pt. 6):1609-1618.
Hutin, Y. J. F., V. Pool, E. H. Cramer, O. V. Nainan, J. Weth, I. T. Williams, S. T. Goldstein, K. F. Gensheimer, B. P. Pell, C. N. Shapiro, M. J. Alter, and H. S. Margolis. 1999. A multistate, food-borne outbreak of hepatitis A. New England Journal of Medicine 340(8):595-602.
Iwai, M., S. Hasegawa, M. Obara, K. Nakamura, E. Horimoto, T. Takizawa, T. Kurata, S. Sogen, and K. Shiraki. 2009. Continuous presence of noroviruses and sapoviruses in raw sewage reflects infections among inhabitants of Toyama, Japan (2006 to 2008). Applied and Environmental Microbiology 75(5):1264-1270.
Iwamoto, M., T. Ayers, B. E. Mahon, and D. L. Swerdlow. 2010. Epidemiology of seafood-associated infections in the United States. Clinical Microbiology Reviews 23(2):399-411.
Jacobsen, K. H., and S. T. Wiersma. 2010. Hepatitis A virus seroprevalence by age and world region, 1990 and 2005. Vaccine 28(41):6653-6657.
Jansen, A., K. Stark, J. Kunkel, E. Schreier, R. Ignatius, O. Liesenfeld, D. Werber, U. B. Göbel, M. Zeitz, and T. Schneider. 2008. Aetiology of community-acquired, acute gastroenteritis in hospitalised adults: A prospective cohort study. BMC Infectious Diseases 8:143.
Johne, R., A. Plenge-Bönig, M. Hess, R. G. Ulrich, J. Reetz, and A. Schielke. 2010. Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. Journal of General Virology 91:750-758.
Jones, K. E., N. G. Patel, M. A. Levy, A. Storeygard, D. Balk, J. L. Gittleman, and P. Daszak. 2008. Global trends in emerging infectious diseases. Nature 451(7181):990-993.
Kamar, N., C. Garrouste, E. B. Haagsma, V. Garrigue, S. Pischke, C. Chauvet, J. Dumortier, A. Cannesson, E. Cassuto-Viguier, E. Thervet, F. Conti, P. Lebray, H. R. Dalton, R. Santella, N. Kanaan, M. Essig, C. Mousson, S. Radenne, A. M. Roque-Afonso, J. Izopet, and L. Rostaing. 2011. Factors associated with chronic hepatitis in patients with hepatitis E virus infection who have received solid organ transplants. Gastroenterology 140(5):1481-1489.
Khuroo, M. S., and M. S. Khuroo. 2008. Hepatitis E virus. Current Opinion in Infectious Diseases 21(5):539-543.
Kirkwood, C. D., R. Clark, N. Bogdanovic-Sakran, and R. F. Bishop. 2005. A 5-year study of the prevalence and genetic diversity of human caliciviruses associated with sporadic cases of acute gastroenteritis in young children admitted to hospital in Melbourne, Australia (1998-2002). Journal of Medical Virology 77(1):96-101.
Klevens, R. M., J. T. Miller, K. Iqbal, A. Thomas, E. M. Rizzo, H. Hanson, K. Sweet, Q. Phan, A. Cronquist, Y. Khudyakov, G. L. Xia, and P. Spradling. 2010. The evolving epidemiology of hepatitis a in the United States: Incidence and molecular epidemiology from population-based surveillance, 2005-2007. Archives of Internal Medicine 170(20):1811-1818.
Kokkinos, P., P. Ziros, S. Filippidou, I. Mpampounakis, and A. Vantarakis. 2010. Molecular characterization of hepatitis A virus isolates from environmental and clinical samples in Greece. Virology Journal 7:235.
Koopmans, M., and E. Duizer. 2004. Foodborne viruses: An emerging problem. International Journal of Food Microbiology 90:23-41.
Koopmans, M., H. Vennema, H. Heersma, E. van Strien, Y. van Duynhoven, D. Brown, M. Reacher, and B. Lopman. 2003. Early identification of common-source foodborne virus outbreaks in Europe. Emerging Infectious Diseases 9:1136-1142.
Kroneman, A., L. Verhoef, J. Harris, H. Vennema, E. Duizer, Y. van Duynhoven, J. Gray, M. Iturriza, B. Böttiger, G. Falkenhorst, C. Johnsen, C. H. von Bonsdorff, L. Maunula, M. Kuusi, P. Pothier, A. Gallay, E. Schreier, M. Höhne, J. Koch, G. Szücs, G. Reuter, K. Krisztalovics, M. Lynch, P. McKeown, B. Foley, S. Coughlan, F. M. Ruggeri, I. Di Bartolo, K. Vainio, E. Isakbaeva, M. Poljsak-Prijatelj, A. H. Grom, J. Z. Mijovski, A. Bosch, J. Buesa, A. S. Fauquier, G. Hernandéz-Pezzi, K. O. Hedlund, and M. Koopmans. 2008a. Analysis of integrated virological and epidemiological reports of norovirus outbreaks collected within the Foodborne Viruses in Europe network from 1 July 2001 to 30 June 2006. Journal of Clinical Microbiology 46(9):2959-2965.
Kroneman, A., J. Harris, H. Vennema, E. Duizer, Y. van Duynhoven, J. Gray, M. Iturriza, B. Böttiger, G. Falkenhorst, C. Johnsen, C. H. von Bonsdorff, L. Maunula, M. Kuusi, P. Pothier, A. Gallay, E. Schreier, J. Koch, G. Szücs, G. Reuter, K. Krisztalovics, M. Lynch, P. McKeown, B. Foley, S. Coughlan, F. M. Ruggeri, I. Di Bartolo, K. Vainio, E. Isakbaeva, M. Poljsak-Prijatelj, A. H. Grom, A. Bosch, J. Buesa, A. S. Fauquier, G. Hernandéz-Pezzi, K. O. Hedlund, and M. Koopmans. 2008b. Data quality of 5 years of central norovirus outbreak reporting in the European Network for food-borne viruses. Journal of Public Health 30(1):82-90.
Kroneman, A., H. Vennema, K. Deforche, H. v d Avoort, S. Peñaranda, M. S. Oberste, J. Vinjé, and M. Koopmans. 2011. An automated genotyping tool for enteroviruses and noroviruses. Journal of Clinical Virology 51(2):121-125.
Le Guyader, F. S., F. Bon, D. DeMedici, S. Parnaudeau, A. Bertone, S. Crudeli, A. Doyle, M. Zidane, E. Suffredini, E. Kohli, F. Maddalo, M. Monini, A. Gallay, M. Pommepuy, P. Pothier, and F. M. Ruggeri. 2006a. Detection of multiple noroviruses associated with an international gastroenteritis outbreak linked to oyster consumption. Journal of Clinical Microbiology 44:3878-3882.
Le Guyader, F., F. Loisy, R. L. Atmar, A. M. Hutson, M. K. Estes, N. Ruvoën-Clouet, M. Pommepuy, and J. Le Pendu. 2006b. Norwalk virus-specific binding to oyster digestive tissues. Emerging Infectious Diseases 12(6):931-936.
Le Guyader, F. S., S. Parnaudeau, J. Schaeffer, A. Bosch, F. Loisy, M. Pommepuy, and R. L. Atmar. 2008a. Detection and quantification of noroviruses in shellfish. Applied and Environmental Microbiology 75(3):618-624.
Le Guyader, F. S., J. C. Le Saux, K. Ambert-Balay, J. Krol, O. Serais, S. Parnaudeau, H. Giraudon, G. Delmas, M. Pommepuy, P. Pothier, and R. L. Atmar. 2008b. Aichi virus, norovirus, astrovirus, enterovirus, and rotavirus involved in clinical cases from a French oyster-related gastroenteritis outbreak. Journal of Clinical Microbiology 46(12):4011-4017.
Leung, G. M., W. W. Lim, L. M. Ho, T. H. Lam, A. C. Ghani, C. A. Donnelly, C. Fraser, S. Riley, N. M. Ferguson, R. M. Anderson, and A. J. Hedley. 2006. Seroprevalence of IgG antibodies to SARS-coronavirus in asymptomatic or subclinical population groups. Epidemiology & Infection 134(2):211-221.
Leung, W. K., K. F. To, P. K. Chan, H. L. Chan, A. K. Wu, N. Lee, K. Y. Yuen, and J. J. Sung. 2003. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology 125:1011-1017.
Lewis, H. C., O. Wichmann, and E. Duizer. 2010. Transmission routes and risk factors for autochthonous hepatitis E virus infection in Europe: A systematic review. Epidemiology & Infection 138(2):145-166.
Li, D., L. Baert, E. Van Coillie, and M. Uyttendaele. 2011. Critical studies on binding-based RT-PCR detection of infectious noroviruses. Journal of Virological Methods 177(2):153-159.
Li, T. C., K. Chijiwa, N. Sera, T. Ishibashi, Y. Etoh, Y. Shinohara, Y. Kurata, M. Ishida, S. Sakamoto, N. Takeda, and T. Miyamura. 2005. Hepatitis E virus transmission from wild boar meat. Emerging Infectious Diseases 11:1958-1960.
Lindesmith, L. C., E. F. Donaldson, A. D. Lobue, J. L. Cannon, D. P. Zheng, J. Vinje, and R. S. Baric. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Medicine 5(2):e31.
Liu, P., L. Li, L. Wang, Q. Bu, H. Fu, J. Han, Y. Zhu, F. Lu, and H. Zhuang. 2012. Phylogenetic analysis of 626 hepatitis E virus (HEV) isolates from humans and animals in China (1986-2011) showing genotype diversity and zoonotic transmission. Infection, Genetics and Evolution 12(2):428-434.
Lochridge, V. P., K. L. Jutila, J. W. Graff, and M. E. Hardy. 2005. Epitopes in the P2 domain of noro-virus VP1 recognized by monoclonal antibodies that block cell interactions. Journal of General Virology 86(Pt. 10):2799-2806.
Lopman, B., H. Vennema, E. Kohli, P. Pothier, A. Sanchez, A. Negredo, J. Buesa, E. Schreier, M. Reacher, D. Brown, J. Gray, M. Iturriza, C. Gallimore, B. Bottiger, K. O. Hedlund, M. Torvén, C. H. von Bonsdorff, L. Maunula, M. Poljsak-Prijatelj, J. Zimsek, G. Reuter, G. Szücs, B. Melegh, L. Svennson, Y. van Duijnhoven, and M. Koopmans. 2004. Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet 363(9410):682-688.
Lowther, J. A., J. M. Avant, K. Gizynski, R. E. Rangdale, and D. N. Lees. 2010. Comparison between quantitative real-time reverse transcription PCR results for norovirus in oysters and self-reported gastroenteric illness in restaurant customers. Journal of Food Protection 73(2):305-311.
Lu, L., C. Li, and C. H. Hagedorn. 2006. Phylogenetic analysis of global hepatitis E virus sequences: Genetic diversity, subtypes and zoonosis. Reviews in Medical Virology 16(1):5-36.
Luby, S. P., M. Rahman, M. J. Hossain, L. S. Blum, M. M. Husain, E. Gurley, R. Khan, B. N. Ahmed, S. Rahman, N. Nahar, E. Kenah, J. A. Comer, and T. G. Ksiazek. 2006. Foodborne transmission of Nipah virus, Bangladesh. Emerging Infectious Diseases 12(12):1888-1894.
Malek, M., E. Barzilay, A. Kramer, B. Camp, L. A. Jaykus, B. Escudero-Abarca, G. Derrick, P. White, C. Gerba, C. Higgins, J. Vinje, R. Glass, M. Lynch, and M. A. Widdowson. 2009. Outbreak of norovirus infection among river rafters associated with packaged delicatessen meat, Grand Canyon, 2005. Clinical Infectious Diseases 48(1):31-37.
Marionneau, S., F. Airaud, N. V. Bovin, J. Le Pendu, and N. Ruvoën-Clouet. 2005. Influence of the combined ABO, FUT2, and FUT3 polymorphism on susceptibility to Norwalk virus attachment. Journal of Infectious Diseases 192(6):1071-1077.
Martella, V., M. Campolo, E. Lorusso, P. Cavicchio, M. Camero, A. L. Bellacicco, N. Decaro, G. Elia, G. Greco, M. Corrente, C. Desario, S. Arista, K. Banyai, M. Koopmans, and C. Buonavoglia. 2007. Norovirus in captive lion cub (Panthera leo). Emerging Infectious Diseases 13(7):1071-1073.
Martella, V., E. Lorusso, N. Decaro, G. Elia, A. Radogna, M. D’Abramo, C. Desario, A. Cavalli, M. Corrente, M. Camero, C. A. Germinario, K. Bányai, B. Di Martino, F. Marsilio, L. E. Carmichael, and C. Buonavoglia. 2008. Detection and molecular characterization of a canine norovirus. Emerging Infectious Diseases 14(8):1306-1308.
Martella, V., P. Pinto, and C. Buonavoglia. 2011. Canine noroviruses. Veterinary Clinics of North America: Small Animal Practice 41(6):1171-1181.
McKinney, K. R., Y. Y. Gong, and T. G. Lewis. 2006. Environmental transmission of SARS at Amoy Gardens. Journal of Environmental Health 68:26-30.
Miranda, M. E., and N. L. Miranda. 2011. Reston ebolavirus in humans and animals in the Philippines: A review. Journal of Infectious Diseases 204(Suppl. 3):S757-S760.
Mohd Hanafiah, K., K. H. Jacobsen, and S. T. Wiersma. 2011. Challenges to mapping the health risk of hepatitis A virus infection. International Journal of Health Geographics 10:57.
Munné, M. S., S. Vladimirsky, L. Otegui, S. Soto, L. Brajterman, R. Castro, M. C. Velasco, A. Bonnano, E. Fernández, C. Remondegui, C. Passeggi, C. Rodríguez, M. Pizarro, A. Fabre, R. Moreiro, J. Quarleri, and J. E. González. 2007. Molecular characterization of hepatitis A virus isolates from Argentina. Journal of Medical Virology 79(7):887-894.
Myrmel, M., E. M. Berg, E. Rimstad, and B. Grinde. 2004. Detection of enteric viruses in shellfish from the Norwegian coast. Applied and Environmental Microbiology 70(5):2678-2684.
Myrmel, M., E. M. Berg, B. Grinde, and E. Rimstad. 2006. Enteric viruses in inlet and outlet samples from sewage treatment plants. Journal of Water and Health 4(2):197-209.
Nakamura, S., C. S. Yang, N. Sakon, M. Ueda, T. Tougan, A. Yamashita, N. Goto, K. Takahashi, T. Yasunaga, K. Ikuta, T. Mizutani, Y. Okamoto, M. Tagami, R. Morita, N. Maeda, J. Kawai, Y. Hayashizaki, Y. Nagai, T. Horii, T. Iida, and T. Nakaya. 2009. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS One 4(1):e4219.
Nejati, A., M. Makvandi, A. Samarbafzadeh, N. Neisi, and H. Moradzadegan. 2012. Molecular epidemiology of hepatitis A virus in patients in the Ahwaz region of Iran. Journal of Medical Virology 84(4):582-586, doi: 10.1002/jmv.23238.
Newell, D. G., M. Koopmans, L. Verhoef, E. Duizer, A. Aidara-Kane, H. Sprong, M. Opsteegh, M. Langelaar, J. Threfall, F. Scheutz, J. van der Giessen, and H. Kruse. 2010. Food-borne diseases—the challenges of 20 years ago still persist while new ones continue to emerge. International Journal of Food Microbiology 139(Suppl. 1):S3-15.
Nishida, T., O. Nishio, M. Kato, T. Chuma, H. Kato, H. Iwata, and H. Kimura. 2007. Genotyping and quantitation of noroviruses in oysters from two distinct sea areas in Japan. Microbiology and Immunology 51(2):177-184.
Nordgren, J., A. Matussek, A. Mattsson, L. Svensson, and P. E. Lindgren. 2009. Prevalence of norovirus and factors influencing virus concentrations during one year in a full-scale wastewater treatment plant. Water Research 43(4):1117-1125.
Ntafis, V., E. Xylouri, A. Radogna, C. Buonavoglia, and V. Martella. 2010. Outbreak of canine noro-virus infection in young dogs. Journal of Clinical Microbiology 48(7):2605-2608.
Okabayashi, T., S. Yokota, H. Ohkoshi, Y. Ohuchi, M. Yoshida, K. Kikuchi, K. Yano, and N. Fujii. 2008. Occurrence of norovirus infections unrelated to norovirus outbreaks in an asymptomatic food handler population. Journal of Clinical Microbiology 46(6):1985-1988.
Olesen, B., J. Neimann, B. Böttiger, S. Ethelberg, P. Schiellerup, C. Jensen, M. Helms, F. Scheutz, K. E. Olsen, K. Krogfel, E. Petersen, K. Mølbak, and P. Gerner-Smidt. 2005. Etiology of diarrhea in young children in Denmark: A case-control study. Journal of Clinical Microbiology 43(8):3636-3641.
Oliver, S. L., E. Asobayire, A. M. Dastjerdi, and J. C. Bridger. 2006. Genomic characterization of the unclassified bovine enteric virus Newbury agent-1(Newbury1) endorses a new genus in the family Caliciviridae. Virology 350(1):240-250.
OzFoodNet Working Group. 2009. Monitoring the incidence and causes of diseases potentially transmitted by food in Australia: Annual report of the OzFoodNet Network. Communicable Diseases Intelligence 34(4):396-426.
Parashar, U. D., L. M. Sunn, F. Ong, A. W. Mounts, M. T. Arif, T. G. Ksiazek, M. A. Kamaluddin, A. N. Mustafa, H. Kaur, L. M. Ding, G. Othman, H. M. Radzi, P. T. Kitsutani, P. C. Stockton, J. Arokiasamy, H. E. Gary, Jr., and L. J. Anderson. 2000. Case-control study of risk factors for human infection with a new zoonotic paramyxovirus, Nipah virus, during a 1998-1999 outbreak of severe encephalitis in Malaysia. Journal of Infectious Disease 181:1755-1759.
Patel, M. M., M. A. Widdowson, R. I. Glass, K. Akazawa, J. Vinjé, and U. D. Parashar. 2008. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerging Infectious Diseases 14(8):1224-1231.
Peeler, E. J., R. Allan Reese, D. L. Cheslett, F. Geoghegan, A. Power, and M. A. Thrush. 2012. Investigation of mortality in Pacific oysters associated with Ostreid herpesvirus-1μVar in the Republic of Ireland in 2009. Preventive Veterinary Medicine 105(1):136-143.
Pérez-Sautu, U., M. I. Costafreda, J. Lite, R. Sala, I. Barrabeig, A. Bosch, and R. M. Pintó. 2011. Molecular epidemiology of hepatitis A virus infections in Catalonia, Spain, 2005-2009: Circulation of newly emerging strains. Journal of Clinical Virology 52(2):98-102.
Petrignani, M., M. Harms, L. Verhoef, R. van Hunen, C. Swaan, J. van Steenbergen, I. Boxman, R. Peran i Sala, H. J. Ober, H. Vennema, M. Koopmans, and M. van Pelt. 2010. Update: A food-borne outbreak of hepatitis A in the Netherlands related to semi-dried tomatoes in oil, January-February 2010. Eurosurveillance 15(20):19572.
Phan, T. G., K. Kaneshi, Y. Ueda, S. Nakaya, S. Nishimura, A. Yamamoto, K. Sugita, S. Takanashi, S. Okitsu, and H. Ushijima. 2007. Genetic heterogeneity, evolution, and recombination in noroviruses. Journal of Medical Virology 79(9):1388-1400.
Pinto, R. M., M. I. Costafreda, and A. Bosch. 2009. Risk assessment in shellfish-borne outbreaks of hepatitis A. Applied and Environmental Microbiology 75:7350-7355.
Pommepuy, M., F. Dumas, M. P. Caprais, P. Camus, C. Le Mennec, S. Parnaudeau, L. Haugarreau, B. Sarrette, P. Vilagines, P. Pothier, E. Kholi, and F. Le Guyader. 2004. Sewage impact on shellfish microbial contamination. Water Science and Technology 50(1):117-124.
Purcell, R. H., and S. U. Emerson. 2001. Hepatitis E virus. In Fields’ Virology, edited by D. M. Knipe and P. M. Howe. New York: Raven Press. Pp. 3051-3061.
Reuter, G., H. Vennema, M. Koopmans, and G. Szücs. 2006. Epidemic spread of recombinant noroviruses with four capsid types in Hungary. Journal of Clinical Virology 35(1):84-88.
Rimoin, A. W., P. M. Mulembakani, S. C. Johnston, J. O. Lloyd Smith, N. K. Kisalu, T. L. Kinkela, S. Blumberg, H. A. Thomassen, B. L. Pike, J. N. Fair, N. D. Wolfe, R. L. Shongo, B. S. Graham, P. Formenty, E. Okitolonda, L. E. Hensley, H. Meyer, L. L. Wright, and J. J. Muyembe. 2010. Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proceedings of the National Academy of Sciences of the United States of America 107(37):16262-16267.
Robertson, B. H., R. W. Jansen, B. Khanna, A. Totsuka, O. V. Nainan, G. Siegl, A. Widell, H. S. Margolis, S. Isomura, and K. Ito. 1992. Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. Journal of General Virology 73:1365-1377.
Rosario, K., C. Nilsson, Y. W. Lim, Y. Ruan, and M. Breitbart. 2009. Metagenomic analysis of viruses in reclaimed water. Environmental Microbiology 11(11):2806-2820.
Rutjes, S. A., F. Lodder-Verschoor, W. H. van der Poel, Y. T. van Duijnhoven, and A. M. de Roda Husman. 2006. Detection of noroviruses in foods: A study on virus extraction procedures in foods implicated in outbreaks of human gastroenteritis. Journal of Food Protection 69(8):1949-1956.
Rydell, G. E., E. Kindberg, G. Larson, and L. Svensson. 2011. Susceptibility to winter vomiting disease: A sweet matter. Reviews in Medical Virology 21(6):370-382.
Sanchez, G., R. M. Pinto, H. Vanaclocha, and A. Bosch. 2002. Molecular characterization of hepatitis A virus isolates from a transcontinental shellfish-borne outbreak. Journal of Clinical Mocrobiology 40:4148-1455.
Scallan, E., R. M. Hoekstra, F. J. Angulo, R. V. Tauxe, M. A. Widdowson, S. L. Roy, J. L. Jones, and P. M. Griffin. 2011. Foodborne illness acquired in the United States—major pathogens. Emerging Infectious Diseases 17(1):7-15.
Schmid, D., H. P. Stüger, I. Lederer, A. M. Pichler, G. Kainz-Arnfelser, E. Schreier, and F. Allerberger. 2007. A foodborne norovirus outbreak due to manually prepared salad, Austria 2006. Infection 35(4):232-239.
Shieh, Y. C., R. S. Baric, J. W. Woods, and K. R. Calci. 2003. Molecular surveillance of enterovirus and norwalk-like virus in oysters relocated to a municipal-sewage-impacted gulf estuary. Applied and Environmental Microbiology 69(12):7130-7136.
Shieh, Y. C., Y. E. Khudyakov, G. Xia, L. M. Ganova-Raeva, F. M. Khambaty, and J. W. Woods. 2007. Molecular confirmation of oysters as the vector for hepatitis A in a 2005 multistate outbreak. Journal of Food Protection 70:145-150.
Siebenga, J. J., H. Vennema, B. Renckens, E. de Bruin, B. van der Veer, R. J. Siezen, and M. Koopmans. 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. Journal of Virology 81(18):9932-9941.
Siebenga, J. J., M. F. Beersma, H. Vennema, P. van Biezen, N. J. Hartwig, and M. Koopmans. 2008. High prevalence of prolonged norovirus shedding and illness among hospitalized patients: A model for in vivo molecular evolution. Journal of Infectious Diseases 198(7):994-1001.
Siebenga, J. J., H. Vennema, D. P. Zheng, J. Vinjé, B. E. Lee, X. L. Pang, E. C. Ho, W. Lim, A. Choudekar, S. Broor, T. Halperin, N. B. Rasool, J. Hewitt, G. E. Greening, M. Jin, Z. J. Duan, Y. Lucero, M. O’Ryan, M. Hoehne, E. Schreier, R. M. Ratcliff, P. A. White, N. Iritani, G. Reuter, and M. Koopmans. 2009. Norovirus illness is a global problem: Emergence and spread of norovirus GII.4 variants, 2001-2007. Journal of Infectious Diseases 200(5):802-812.
Siebenga, J. J., P. Lemey, S. L. Kosakovsky Pond, A. Rambaut, H. Vennema, and M. Koopmans. 2010. Phylodynamic reconstruction reveals norovirus GII4 epidemic expansions and their molecular determinants. PLoS Pathogens 6:e1000884.
Smiley, J. R., A. E. Hoet, M. Tråvén, H. Tsunemitsu, and L. J. Saif. 2003. Reverse transcription-PCR assays for detection of bovine enteric caliciviruses (BEC) and analysis of the genetic relationships among BEC and human caliciviruses. Journal of Clinical Microbiology 41(7):3089-3099.
Souza, M., M. S. Azevedo, K. Jung, S. Cheetham, and L. J. Saif. 2007. Pathogenesis and immune responses in gnotobiotic calves after infection with the genogroup II.4-HS66 strain of human norovirus. Journal of Virology 82(4):1777-1786.
Stals, A., L. Baert, V. Jasson, E. Van Coillie, and M. Uyttendaele. 2011. Screening of fruit products for norovirus and the difficulty of interpreting positive PCR results. Journal of Food Protection 74(3):425-431.
Strawn, L. K., K. R. Schneider, and M. D. Danyluk. 2011. Microbial safety of tropical fruits. Critical Reviews in Food Science and Nutrition 51(2):132-145.
Suffredini, E., C. Corrain, G. Arcangeli, L. Fasolato, A. Manfrin, E. Rossetti, E. Biazzi, R. Mioni, E. Pavoni, M. N. Losio, G. Sanavio, and L. Croci. 2008. Occurrence of enteric viruses in shellfish and relation to climatic-environmental factors. Letters in Applied Microbiology 47(5):467-474.
Sukhrie, F. H., M. F. Beersma, A. Wong, B. van der Veer, H. Vennema, J. Bogerman, and M. Koopmans. 2010. Using molecular epidemiology to trace transmission of nosocomial norovirus infection. Journal of Clinical Microbiology 49(2):602-606.
Sulbaran, Y., C. R. Gutierrez, B. Marquez, D. Rojas, D. Sanchez, J. Navas, E. Rovallo, and F. H. Pujol. 2010. Hepatitis A virus genetic diversity in Venezuela: Exclusive circulation of sub-genotype IA and evidence of quasispecies distribution in the isolates. Journal of Medical Virology 82(11):1829-1834.
Svraka, S., K. Rosario, E. Duizer, H. van der Avoort, M. Breitbart, and M. Koopmans. 2010. Metagenomic sequencing for virus identification in a public-health setting. Journal of General Virology 91(Pt. 11):2846-2856.
Symes, S. J., I. C. Gunesekere, J. A. Marshall, and P. J. Wright. 2007. Norovirus mixed infection in an oyster-associated outbreak: An opportunity for recombination. Archives of Virology 152(6):1075-1086.
Tam, C. C., S. J. O’Brien, D. S. Tompkins, F. J. Bolton, L. Berry, J. Dodds, D. Choudhury, F. Halstead, M. Iturriza-Gómara, K. Mather, G. Rait, A. Ridge, L. C. Rodrigues, J. Wain, B. Wood, and J. J. Gray; the IID2 Study Executive Committee. 2012. Changes in causes of acute gastroenteritis in the United Kingdom over 15 years: Microbiologic findings from 2 prospective, population-based studies of infectious intestinal disease. Clinical Infectious Diseases 54(9):1275-1286, doi: 10.1093/cid/cis028.
Tan, M., and X. Jiang. 2011. Norovirus-host interaction: Multi-selections by human histo-blood group antigens. Trends in Microbiology 19(8):382-388.
Tei, S., N. Kitajima, K. Takahashi, and S. Mishiro. 2003. Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet 362(9381):371-373.
Teo, C. G. 2009. Much meat, much malady: Changing perceptions of the epidemiology of hepatitis E. Clinical Microbiology and Infection 16:24-32.
Tessé, S., B. Lioure, L. Fornecker, M. J. Wendling, F. Stoll-Keller, C. Bigaillon, and E. Nicand. 2012. Circulation of genotype 4 hepatitis E virus in Europe: First autochthonous hepatitis E infection in France. Journal of Clinical Virology 54(2):197-200.
Teunis, P. F., C. L. Moe, P. Liu, S. E. Miller, L. Lindesmith, R. S. Baric, J. Le Pendu, and R. L. Calderon. 2008. Norwalk virus: How infectious is it? Journal of Medical Virology 80(8):1468-1476.
Todd, E. C., J. D. Greig, C. A. Bartleson, and B. S. Michaels. 2008. Outbreaks where food workers have been implicated in the spread of foodborne disease. Part 4. Infective doses and pathogen carriage. Journal of Food Protection 71(11):2339-2373.
Tompkins, D. S., M. J. Hudson, H. R. Smith, R. P. Eglin, J. G. Wheeler, M. M. Brett, R. J. Owen, J. S. Brazier, P. Cumberland, V. King, and P. E. Cook. 1999. A study of infectious intestinal disease in England: Microbiological findings in cases and controls. Communicable Diseases and Public Health 2(2):108-113; Erratum. 1999. Communicable Diseases and Public Health 2(3):222.
Turcios, R. M., M. A. Widdowson, A. C. Sulka, P. S. Mead, and R. I. Glass. 2006. Reevaluation of epidemiological criteria for identifying outbreaks of acute gastroenteritis due to norovirus: United States, 1998-2000. Clinical Infectious Diseases 42(7):964-969.
Ueki, Y., M. Shoji, A. Suto, T. Tanabe, Y. Okimura, Y. Kikuchi, N. Saito, D. Sano, and T. Omura. 2007. Persistence of caliciviruses in artificially contaminated oysters during depuration. Applied and Environmental Microbiology 73(17):5698-5701.
van Asten, L., J. Siebenga, C. van den Wijngaard, R. Verheij, H. van Vliet, M. Kretzschmar, H. Boshuizen, W. van Pelt, and M. Koopmans. 2011. Unspecified gastroenteritis illness and deaths in the elderly associated with norovirus epidemics. Epidemiology 22(3):336-343.
Van Damme, P., G. Leroux-Roels, P. Crasta, M. Messier, J. M. Jacquet, and K. Van Herck. 2011. Antibody persistence and immune memory in adults, 15 years after a three-dose schedule of a combined hepatitis A and B vaccine. Journal of Medical Virology 84(1):11-17.
van den Berg, H., W. Lodder, W. van der Poel, H. Vennema, and A. M. de Roda Husman. 2005. Genetic diversity of noroviruses in raw and treated sewage water. Research in Microbiology 156(4):532-540.
van der Sanden, S., M. Koopmans, G. Uslu, and H. van der Avoort; Dutch Working Group for Clinical Virology. 2009. Epidemiology of enterovirus 71 in the Netherlands, 1963 to 2008. Journal of Clinical Microbiology 47(9):2826-2833.
Verhoef, L., E. Depoortere, I. Boxman, E. Duizer, Y. van Duynhoven, J. Harris, C. Johnsen, A. Kroneman, S. Le Guyader, W. Lim, L. Maunula, H. Meldal, R. Ratcliff, G. Reuter, E. Schreier, J. Siebenga, K. Vainio, C. Varela, H. Vennema, and M. Koopmans; Food Borne Viruses in Europe Network. 2008. Emergence of new norovirus variants on spring cruise ships and prediction of winter epidemics. Emerging Infectious Diseases 14(2):238-243.
Verhoef, L., H. Vennema, W. van Pelt, D. Lees, H. Boshuizen, K. Henshilwood, and M. Koopmans; Food-Borne Viruses in Europe Network. 2010. Use of norovirus genotype profiles to differentiate origins of foodborne outbreaks. Emerging Infectious Diseases 16:617-624.
Verhoef, L., R. D. Kouyos, H. Vennema, A. Kroneman, J. Siebenga, W. van Pelt, and M. Koopmans; Foodborne Viruses in Europe Network. 2011. An integrated approach to identifying international foodborne norovirus outbreaks. Emerging Infectious Diseases 17(3):412-418.
Victoria, M., F. R. Guimarães, T. M. Fumian, F. F. Ferreira, C. B. Vieira, T. Shubo, J. P. Leite, and M. P. Miagostovich. 2010. One year monitoring of norovirus in a sewage treatment plant in Rio de Janeiro, Brazil. Journal of Water and Health 8(1):158-165.
Wang, M., M. Yan, H. Xu, W. Liang, B. Kan, B. Zheng, H. Chen, H. Zheng, Y. Xu, E. Zhang, H. Wang, J. Ye, G. Li, M. Li, Z. Cui, Y. F. Liu, R. T. Guo, X. N. Liu, L. H. Zhan, D. H. Zhou, A. Zhao, R. Hai, D. Yu, Y. Guan, and J. Xu. 2005a. SARS-CoV infection in a restaurant from palm civet. Emerging Infectious Diseases 11(12):1860-1865.
Wang, Q. H., M. G. Han, S. Cheetham, M. Souza, J. A. Funk, and L. J. Saif. 2005b. Porcine noroviruses related to human noroviruses. Emerging Infectious Diseases 11(12):1874-1881.
Wang, Q. H., M. Souza, J. A. Funk, W. Zhang, and L. J. Saif. 2006. Prevalence of noroviruses and sapoviruses in swine of various ages determined by reverse transcription-PCR and microwell hybridization assays. Journal of Clinical Microbiology 44(6):2057-2062.
Webby, R. J., K. S. Carville, M. D. Kirk, G. Greening, R. M. Ratcliff, S. K. Crerar, K. Dempsey, M. Sarna, R. Stafford, M. Patel, and G. Hall. 2007. Internationally distributed frozen oyster meat causing multiple outbreaks of norovirus infection in Australia. Clinical Infectious Diseases 44(8):1026-1031.
Westhoff, T. H., M. Vergoulidou, C. Loddenkemper, S. Schwartz, J. Hofmann, T. Schneider, W. Zidek, and M. van der Giet. 2009. Chronic norovirus infection in renal transplant recipients. Nephrology, Dialysis, and Transplantation 24(3):1051-1053.
Wheeler, C., T. M. Vogt, G. L. Armstrong, G. Vaughan, A. Weltman, O. V. Nainan, V. Dato, G. Xia, K. Waller, J. Amon, T. M. Lee, A. Highbaugh-Battle, C. Hembree, S. Evenson, M. A. Ruta, I. T. Williams, A. E. Fiore, and B. P. Bell. 2005. An outbreak of hepatitis A associated with green onions. New England Journal of Medicine 353:890-897.
Wheeler, J. G., D. Sethi, J. M. Cowden, P. G. Wall, L. C. Rodrigues, D. S. Tompkins, M. J. Hudson, and P. J. Roderick. 1999. Study of infectious intestinal disease in England: Rates in the community, presenting to general practice, and reported to national surveillance. The Infectious Intestinal Disease Study Executive. British Medical Journal 318(7190):1046-1050.
WHO (World Health Organization). 2008. Viruses in food: Scientific advice to support risk management. Microbiological Risk Assessment Series, No. 13. http://www.who.int/foodsafety/publications/micro/mra13/en/index.html (accessed March 28, 2012).
Wichmann, O., S. Schimanski, J. Koch, M. Kohler, C. Rothe, A. Plentz, W. Jilg, and K. Stark. 2008. Phylogenetic and case control study on hepatitis E virus infection in Germany. Journal of Infectious Diseases 198(12):1732-1741.
Wobus, C. E., L. B. Thackray, and H. W. Virgin, 4th. 2006. Murine norovirus: A model system to study norovirus biology and pathogenesis. Journal of Virology 80(11):5104-5112.
Wolf, S., W. Williamson, J. Hewitt, S. Lin, M. Rivera-Aban, A. Ball, P. Scholes, M. Savill, and G. E. Greening. 2009. Molecular detection of norovirus in sheep and pigs in New Zealand farms. Veterinary Microbiology 133(1-2):184-189.
Wolf, S., J. Hewitt, and G. E. Greening. 2010. Viral multiplex quantitative PCR assays for tracking sources of fecal contamination. Applied and Environmental Microbiology 76(5):1388-1394.
Yoon, Y. K., J. E. Yeon, J. H. Kim, H. S. Sim, J. Y. Kim, D. W. Park, J. W. Sohn, B. C. Chun, and M. J. Kim. 2011. Comparative analysis of disease severity between genotypes IA and IIIA of hepatitis A virus. Journal of Medical Virology 83(8):1308-1314.
Yun, H., S. Kim, H. Lee, K. S. Byun, S. Y. Kwon, H. J. Yim, Y. S. Lim, S. H. Jeong, and Y. Jee. 2008. Genetic analysis of HAV strains isolated from patients with acute hepatitis in Korea, 2005-2006. Journal of Medical Virology 80(5):777-784.
Zhao, C., Z. Ma, T. J. Harrison, R. Feng, C. Zhang, Z. Qiao, J. Fan, H. Ma, and M. Li. 2009. A novel genotype of hepatitis E virus prevalent among farmed rabbits in China. Journal of Medical Virology 81:1371-1379.
Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2005. Norovirus classification and proposed strain nomenclature. Virology 346(2):312-323.
MICROBE HUNTING AND PATHOGEN DISCOVERY16
Nicole C. Arrigo17and W. Ian Lipkin17
Introduction to Pathogen Discovery
Over the past few decades, numerous factors have contributed to a dramatic increase in the rate of microbial and pathogen discovery. The globalization of travel and trade, changes in demographics and land use, susceptibility to opportunistic organisms associated with immunosuppression, and climate change have all contributed to the physical emergence and reemergence of novel and known microbial pathogens (Morse, 1995). Advanced molecular technologies, such as MassTag polymerase chain reaction (PCR) (Briese et al., 2005), microbial micro-arrays (Cox-Foster et al., 2007; Palacios et al., 2007; Wang et al., 2002), and unbiased high-throughput sequencing (Cox-Foster et al., 2007), have enabled efficient microbial surveillance and detection. As a result of technological advancements, our understanding of sample quality has increased, and specimen collection has become more sophisticated and comprehensive. Dramatic improvements in bioinformatics expertise and computing power have enabled the creation and management of databases needed to compare and distinguish genetic sequences between host and microbe. Finally, our models for pathogenesis embrace increas-
_______________
16 Portions adapted and reprinted with proper permission from two of the author’s previous publications: (Lipkin, 2010) Copyright © American Society for Microbiology, Lipkin, W. I. 2010. Microbe hunting. Microbiology and Molecular Biology Reviews 74(3):363-377, doi:10.1128/MMBR.00007-10; and (Lipkin, 2008) Lipkin, W. I. 2008. Pathogen discovery. PLoS Pathogens 4(4):e1000002, doi:10.1371/journal.ppat.1000002.
17 Center for Infection and Immunity, Mailman School of Public Health, Columbia University, New York, NY.
ingly complex mechanisms that consider host—microbe—timing interactions in both acute and chronic diseases.
This paper summarizes and compares methods now in use and suggests possible directions in which we might travel over the next few years, providing examples of discoveries from our research group that stress the importance of the One Health approach to the topic of food supply and safety.
Diversity in the Microbial Universe
The introduction of cultivation-independent methods for microbial discovery and surveillance has dramatically altered our view of the breadth and diversity of the microbial world. Not only can we now detect and characterize disease agents for which we have no culture system, we can also more rapidly survey ourselves and the larger biosphere. These advances have enabled extraordinary revelations, including the extent to which humans represent microbial vessels. While the number of cells of the human body has been estimated to be 1013, our bacterial passengers on internal and external surfaces are estimated to number at least 1014, or ten times the number of microbial cells to host cells (Savage, 1977). 16S rRNA gene analyses of the oropharynx (Aas et al., 2005), esophagus (Pei et al., 2004), stomach (Bik et al., 2006), intestine and colon (Eckburg et al., 2005), vagina (Oakley et al., 2008), and skin (Gao et al., 2007) indicate differences in human bacterial microflora by anatomical location, individual, and area of residence. This dynamic bacterial composition can also vary over time and can be modified as a function of diet (including the use of probiotics), antibiotics (Hoban, 2003), hygiene, and, in the instance of intestinal microflora, surgical interventions such as bypass procedures (Zhang et al., 2009). The mouth alone has been shown to harbor more than 600 species of bacteria (Paster et al., 2001), and recent improvements in throughput, reductions in costs, and investments in metagenomic sequencing will predictably drive this figure much higher. Environmental sampling has also revealed bacteria and fungi that thrive in extreme temperatures and in the presence of radioactivity, organic compounds, and heavy metals not tolerated by higher organisms (Degryse et al., 1978; Nicholson et al., 2000; Rothschild and Mancinelli, 2001).
Unlike bacteria, viruses do not comprise regions of sequence conservation that enable surveillance and discovery by a method analogous to 16S rRNA gene PCR. Thus, with a few notable exceptions in which agents have been shown to be present because investigators invested in more complex analyses (e.g., subtractive cloning [Lipkin et al., 1990] or consensus PCR using sequences of related agents) based on clues from immunohistochemistry (Nichol et al., 1993), studies of viral diversity have come into their own only more recently with the introduction of high-throughput sequencing. However, even with the use of this technology, we are limited by our capacity to recognize similarities between what we observe for a sample and what is present in a database. The number of vertebrate species
is estimated to be greater than 50,000 (Personal communication, P. Daszak), and if each is associated with only 20 endemic viruses, the vertebrate virome would exceed 1 million. Furthermore, up to 10 percent of the human genome comprises retroviral sequences (Griffiths, 2001).
Virus abundance in aquatic environments is also extremely high, with concentrations estimated at 106 per 1 ml in the deep sea and 108 per 1 ml in coastal waters, for a total of approximately 1030 viruses throughout the world’s oceans (Suttle, 2005). The extent to which these viruses pose threats to human health remains to be determined. Nonetheless, their sheer mass and diversity are staggering, and it is clear that we have only begun to scratch the surface of virus discovery. Figure A10-1 illustrates this point by tracking the annual growth of the viral sequence database vis-à-vis selected seminal discoveries and improvements in sequencing technology since 1982.
Links to Causation
Detecting an organism in a sample is only one step in establishing a causal relationship or understanding how it may be associated with disease, and many have wrestled with the challenge of codifying the process of proving causation. Based on the germ theory of disease of Pasteur, Koch and Loeffler proposed criteria that define a causative relationship between agent and disease: the agent is present in every case of a disease; it is specific for that disease; and it can be propagated in culture and inoculated into a naïve host to cause the same disease. Known as Koch’s postulates, these criteria were modified by Rivers for viruses (Rivers, 1937) and by Fredericks and Relman to reflect the introduction of molecular methods (Fredericks and Relman, 1996). Although fulfillment of Koch’s postulates remains the most persuasive evidence of causation, there are numerous challenges with holding to this standard. Overlap in signs and symptoms due to infection with different agents is common, and results of infection may vary with genetic background, age, nutrition, and previous exposure to similar agents. Furthermore, many agents cannot be cultured and/or there may be no animal model for experimentation. Proving causation is particularly difficult where agents have remote effects or require cofactors for expression. Here, one may resort to a statistical assessment of the strength of epidemiological association based on the presence of the agent or its footprints (nucleic acid, antigen, and, preferably, an immune response) and biological plausibility as indicated by analogy to diseases with other organisms where linkage is persuasive.
Complex Pathogenesis as a Confounder of Microbial Implication
In the most straightforward pathogen discovery expeditions, an agent is present in high concentrations at a site where pathology is readily apparent and organ dysfunction is dramatic. Classical examples include infections with polioviruses
FIGURE A10-1 Growth of the viral sequence database mapped to seminal discoveries and improvements in sequencing technology. EM, electron microscopy. (Image courtesy of Omar Jabado, reproduced with permission.)
SOURCE: Lipkin (2010) Copyright © American Society for Microbiology, Lipkin, W. I. (2010). Microbe Hunting. Microbiology and Molecular Biology Reviews. 74 (3):363-377: doi:10.1128/MMBR.00007-10. Reproduced with permission from American Society for Microbiology.
and motor neuron disease, an influenza virus or Streptococcus pneumoniae and acute respiratory disease, and a rotavirus or Shigella sp. and diarrhea. Viruses may kill cells directly through intracellular replication and lysis, the induction of apoptosis, or autophagy. They may also do so indirectly by presenting antigens that are recognized by cytotoxic T lymphocytes or that become bound to antibodies and trigger the activation of the classical complement cascade. Causal links may be more difficult to establish when damage is indirect, particularly when effects are manifest at sites other than the replication site. Clostridium botulinum and C. tetani bacteria, for example, grow in the skin or the gastro-
intestinal tract and release zinc metalloproteases that have distal effects on motor function by modulating neurotransmitter release (botulism [Segelke et al., 2004] and tetanus [Bruggemann et al., 2003]).
Another example of indirect pathogenesis is Sin Nombre virus infection, a hantavirus that induces the expression of cytokines that in turn promote pulmonary capillary leakage, culminating in an acute respiratory distress syndrome (Mori et al., 1999). Microbes can elicit immune responses that break tolerance to self, resulting in autoimmune disease. A well-known example is group A beta-hemolytic streptococcus (GABHS). GABHS infection of the oropharynx may cause local inflammation or be asymptomatic. In either case, infection in susceptible individuals elicits a humoral immune response that can cause both cardiac valvular damage (rheumatic heart disease) and abnormalities in movement and behavior (Sydenham’s chorea) (Pichichero, 1998). Campylobacter jejuni has been linked to Guillain-Barré syndrome (GBS), an acute demyelinating neuropathy treated by plasmapheresis or the administration of intravenous immunoglobulin (Buchwald et al., 2002). C. jejuni elicits an immune response that cross-reacts with the ganglioside GM1 in host neural tissue (Yuki et al., 2004), and more than 25 percent of individuals with GBS are infected with C. jejuni.
Infection with one organism may increase vulnerability to others. HIV/ AIDS is an extreme example of this occurrence, in which immunosuppression sets the stage for opportunistic infection with Toxoplasma gondii, Pneumocystis jirovecii, human herpesvirus 8, or Cryptococcus neoformans. This phenomenon was previously described in 1908 when von Pirquet reported that measles was associated with a loss of delayed-type hypersensitivity to tuberculin antigen and suggested that impaired immunity might explain the dissemination of tuberculosis in individuals with measles (von Pirquet, 1908). Infection with one microbe may also directly facilitate local invasion with another. S. pneumoniae invasion in influenza, for example, is linked to damage to respiratory tract epithelium and is correlated with the sialidase activity of influenza virus neuraminidase (McCullers and Bartmess, 2003).
The duration of pathogenesis is also a complicating factor; some have both acute and long-term effects. Viruses can express gene products (oncoproteins) that impair cell cycle regulation (Weinberg, 1995) or integrate into the host genome (zur Hausen, 2002) to promote neoplasia. Inflammation associated with persistent bacterial, parasitic, or viral infection has also been implicated in cancer (Mantovani et al., 2008). During vulnerable periods of embryogenesis, any of a variety of agents may cause similar types of structural damage to the central nervous or cardiovascular system, damage that continues long after the infection has cleared. In TORCH syndrome, for example, neurological effects of prenatal infection with T. gondii, rubella virus, cytomegalovirus, or herpes simplex virus cannot be distinguished by clinical criteria. In some animal models of autism, schizophrenia, and attention deficit hyperactivity disorder, the neurological effects of prenatal infection with RNA viruses and Gram-negative bacteria
can be recreated by using the double-stranded RNA virus mimic, polyinosine/ cytosine (De Miranda et al., 2009), and lipopolysaccharide (Cai et al., 2000), respectively. It is unknown whether sequelae in these examples are mediated by the loss of somatic or stem cells (or both), by altered signaling that impedes the trafficking of cells to their appropriate destinations, or by another mechanism. As these examples demonstrate, it may be that any heuristic is perhaps too stringent if it requires an exclusive relationship between a pathogen and a specific outcome.
Strategies for Pathogen Discovery
Although reviews on pathogen surveillance and discovery typically focus on the latest molecular technologies, it is important to emphasize the pivotal roles of clinical acumen, pathology, serology, and classical culture techniques. Clinicians, veterinarians, and epidemiologists are the prime movers in pathogen discovery. They recognize the appearance of new syndromes, collect materials for investigation, and persuade their colleagues to take up the search. When possible, a comprehensive collection of biological specimens allows for the complementation of various diagnostic and discovery techniques. For example, the use of anatomic pathology can be instrumental in directing downstream molecular work. The discoveries of Nipah virus (Paton et al., 1999) and West Nile virus (Briese et al., 1999; Shieh et al., 2000) were facilitated by demonstration of henipavirus and flavivirus proteins in tissues, which allowed focused genetic analyses. In addition to serology, classical virological methods, such as tissue culture, proved pivotal in the 2003 severe acute respiratory syndrome (SARS) outbreak (Challoner et al., 1995). Propagation of the virus in tissue culture enabled its rapid characterization by a variety of molecular and morphological techniques, including consensus and random PCR, cloning, microarray, and electron microscopy.
Isolation and Visualization of Infectious Agents
Microbe hunters employ a wide range of media and tissue culture systems, including complex organotypic cultures (Braun et al., 2006; Honer zu Bentrup et al., 2006), to isolate and grow prokaryotic and eukaryotic organisms. When these efforts fail, alternative strategies may include inoculation of immature or genetically modified higher organisms that possess innate immune responses that are inefficient or disabled (e.g., newborn [Bowen et al., 1977] and knockout [Glaser et al., 2007] mice) or transgenes that are introduced to express products essential to the entry or replication of viruses (Martina et al., 2006; Ren et al., 1990) or prions (Scott et al., 1989). The choice of an in vitro versus an in vivo strategy for the isolation of infectious agents can have a profound impact on what one can find. For example, whereas the surveillance of human stool for enteroviruses by the inoculation of suckling mice favors detection of human enterovirus type A, tissue culture favors the detection of human enterovirus type B (Witso et al., 2007).
Although isolation of the agent is highly encouraged (Arrigo et al., 2012), it is not always possible. If the sequence of the pathogen candidate is known, genomic reconstruction can circumvent the need for a viable isolate ( Handelsman, 2004). This approach has enabled a new field of archaeovirology wherein infectious retroviruses have been built from endogenous retroviral sequences (Lower et al., 1996), and the 1918 pandemic influenza strain was rebuilt and analyzed for pathogenetic properties (Tumpey et al., 2005).
When an agent cannot be isolated, propagated, or studied through reconstruction, one may nonetheless find evidence of its presence by imaging it morphologically via light or electron microscopy or imaging its proteins or nucleic acids through immunohistochemistry or in situ hybridization, respectively. In some instances, a candidate agent is sufficiently similar to known ones such that available antibodies to the latter are cross-reactive with the former. Indeed, immunohistochemistry has been used not only to confirm the presence of an agent or determine its anatomic distribution, but also as a clue to its identity. Prominent examples include the identification of Sin Nombre virus (Chizhikov et al., 1995), Nipah virus (Paton et al., 1999), and West Nile virus (Briese et al., 1999), for which the screening of tissues from victims of unrecognized infectious diseases with broadly reactive sera led investigators to focus on candidate viral families by consensus PCR.
Molecular Methods
The advent of methods of detecting and cloning nucleic acids of microbial pathogens directly from clinical specimens ushered in a new era in pathogen discovery. Over the past two decades, subtractive cloning, expression cloning, consensus PCR, and high-throughput sequencing resulted in identification of novel agents associated with both acute and chronic diseases, including Borna disease virus, hepatitis C virus, Sin Nombre virus, HHV-6, HHV-8, Bartonella henselae, Tropheryma whippelii, Nipah virus, SARS coronavirus, and Israel Acute Paralysis virus (Challoner et al., 1995; Chang et al., 1994; Cox-Foster et al., 2007; Lipkin et al., 1990; Nichol et al., 1993; Paton et al., 1999; Peiris et al., 2003; Relman et al., 1990, 1992; VandeWoude et al., 1990).
Singleplex Assays The most common singleplex assays employed in clinical microbiology and microbial surveillance are conventional or quantitative PCR assays. The DNA products of conventional PCR are visualized via ethidium bromide—stained agarose gels, while DNA strand replication in quantitative PCR results in cleavage of a fluorescence-labeled oligonucleotide probe bound to a sequence between the forward and reverse nucleotide primers. Equipment needs are simple (thermal cycler, fluorescent reader, and laptop computer), and rugged instruments have been implemented for field use with battery power. Loop- mediated isothermal amplification (LAMP) does not require programmable
thermal cyclers (Hagiwara et al., 2007; Notomi et al., 2000; Shirato et al., 2007). In laboratory settings, LAMP products are also detected via ethidium bromide-stained agarose gels. However, in the field, changes in the turbidity of the amplification solution may be sufficient, and assays in which the accumulation of product can be detected by eye have been described (Jayawardena et al., 2007).
The most sensitive assays are those for which primers and/or probes perfectly match a single genetic target. Fluorescence reporter-based TaqMan or molecular beacon singleplex PCR assays, for example, typically have detection thresholds of <10 RNA molecules. Although ideal for detecting the presence of a specific agent and for quantitating burden (Heid et al., 1996; Tyagi and Kramer, 1996), these assays may nonetheless fail with RNA viruses characterized by high mutation rates and genetic variability. Consensus PCR assays that use degenerate primers are less likely to be confounded by sequence divergence, but they are less sensitive than specific PCR assays. Furthermore, given that many potential pathogens can overlap in clinical presentation, unless one has the sample mass, resources, and time to invest in many singleplex assays for different agents, there is the risk that a spurious candidate or candidates will be selected. Bacterial 16S rRNA gene assays are increasingly particularly powerful tools, with such seminal contributions as the discovery of Tropheryma whippeli (Relman et al., 1992), and are becoming more powerful with the introduction of sequencing technologies that enable the description of microbial communities.
Nested PCR, in which two amplification reactions are pursued sequentially with either one (heminested) or two (fully nested) primers located 3’ with respect to the original primer set, may be more sensitive than fluorescence reporter-based singleplex assays. However, because the original reaction vessels must be opened to add reagents for the second nested reaction, the risk for contamination is high, even in laboratories with scrupulous experimental hygiene.
Multiplex Assays Signs and symptoms of disease are rarely pathognomonic of a single agent, particularly early in the course of an illness. Multiplex assays may be helpful in such situations because they may be used to entertain many hypotheses simultaneously. The number of candidates considered ranges from 10 to 100 with multiplex PCR, to thousands with microarrays, and to the entire tree of life with unbiased high-throughput sequencing. In multiplex assays many genetic targets compete for assay components (e.g., nucleotides, polymerases, and dyes), in some instances with variable efficiencies. Thus, multiplex assays tend to be less sensitive than singleplex assays.
Multiplex PCR Assays Multiplex PCR assays are more difficult to establish because primer sets may differ in optimal reaction conditions (e.g., annealing temperature and magnesium concentration). Furthermore, complex primer mixtures are more likely to result in primer-primer interactions that reduce the assay sensitivity and/or specificity. To enable multiplex primer design, we developed
Greene SCPrimer, a software program that automates consensus primer design over a multiple-sequence alignment and allows users to specify the primer length, melting temperature, and degree of degeneracy (Jabado et al., 2006).
Gel-based multiplex PCR assays, wherein products are distinguished by mass, can detect as many as 10 distinct targets (Casas et al., 1997; Templeton et al., 2004). Fluorescence reporter—based multiplex assays are more sensitive but are limited by the number of fluorescent emission peaks that can be unequivocally separated. At present, up to four fluorescent reporter dyes are detected simultaneously. “Sloppy molecular beacons” can address this limitation in part by binding to related targets at different melting temperatures; however, they cannot detect targets that differ by more than a few nucleotides and, thus, their applications are limited.
Two platforms that combine PCR and mass spectroscopy (MS) for the sensitive, simultaneous detection of several targets have been established. The Ibis T5000 biosensor system uses matrix-assisted laser desorption-ionization (MALDI) MS to directly measure the molecular weights of PCR products obtained in an experimental sample and to compare them with a database of known or predicted product weights (Hofstadler et al., 2005; Sampath et al., 2007; Van Ert et al., 2004). MassTag PCR uses atmospheric pressure chemical ionization (APCI) MS to detect molecular weight reporter tags attached to PCR primers. Whereas the Ibis system is confined to specialized laboratories, MassTag PCR can be performed by using smaller, less expensive instruments and does not require sophisticated operators. The Ibis system has an advantage in that it can detect novel variants of known organisms via a divergent product weight; nonetheless, like MassTag PCR, it too requires sequencing for a detailed characterization. Syndrome-specific MassTag PCR panels have been established for the detection of viruses, bacteria, fungi, and parasites associated with acute respiratory diseases, diarrheas, encephalitides/meningitides, and hemorrhagic fevers (Briese et al., 2005; Lamson et al., 2006; Palacios et al., 2006).
The Bio-Plex (also known as Luminex) is a multiplex platform that employs flow cytometry to detect PCR amplification products bound to matching oligo-nucleotides on fluorescent beads (Brunstein and Thomas, 2006; Han et al., 2006; Li et al., 2007). Assay panels that allow the detection of up to 50 genetic targets simultaneously have been developed.
Although multiplex PCR methods are designed to detect known agents, they can nonetheless facilitate pathogen discovery. For example, the use of MassTag PCR to investigate influenza-like illness in New York State revealed the presence of a novel rhinovirus clade. This discovery enabled follow-up studies across the globe wherein this novel genetic clade was implicated not only in influenza-like illnesses but also in asthma, pediatric pneumonia, and otitis media (Blomqvist et al., 2009; Briese et al., 2008; Dominguez et al., 2008; Khetsuriani et al., 2008; Kistler et al., 2007; Lau et al., 2007; Lee et al., 2007; Renwick et al., 2007; Savolainen-Kopra et al., 2009).
Microarrays Microarray technology has been used to develop assays that comprise hundreds to millions of genetic probes for applications in diagnostics, screening, pathogen identification, and discovery. Probes can be designed to discriminate differences in related sequences of known agents with the purpose of speciation. An example of this application is respiratory virus resequencing arrays, where specific genetic targets are amplified by multiplex consensus PCR and the resultant products are hybridized to oligonucleotide probes less than 25 nucleotides in length (Chiu et al., 2006; Lin et al., 2007; Wong et al., 2004). These arrays are easily implemented when one considers only a limited number of known agents. However, because the signal is dependent on precise complementarity between probes and their genetic targets, these arrays are not ideal for pathogen discovery. In contrast, arrays comprising longer probes (e.g., >60 nucleotides) are more tolerant of sequence mismatches and may detect agents more divergent than those presently known.
Two longer probe array platforms that are in common use are the GreeneChip and the Virochip (Palacios et al., 2007; Wang et al., 2002). Although they differ in design, both employ random amplification strategies to allow an unbiased detection of microbial targets, which is critical to exploiting the broad probe repertoire of these arrays. Recently, Lawrence Livermore National Laboratory developed the Lawrence Livermore Microbial Detection Array, which incorporated into its first design all available viral and bacterial sequences to detect 2,000 viral and 900 bacterial species. A newer version will expand this capability with the goal of detecting nearly 6,000 viruses, 15,000 bacteria, and fungi and protozoa (Gardner et al., 2010). A challenge with each of these platforms is that host and microbe sequences are amplified with similar efficiencies, reducing sensitivity for microbial detection in tissues rich in host genetic material. Host DNA can be eliminated by enzymatic digestion; however, host rRNA remains a major confounder, making these platforms most successful with acellular template sources, such as virus cell culture supernatant, serum, plasma, cerebrospinal fluid, or urine. Methods for depleting host rRNA prior to amplification through subtraction or the use of random primers selected for the lack of complementarity to rRNA have been described (Armour et al., 2009). Whether these interventions will sufficiently enhance sensitivity to enable pathogen discovery in tissues remains to be determined.
At present, hybridization to probes representing pathogen targets is detected by binding of a fluorescent label. However, platforms that will detect hybridization as changes in electrical conductance are in development, which may enhance both ease of use and sensitivity. During a Marburg virus outbreak, the GreeneChip panmicrobial array implicated Plasmodium falciparum in a fatal case of hemorrhagic fever that was not resolved using standard diagnostic methods (Palacios et al., 2007), and a variant of the GreeneChip array facilitated the discovery of Ebola virus Reston in a porcine respiratory illness outbreak in the Philippines (Barrette et al., 2009). The Virochip was also successfully employed in the characterization of the SARS coronavirus in 2003 (Wang et al., 2002).
Unbiased High-Throughput Sequencing The power of unbiased high-throughput sequencing has enabled unique advances in microbial surveillance and discovery. Applications include metagenomic characterization of environmental and clinical samples, rapid and comprehensive sequence analysis of microbial strains and isolates, and pathogen discovery. Unlike consensus PCR or microarray methods, whereby investigators are limited by known sequence information and must choose the pathogens to be considered in an experiment, high-throughput sequencing can be unbiased, providing an opportunity to inventory the entire tree of life. Research in our laboratory has chiefly used the 454 Life Sciences pyrosequencing system; however, applications and principles are similar across platforms, including Illumina sequencing by synthesis (SBS) technology and Life Technologies’ Ion Torrent semiconductor sequencing technology. The Ion Torrent Personal Genome Machine™ was recently used to sequence the novel Shiga toxin—producing Escherichia coli variant responsible for an outbreak with unusually high mortality in Germany in June 2011 (Mellmann et al., 2011). With whole-genome results in only 2 hours, this situation demonstrates the powerful potential of advancements in this technology not only for basic research, but also as a valuable public health tool.
While our laboratory primarily focuses use of unbiased high-throughput sequencing on pathogen discovery, we have also employed primers designed to amplify phyla (e.g., 16S rRNA gene analyses of gastrointestinal flora) or specific viruses (e.g., characterizations of influenza or dengue virus isolates). Similar to obstacles faced with applying microarray technology to unbiased PCR amplification strategies, host nucleic acid can be a critical impediment to the sensitivity of unbiased high-throughput sequencing. The same caveats and potential solutions also apply. After amplification and sequencing, raw sequence reads are clustered into nonredundant sequence sets. Unique sequence reads are assembled into contiguous sequences, which are then compared to databases using programs that examine homology at the nucleotide and amino acid levels considering all six potential reading frames. A truly novel pathogen might elude this level of analysis, thus our laboratory and others are exploring the implementation of additional characteristics to aid in pathogen discovery, including relative nucleotide composition or predicted secondary or tertiary structures.
A Staged Strategy for Pathogen Detection and Discovery
A staged investment strategy for pathogen discovery is important to contain costs, reduce extraneous efforts, and conserve valuable sample materials. When epidemiology, serology, and/or pathology suggest one or a few candidates, singleplex PCR is an ideal approach. Where no such clues pertain or singleplex assays are negative, syndromic multiplex PCR assays allow rapid examination of up to 30 candidates with only a modest increase in time and expense. Microarrays are the next step indicated if multiplex PCR fails to provide a result. Because
each of these methods requires that an agent be related to those already known, novel or distantly related agents may require subtractive cloning or unbiased high-throughput sequencing. Irrespective of the route that results in identification of a candidate, subsequent steps include quantitation of pathogen burden in affected hosts and unaffected controls, detailed characterization of the pathogen for features that may contribute to virulence or provide clues to provenance, and serology to indicate acute versus convalescent infection and examine the prevalence of infection over time and geography. Figure A10-2 depicts the complementary and parallel or progressive use of multiple techniques in a typical strategy for pathogen discovery.
Examples of Pathogen Discovery Related to Food Safety and One Health
As population increases and globalization and trade expand, the modern food supply chain becomes more diverse and complex. In the past few years, our research group has detected a number of novel and known pathogens in animal species that serve as food products, including salmon (Palacios et al., 2010), turkey (Honkavuori et al., 2011), and imported bush meat (Smith et al., 2012). While a direct threat to human health through consumption is not clear, the following examples demonstrate the application of pathogen discovery to monitoring food safety and emphasize the importance of pursuing esoteric events in consumable animal species or those that could potentially threaten independent or commercial food sources.
Salmon: Heart and Skeletal Muscle Inflammation of Farmed Salmon Is Associated with Infection with a Novel Reovirus
Abstract18
Atlantic salmon (Salmo salar L.) mariculture has been associated with epidemics of infectious diseases that threaten not only local production, but also wild fish coming into close proximity to marine pens and fish escaping from them. Heart and skeletal muscle inflammation (HSMI) is a frequently fatal disease of farmed Atlantic salmon. First recognized in one farm in Norway in 1999, HSMI was subsequently implicated in outbreaks in other farms in Norway and the United Kingdom. Although pathology and disease transmission studies indicated an infectious basis, efforts to identify an agent were unsuccessful. Here we provide evidence that HSMI is associated with infection with piscine reovirus (PRV). PRV is a novel reovirus identified by unbiased high-throughput DNA sequencing and a bioinformatics program focused on nucleotide frequency as well as sequence alignment and motif analyses. Formal implication of PRV in HSMI will
_______________
18 Abstract reprinted with proper citation and open access to Palacios et al. (2010).
FIGURE A10-2 Staged strategy for pathogen discovery and link to causation. In the molecular era of pathogen discovery, culture and molecular methods are pursued in parallel until an agent is detected, isolated, and characterized. +, positive result; -, negative result. ssRNA, single-stranded RNA; dsRNA, double-stranded RNA.
SOURCE: Lipkin (2010) Copyright © American Society for Microbiology, Lipkin, W. I. (2010). Microbe Hunting. Microbiology and Molecular Biology Reviews. 74 (3):363-377: doi:10.1128/MMBR.00007-10. Reproduced with permission from American Society for Microbiology.
require isolation in cell culture and fulfillment of Koch’s postulates, or prevention or modification of disease through use of specific drugs or vaccines. Nonetheless, as our data indicate that a causal relationship is plausible, measures must be taken to control PRV not only because it threatens domestic salmon production but also due to the potential for transmission to wild salmon populations.
Poultry: The Discovery of a Novel Picornavirus in Turkey Poults with Hepatitis in California, U.S.A
Abstract19
To identify a candidate etiologic agent for turkey viral hepatitis, we analyzed samples from diseased turkey poults from 8 commercial flocks in California, USA, that were collected during 2008-2010. High-throughput pyrosequencing of RNA from livers of poults with turkey viral hepatitis (TVH) revealed picornavirus sequences. Subsequent cloning of the ˜9-kb genome showed an organization similar to that of picornaviruses with conservation of motifs within the P1, P2, and P3 genome regions, but also unique features, including a 1.2-kb sequence of unknown function at the junction of P1 and P2 regions. Real-time PCR confirmed viral RNA in liver, bile, intestine, serum, and cloacal swab specimens from diseased poults. Analysis of liver by in situ hybridization with viral probes and immunohistochemical testing of serum demonstrated viral nucleic acid and protein in livers of diseased poults. Molecular, anatomic, and immunologic evidence suggests that TVH is caused by a novel picornavirus, tentatively named turkey hepatitis virus.
Bushmeat: The Identification of Zoonotic Viruses Associated with Illegally Imported Wildlife Products
Abstract20
The global trade in wildlife has historically contributed to the emergence and spread of infectious diseases. The United States is the world’s largest importer of wildlife and wildlife products, yet minimal pathogen surveillance has precluded assessment of the health risks posed by this practice. This report details the findings of a pilot project to establish surveillance methodology for zoonotic agents in confiscated wildlife products. Initial findings from samples collected at several international airports identified parts originating from nonhuman primate (NHP) and rodent species, including baboon, chimpanzee, mangabey, guenon, green monkey, cane rat and rat. Pathogen screening identified retroviruses (simian foamy virus) and/or herpesviruses (cytomegalovirus and lymphocryptovirus) in the NHP samples. These results are the first demonstration that illegal bushmeat importation into the United States could act as a conduit for pathogen spread, and suggest that implementation of disease surveillance of the wildlife trade will help facilitate prevention of disease emergence.
_______________
19 Abstract reprinted with proper citation and open access to Honkavuori et al. (2011).
20 Abstract reprinted with proper citation and open access to Smith et al. (2012).
Future Perspectives
Molecular platforms are rapidly evolving, with enhancements in sensitivity and throughput at a lower cost. Such improvements are facilitating the decentralization of technology such that studies now restricted to a few specialized laboratories will soon be feasible on a global scale and to a broader industry base. This technology transfer will, in turn, circumvent logistical and political issues relating to specimen transfer that can delay informed responses to outbreaks of acute disease, which is a particularly important issue when considering food safety.
With some mature technology, such as multiplex PCR, advances are likely to be incremental. In contrast, microarray technology is less advanced, and predictable, near-term improvements include higher-density arrays, automation, microfluidic sample processing, and alternatives to imaging of results, such as the direct measurement of conductance changes associated with hybridization. Unbiased high-throughput sequencing technology is expected and currently progressing rapidly, and a corresponding need for advancements in data management and bioinformatics are becoming increasingly important with the growing complexity of each of these platforms.
Although significant, this article did not address the emerging fields of proteomics and host response profiling, nor did it discuss new platforms for serology. It is conceivable that biomarkers will be found that are specific for classes of infectious agents and/or provide insights that can guide clinical management. Although less advanced, there are also efforts to develop high-density serological arrays capable of depicting previous microbial exposures to a wide range of pathogens. There is also an increasing appreciation for individualistic responses to infectious agents based on differences in genetic and epigenetic factors, nutritional status, age, exposure history, and simultaneous infections with other microbes. Thus, it is anticipated that many substantive advances may come not from technical improvements but from investments in prospective serial sampling and a shifting perspective that many diseases reflect a more complex and temporal intersection of genes and the environment.
Acknowledgments
The authors gratefully acknowledge generous support from National Institutes of Health awards AI57158 (Northeast Biodefense Center-Lipkin) and AI079231, and the Defense Threat Reduction Agency.
References
Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. Journal of Clinical Microbiology 43:5721-5732.
Armour, C. D., J. C. Castle, R. Chen, T. Babak, P. Loerch, S. Jackson, J. K. Shah, J. Dey, C. A. Rohl, J. M. Johnson, and C. K. Raymond. 2009. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nature Methods 6:647-649.
Arrigo, N. C., T. Briese, C. H. Calisher, M. A. Drebot, B. Hjelle, J. W. LeDuc, A. M. Powers, P. M. Repik, J. T. Roehrig, C. S. Schmaljohn, R. B. Tesh, and S. C. Weaver. 2012. Recommendations for publication of viral genetic data and sample access for novel viruses and strains. American Journal of Tropical Medicine and Hygiene 86:189-191.
Barrette, R. W., S. A. Metwally, J. M. Rowland, L. Xu, S. R. Zaki, S. T. Nichol, P. E. Rollin, J. S. Towner, W. J. Shieh, B. Batten, T. K. Sealy, C. Carrillo, K. E. Moran, A. J. Bracht, G. A. Mayr, M. Sirios-Cruz, D. P. Catbagan, E. A. Lautner, T. G. Ksiazek, W. R. White, and M. T. McIntosh. 2009. Discovery of swine as a host for the reston ebolavirus. Science 325:204-206.
Bik, E. M., P. B. Eckburg, S. R. Gill, K. E. Nelson, E. A. Purdom, F. Francois, G. Perez-Perez, M. J. Blaser, and D. A. Relman. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proceedings of the National Academy of Sciences of the United States of America 103:732-737.
Blomqvist, S., C. Savolainen-Kopra, A. Paananen, T. Hovi, and M. Roivainen. 2009. Molecular characterization of human rhinovirus field strains isolated during surveillance of enteroviruses. Journal of General Virology 90:1371-1381.
Bowen, E. T., G. Lloyd, W. J. Harris, G. S. Platt, A. Baskerville, and E. E. Vella. 1977. Viral haemorrhagic fever in southern Sudan and northern Zaire. Preliminary studies on the aetiological agent. Lancet 1:571-573.
Braun, E., T. Zimmerman, T. B. Hur, E. Reinhartz, Y. Fellig, A. Panet, and I. Steiner. 2006. Neurotropism of herpes simplex virus type 1 in brain organ cultures. Journal of General Virology 87:2827-2837.
Briese, T., X. Y. Jia, C. Huang, L. J. Grady, and W. I. Lipkin. 1999. Identification of a kunjin/West Nile-like flavivirus in brains of patients with new york encephalitis. Lancet 354:1261-1262.
Briese, T., G. Palacios, M. Kokoris, O. Jabado, Z. Liu, N. Renwick, V. Kapoor, I. Casas, F. Pozo, R. Limberger, P. Perez-Brena, J. Ju, and W. I. Lipkin. 2005. Diagnostic system for rapid and sensitive differential detection of pathogens. Emerging Infectious Diseases 11:310-313.
Briese, T., N. Renwick, M. Venter, R. G. Jarman, D. Ghosh, S. Kondgen, S. K. Shrestha, A. M. Hoegh, I. Casas, E. V. Adjogoua, C. Akoua-Koffi, K. S. Myint, D. T. Williams, G. Chidlow, R. van den Berg, C. Calvo, O. Koch, G. Palacios, V. Kapoor, J. Villari, S. R. Dominguez, K. V. Holmes, G. Harnett, D. Smith, J. S. Mackenzie, H. Ellerbrok, B. Schweiger, K. Schonning, M. S. Chadha, F. H. Leendertz, A. C. Mishra, R. V. Gibbons, E. C. Holmes, and W. I. Lipkin. 2008. Global distribution of novel rhinovirus genotype. Emerging Infectious Diseases 14:944-947.
Bruggemann, H., S. Baumer, W. F. Fricke, A. Wiezer, H. Liesegang, I. Decker, C. Herzberg, R. Martinez-Arias, R. Merkl, A. Henne, and G. Gottschalk. 2003. The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proceedings of the National Academy of Sciences of the United States of America 100:1316-1321.
Brunstein, J., and E. Thomas. 2006. Direct screening of clinical specimens for multiple respiratory pathogens using the genaco respiratory panels 1 and 2. Diagnostic Molecular Pathology: The American Journal of Surgical Pathology, Part B 15:169-173.
Buchwald, B., R. Ahangari, A. Weishaupt, and K. V. Toyka. 2002. Intravenous immunoglobulins neutralize blocking antibodies in Guillain-Barre syndrome. Annals of Neurology 51:673-680.
Cai, Z., Z. L. Pan, Y. Pang, O. B. Evans, and P. G. Rhodes. 2000. Cytokine induction in fetal rat brains and brain injury in neonatal rats after maternal lipopolysaccharide administration. Pediatric Research 47:64-72.
Casas, I., A. Tenorio, J. M. Echevarria, P. E. Klapper, and G. M. Cleator. 1997. Detection of enteroviral RNA and specific DNA of herpesviruses by multiplex genome amplification. Journal of Virological Methods 66:39-50.
Challoner, P. B., K. T. Smith, J. D. Parker, D. L. MacLeod, S. N. Coulter, T. M. Rose, E. R. Schultz, J. L. Bennett, R. L. Garber, and M. Chang. 1995. Plaque-associated expression of human herpesvirus 6 in multiple sclerosis. Proceedings of the National Academy of Sciences of the United States of America 92:7440-7444.
Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 266:1865-1869.
Chiu, C. Y., S. Rouskin, A. Koshy, A. Urisman, K. Fischer, S. Yagi, D. Schnurr, P. B. Eckburg, L. S. Tompkins, B. G. Blackburn, J. D. Merker, B. K. Patterson, D. Ganem, and J. L. DeRisi. 2006. Microarray detection of human parainfluenzavirus 4 infection associated with respiratory failure in an immunocompetent adult. Clinical Infectious Diseases 43:e71-76.
Chizhikov, V. E., C. F. Spiropoulou, S. P. Morzunov, M. C. Monroe, C. J. Peters, and S. T. Nichol. 1995. Complete genetic characterization and analysis of isolation of Sin Nombre virus. Journal of Virology 69:8132-8136.
Cox-Foster, D. L., S. Conlan, E. C. Holmes, G. Palacios, J. D. Evans, N. A. Moran, P. L. Quan, T. Briese, M. Hornig, D. M. Geiser, V. Martinson, D. vanEngelsdorp, A. L. Kalkstein, A. Drysdale, J. Hui, J. Zhai, L. Cui, S. K. Hutchison, J. F. Simons, M. Egholm, J. S. Pettis, and W. I. Lipkin. 2007. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318:283-287.
De Miranda, J., K. Yaddanapudi, M. Hornig, and W. I. Lipkin. 2009. Astrocytes recognize intracellular polyinosinic-polycytidylic acid via MDA-5. FASEB Journal 23:1064-1071.
Degryse, E., N. Glansdorff, and A. Pierard. 1978. A comparative analysis of extreme thermophilic bacteria belonging to the genus thermus. Archives of Microbiology 117:189-196.
Dominguez, S. R., T. Briese, G. Palacios, J. Hui, J. Villari, V. Kapoor, R. Tokarz, M. P. Glode, M. S. Anderson, C. C. Robinson, K. V. Holmes, and W. I. Lipkin. 2008. Multiplex MassTag-PCR for respiratory pathogens in pediatric nasopharyngeal washes negative by conventional diagnostic testing shows a high prevalence of viruses belonging to a newly recognized rhinovirus clade. Journal of Clinical Virology 43:219-222.
Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638.
Fredericks, D. N., and D. A. Relman. 1996. Sequence-based identification of microbial pathogens: A reconsideration of Koch’s postulates. Clinical Microbiology Reviews 9:18-33.
Gao, Z., C. H. Tseng, Z. Pei, and M. J. Blaser. 2007. Molecular analysis of human forearm superficial skin bacterial biota. Proceedings of the National Academy of Sciences of the United States of America 104:2927-2932.
Gardner, S. N., C. J. Jaing, K. S. McLoughlin, and T. R. Slezak. 2010. A microbial detection array (MDA) for viral and bacterial detection. BMC Genomics 11:668.
Glaser, L., G. Conenello, J. Paulson, and P. Palese. 2007. Effective replication of human influenza viruses in mice lacking a major alpha 2,6 sialyltransferase. Virus Research 126:9-18.
Griffiths, D. J. 2001. Endogenous retroviruses in the human genome sequence. Genome Biology 2:reviews1017.
Hagiwara, M., H. Sasaki, K. Matsuo, M. Honda, M. Kawase, and H. Nakagawa. 2007. Loop-mediated isothermal amplification method for detection of human papillomavirus type 6, 11, 16, and 18. Journal of Medical Virology 79:605-615.
Han, J., D. C. Swan, S. J. Smith, S. H. Lum, S. E. Sefers, E. R. Unger, and Y. W. Tang. 2006. Simultaneous amplification and identification of 25 human papillomavirus types with templex technology. Journal of Clinical Microbiology 44:4157-4162.
Handelsman, J. 2004. Metagenomics: Application of genomics to uncultured microorganisms. Microbiology and Molecular Biology Reviews 68:669-685.
Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Research 6:986-994.
Hoban, D. J. 2003. Antibiotics and collateral damage. Clinical Cornerstone Suppl. 3:S12-S20.
Hofstadler, S. A., K. A. Sannes-Lowery, and J. C. Hannis. 2005. Analysis of nucleic acids by FTICR MS. Mass Spectrometry Reviews 24:265-285.
Honer zu Bentrup, K., R. Ramamurthy, C. M. Ott, K. Emami, M. Nelman-Gonzalez, J. W. Wilson, E. G. Richter, T. J. Goodwin, J. S. Alexander, D. L. Pierson, N. Pellis, K. L. Buchanan, and C. A. Nickerson. 2006. Three-dimensional organotypic models of human colonic epithelium to study the early stages of enteric salmonellosis. Microbes and Infection / Institut Pasteur 8:1813-1825.
Honkavuori, K. S., H. L. Shivaprasad, T. Briese, C. Street, D. L. Hirschberg, S. K. Hutchison, and W. I. Lipkin. 2011. Novel picornavirus in turkey poults with hepatitis, California, USA. Emerging Infectious Diseases 17:480-487.
Jabado, O. J., G. Palacios, V. Kapoor, J. Hui, N. Renwick, J. Zhai, T. Briese, and W. I. Lipkin. 2006. Greene SCPrimer: A rapid comprehensive tool for designing degenerate primers from multiple sequence alignments. Nucleic Acids Research 34:6605-6611.
Jayawardena, S., C. Y. Cheung, I. Barr, K. H. Chan, H. Chen, Y. Guan, J. S. Peiris, and L. L. Poon. 2007. Loop-mediated isothermal amplification for influenza A (H5N1) virus. Emerging Infectious Diseases 13:899-901.
Khetsuriani, N., X. Lu, W. G. Teague, N. Kazerouni, L. J. Anderson, and D. D. Erdman. 2008. Novel human rhinoviruses and exacerbation of asthma in children. Emerging Infectious Diseases 14:1793-1796.
Kistler, A., P. C. Avila, S. Rouskin, D. Wang, T. Ward, S. Yagi, D. Schnurr, D. Ganem, J. L. DeRisi, and H. A. Boushey. 2007. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. Journal of Infectious Diseases 196:817-825.
Lamson, D., N. Renwick, V. Kapoor, Z. Liu, G. Palacios, J. Ju, A. Dean, K. St George, T. Briese, and W. I. Lipkin. 2006. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004-2005. Journal of Infectious Diseases 194:1398-1402.
Lau, S. K., C. C. Yip, H. W. Tsoi, R. A. Lee, L. Y. So, Y. L. Lau, K. H. Chan, P. C. Woo, and K. Y. Yuen. 2007. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. Journal of Clinical Microbiology 45:3655-3664.
Lee, W. M., C. Kiesner, T. Pappas, I. Lee, K. Grindle, T. Jartti, B. Jakiela, R. F. Lemanske, Jr., P. A. Shult, and J. E. Gern. 2007. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PloS One 2:e966.
Li, H., M. A. McCormac, R. W. Estes, S. E. Sefers, R. K. Dare, J. D. Chappell, D. D. Erdman, P. F. Wright, and Y. W. Tang. 2007. Simultaneous detection and high-throughput identification of a panel of RNA viruses causing respiratory tract infections. Journal of Clinical Microbiology 45:2105-2109.
Lin, B., K. M. Blaney, A. P. Malanoski, A. G. Ligler, J. M. Schnur, D. Metzgar, K. L. Russell, and D. A. Stenger. 2007. Using a resequencing microarray as a multiple respiratory pathogen detection assay. Journal of Clinical Microbiology 45:443-452.
Lipkin, W. I. 2008. Pathogen discovery. PLoS Pathogens 4:e1000002.
———. 2010. Microbe hunting. Microbiology and Molecular Biology Reviews 74:363-377.
Lipkin, W. I., G. H. Travis, K. M. Carbone, and M. C. Wilson. 1990. Isolation and characterization of borna disease agent cdna clones. Proceedings of the National Academy of Sciences of the United States of America 87:4184-4188.
Lower, R., J. Lower, and R. Kurth. 1996. The viruses in all of us: Characteristics and biological significance of human endogenous retrovirus sequences. Proceedings of the National Academy of Sciences of the United States of America 93:5177-5184.
Mantovani, A., P. Allavena, A. Sica, and F. Balkwill. 2008. Cancer-related inflammation. Nature 454:436-444.
Martina, Y., K. T. Marcucci, S. Cherqui, A. Szabo, T. Drysdale, U. Srinivisan, C. A. Wilson, C. Patience, and D. R. Salomon. 2006. Mice transgenic for a human porcine endogenous retrovirus receptor are susceptible to productive viral infection. Journal of Virology 80:3135-3146.
McCullers, J. A., and K. C. Bartmess. 2003. Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. Journal of Infectious Diseases 187:1000-1009.
Mellmann, A., D. Harmsen, C. A. Cummings, E. B. Zentz, S. R. Leopold, A. Rico, K. Prior, R. Szczepanowski, Y. Ji, W. Zhang, S. F. McLaughlin, J. K. Henkhaus, B. Leopold, M. Bielaszewska, R. Prager, P. M. Brzoska, R. L. Moore, S. Guenther, J. M. Rothberg, and H. Karch. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PloS One 6:e22751.
Mori, M., A. L. Rothman, I. Kurane, J. M. Montoya, K. B. Nolte, J. E. Norman, D. C. Waite, F. T. Koster, and F. A. Ennis. 1999. High levels of cytokine-producing cells in the lung tissues of patients with fatal hantavirus pulmonary syndrome. Journal of Infectious Diseases 179:295-302.
Morse, S. S. 1995. Factors in the emergence of infectious diseases. Emerging Infectious Diseases 1:7-15.
Nichol, S. T., C. F. Spiropoulou, S. Morzunov, P. E. Rollin, T. G. Ksiazek, H. Feldmann, A. Sanchez, J. Childs, S. Zaki, and C. J. Peters. 1993. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science 262:914-917.
Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiology and Molecular Biology Reviews 64:548-572.
Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Research 28:E63.
Oakley, B. B., T. L. Fiedler, J. M. Marrazzo, and D. N. Fredricks. 2008. Diversity of human vaginal bacterial communities and associations with clinically defined bacterial vaginosis. Applied and Environmental Microbiology 74:4898-4909.
Palacios, G., T. Briese, V. Kapoor, O. Jabado, Z. Liu, M. Venter, J. Zhai, N. Renwick, A. Grolla, T. W. Geisbert, C. Drosten, J. Towner, J. Ju, J. Paweska, S. T. Nichol, R. Swanepoel, H. Feldmann, P. B. Jahrling, and W. I. Lipkin. 2006. MassTag polymerase chain reaction for differential diagnosis of viral hemorrhagic fever. Emerging Infectious Diseases 12:692-695.
Palacios, G., P. L. Quan, O. J. Jabado, S. Conlan, D. L. Hirschberg, Y. Liu, J. Zhai, N. Renwick, J. Hui, H. Hegyi, A. Grolla, J. E. Strong, J. S. Towner, T. W. Geisbert, P. B. Jahrling, C. Buchen-Osmond, H. Ellerbrok, M. P. Sanchez-Seco, Y. Lussier, P. Formenty, M. S. Nichol, H. Feldmann, T. Briese, and W. I. Lipkin. 2007. Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerging Infectious Diseases 13:73-81.
Palacios, G., M. Lovoll, T. Tengs, M. Hornig, S. Hutchison, J. Hui, R. T. Kongtorp, N. Savji, A. V. Bussetti, A. Solovyov, A. B. Kristoffersen, C. Celone, C. Street, V. Trifonov, D. L. Hirschberg, R. Rabadan, M. Egholm, E. Rimstad, and W. I. Lipkin. 2010. Heart and skeletal muscle inflammation of farmed salmon is associated with infection with a novel reovirus. PloS One 5:e11487.
Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. Journal of Bacteriology 183:3770-3783.
Paton, N. I., Y. S. Leo, S. R. Zaki, A. P. Auchus, K. E. Lee, A. E. Ling, S. K. Chew, B. Ang, P. E. Rollin, T. Umapathi, I. Sng, C. C. Lee, E. Lim, and T. G. Ksiazek. 1999. Outbreak of Nipah-virus infection among abattoir workers in Singapore. Lancet 354:1253-1256.
Pei, Z., E. J. Bini, L. Yang, M. Zhou, F. Francois, and M. J. Blaser. 2004. Bacterial biota in the human distal esophagus. Proceedings of the National Academy of Sciences of the United States of America 101:4250-4255.
Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325.
Pichichero, M. E. 1998. Group a beta-hemolytic streptococcal infections. Pediatrics in Review 19:291-302.
Relman, D. A., J. S. Loutit, T. M. Schmidt, S. Falkow, and L. S. Tompkins. 1990. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. New England Journal of Medicine 323:1573-1580.
Relman, D. A., T. M. Schmidt, R. P. MacDermott, and S. Falkow. 1992. Identification of the uncultured Bacillus of whipple’s disease. New England Journal of Medicine 327:293-301.
Ren, R. B., F. Costantini, E. J. Gorgacz, J. J. Lee, and V. R. Racaniello. 1990. Transgenic mice expressing a human poliovirus receptor: A new model for poliomyelitis. Cell 63:353-362.
Renwick, N., B. Schweiger, V. Kapoor, Z. Liu, J. Villari, R. Bullmann, R. Miething, T. Briese, and W. I. Lipkin. 2007. A recently identified rhinovirus genotype is associated with severe respiratory-tract infection in children in Germany. Journal of Infectious Diseases 196:1754-1760.
Rivers, T. M. 1937. Viruses and Koch’s postulates. Journal of Bacteriology 33:1-12.
Rothschild, L. J., and R. L. Mancinelli. 2001. Life in extreme environments. Nature 409:1092-1101.
Sampath, R., T. A. Hall, C. Massire, F. Li, L. B. Blyn, M. W. Eshoo, S. A. Hofstadler, and D. J. Ecker. 2007. Rapid identification of emerging infectious agents using PCR and electrospray ionization mass spectrometry. Annals of the New York Academy of Sciences 1102:109-120.
Savage, D. C. 1977. Microbial ecology of the gastrointestinal tract. Annual Review of Microbiology 31:107-133.
Savolainen-Kopra, C., S. Blomqvist, T. Kilpi, M. Roivainen, and T. Hovi. 2009. Novel species of human rhinoviruses in acute otitis media. Pediatric Infectious Disease Journal 28:59-61.
Scott, M., D. Foster, C. Mirenda, D. Serban, F. Coufal, M. Walchli, M. Torchia, D. Groth, G. Carlson, S. J. DeArmond, D. Westaway, and S. B. Prusiner. 1989. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell 59:847-857.
Segelke, B., M. Knapp, S. Kadkhodayan, R. Balhorn, and B. Rupp. 2004. Crystal structure of Clostridium botulinum neurotoxin protease in a product-bound state: Evidence for noncanonical zinc protease activity. Proceedings of the National Academy of Sciences of the United States of America 101:6888-6893.
Shieh, W. J., J. Guarner, M. Layton, A. Fine, J. Miller, D. Nash, G. L. Campbell, J. T. Roehrig, D. J. Gubler, and S. R. Zaki. 2000. The role of pathology in an investigation of an outbreak of West Nile encephalitis in New York, 1999. Emerging Infectious Diseases 6:370-372.
Shirato, K., H. Nishimura, M. Saijo, M. Okamoto, M. Noda, M. Tashiro, and F. Taguchi. 2007. Diagnosis of human respiratory syncytial virus infection using reverse transcription loop-mediated isothermal amplification. Journal of Virological Methods 139:78-84.
Smith, K. M., S. J. Anthony, W. M. Switzer, J. H. Epstein, T. Seimon, H. Jia, M. D. Sanchez, T. T. Huynh, G. G. Galland, S. E. Shapiro, J. M. Sleeman, D. McAloose, M. Stuchin, G. Amato, S. O. Kolokotronis, W. I. Lipkin, W. B. Karesh, P. Daszak, and N. Marano. 2012. Zoonotic viruses associated with illegally imported wildlife products. PloS One 7:e29505.
Suttle, C. A. 2005. Viruses in the sea. Nature 437:356-361.
Templeton, K. E., S. A. Scheltinga, M. F. Beersma, A. C. Kroes, and E. C. Claas. 2004. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza A and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. Journal of Clinical Microbiology 42:1564-1569.
Tumpey, T. M., C. F. Basler, P. V. Aguilar, H. Zeng, A. Solorzano, D. E. Swayne, N. J. Cox, J. M. Katz, J. K. Taubenberger, P. Palese, and A. Garcia-Sastre. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310:77-80.
Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: Probes that fluoresce upon hybridization. Nature Biotechnology 14:303-308.
Van Ert, M. N., S. A. Hofstadler, Y. Jiang, J. D. Busch, D. M. Wagner, J. J. Drader, D. J. Ecker, J. C. Hannis, L. Y. Huynh, J. M. Schupp, T. S. Simonson, and P. Keim. 2004. Mass spectrometry provides accurate characterization of two genetic marker types in Bacillus anthracis. BioTechniques 37:642-644, 646, 648 passim.
VandeWoude, S., J. A. Richt, M. C. Zink, R. Rott, O. Narayan, and J. E. Clements. 1990. A borna virus cDNA encoding a protein recognized by antibodies in humans with behavioral diseases. Science 250:1278-1281.
von Pirquet, C. 1908. Das verhalten der kutanen tuberkulin-reaktion warend der masern. Deutsche Medizinische Wochenschrift 34:1297-1300.
Wang, D., L. Coscoy, M. Zylberberg, P. C. Avila, H. A. Boushey, D. Ganem, and J. L. DeRisi. 2002. Microarray-based detection and genotyping of viral pathogens. Proceedings of the National Academy of Sciences of the United States of America 99:15687-15692.
Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330.
Witso, E., G. Palacios, K. S. Ronningen, O. Cinek, D. Janowitz, M. Rewers, B. Grinde, and W. I. Lipkin. 2007. Asymptomatic circulation of HEV71 in Norway. Virus Research 123:19-29.
Wong, C. W., T. J. Albert, V. B. Vega, J. E. Norton, D. J. Cutler, T. A. Richmond, L. W. Stanton, E. T. Liu, and L. D. Miller. 2004. Tracking the evolution of the sars coronavirus using high-throughput, high-density resequencing arrays. Genome Research 14:398-405.
Yuki, N., K. Susuki, M. Koga, Y. Nishimoto, M. Odaka, K. Hirata, K. Taguchi, T. Miyatake, K. Furukawa, T. Kobata, and M. Yamada. 2004. Carbohydrate mimicry between human ganglioside gm1 and Campylobacter jejuni lipooligosaccharide causes Guillain-Barre syndrome. Proceedings of the National Academy of Sciences of the United States of America 101:11404-11409.
Zhang, H., J. K. DiBaise, A. Zuccolo, D. Kudrna, M. Braidotti, Y. Yu, P. Parameswaran, M. D. Crowell, R. Wing, B. E. Rittmann, and R. Krajmalnik-Brown. 2009. Human gut microbiota in obesity and after gastric bypass. Proceedings of the National Academy of Sciences of the United States of America 106:2365-2370.
zur Hausen, H. 2002. Papillomaviruses and cancer: From basic studies to clinical application. Nature Reviews: Cancer 2:342-350.
TRANSMISSION OF HUMAN INFECTION WITH NIPAH VIRUS21
Stephen P. Luby,22,23Emily S. Gurley,22and M. Jahangir Hossain22
Nipah virus (NiV) is a paramyxovirus whose reservoir host is fruit bats of the genus Pteropus. Occasionally the virus is introduced into human populations and causes severe illness characterized by encephalitis or respiratory disease. The first outbreak of NiV was recognized in Malaysia, but 8 outbreaks have been reported from Bangladesh since 2001. The primary pathways of transmission from bats to people in Bangladesh are through
_______________
21 Stephen P. Luby, Emily S. Gurley, M. Jahangir Hossain. 2009. Transmission of Human Infection with Nipah Virus. Clinical Infectious Diseases, 49(11):1743-8.
Reprinted by permission of Oxford University Press.
22 International Centre for Diarrheal Diseases Research, Bangladesh, Dhaka, Bangladesh.
23 Centers for Disease Control and Prevention, Division of Emerging Infections and Surveillance Services, Atlanta, Georgia.
Received 1 May 2009; accepted 19 July 2009; electronically published 2 November 2009. Reprints and correspondence: Dr Stephen P. Luby, International Centre for Diarrheal Diseases Research, Bangladesh, GPO Box 128, Mohakhali, Dhaka 1212 Bangladesh (sluby@icddrb.org).
Clinical Infectious Diseases 2009; 49:000–000
2009 by the Infectious Diseases Society of America. All rights reserved.
1058-4838/2009/4911-00XX$15.00
DOI: 10.1086/647951
contamination of raw date palm sap by bats with subsequent consumption by humans and through infection of domestic animals (cattle, pigs, and goats), presumably from consumption of food contaminated with bat saliva or urine with subsequent transmission to people. Approximately one-half of recognized Nipah case patients in Bangladesh developed their disease following person-to-person transmission of the virus. Efforts to prevent transmission should focus on decreasing bat access to date palm sap and reducing family members’ and friends’ exposure to infected patients’ saliva.
Human Nipah virus (NiV) infection was first recognized in a large outbreak of 276 reported cases in peninsular Malaysia and Singapore from September 1998 through May 1999 (Chua, 2003; Chua et al., 2000; Paton et al., 1999). Most patients had contact with sick pigs (Parashar et al., 2000). Patients presented primarily with encephalitis; 39% died (Chua, 2003; Goh et al., 2000). Autopsy studies noted diffuse vasculitis most prominently involving the central nervous system with intense immunostaining of endothelial cells with anti—Nipah virus hyperimmune serum (Chua et al., 2000). The virus, a member of the recently designated genus Henipavirus, within the family Paramyxoviridae, was first isolated from a patient from Sungai Nipah village (Chua, 2003; Chua et al., 2000). The human outbreak of Nipah infection ceased after widespread deployment of personal protective equipment to people contacting sick pigs, restriction on livestock movements, and culling over 900,000 pigs (Uppal, 2000).
Large fruit bats of the genus Pteropus appear to be the natural reservoir of NiV. In Malaysia the seroprevalence of neutralizing antibodies to NiV in colonies of Pteropus vampyrus and Pteropus hypomelanus ranged from 7% to 58% (Yob et al., 2001; Daszak et al., 2006). Antibodies against henipaviruses have been identified in Pteropus bats wherever they have been tested including Cambodia, Thailand, India, Bangladesh, and Madagascar (Epstein et al., 2004; Hsu et al., 2004; Iehle et al., 2007; Reynes et al., 2005; Wacharapluesadee et al., 2005). NiV was isolated from urine specimens collected underneath a P. hypomelanus roost and from partially eaten fruit dropped during feeding activity in Malaysia (Chua et al., 2002), from urine collected underneath a Pteropus lylei roost in Cambodia (Reynes et al., 2005), and from saliva and urine of P. lylei in Thailand ( Wacharapluesadee, 2005). Experimental infection of Pteropus bats with NiV does not cause illness in the bats (Middleton et al., 2007). Surveys of rodents and other animals have not identified other wildlife reservoirs for NiV (Hsu et al., 2004; Yob et al., 2001). Over 50 species of Pteropus bats live in South and South East Asia (Figure A11-1) (Nowak, 1994). Pteropus giganteus, the only Pteropus species found in Bangladesh, is widely distributed across the country and frequently has antibody to NiV (Bates and Harrison, 1997; Hsu et al., 2004).
The growth of large intensively managed commercial pig farms in Malaysia with fruit trees on the farm created an environment where bats could drop partially eaten fruit contaminated with NiV laden bat saliva into pig stalls. The pigs
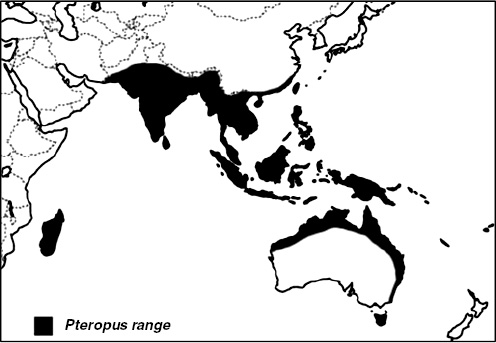
FIGURE A11-1 Range of Pteropus bats based on RM Nowak (Nowak, 1994).
could eat the fruit, become infected with NiV, and efficiently transmit virus to other pigs because of the dense pig population on the farms, frequent respiratory shedding of the virus among infected pigs (Middleton et al., 2002), and the pigs’ high birth rate that regularly brought newly susceptible young pigs into the population at risk (Epstein et al., 2006).
Recurrent Outbreaks of NiV Infection in Bangladesh
In the 10 years following the Nipah outbreak in Malaysia, no further human cases of NiV infection have been reported from Malaysia, but 8 human outbreaks of NiV infection in Bangladesh were reported from 2001 through 2008, all occurring between December and May (Gurley et al., 2007; Hsu et al., 2004; ICDDRB, 2007; ICDDRB, 2008; Luby et al., 2006; Montgomery et al., 2008). A total of 135 human cases of Nipah infection in Bangladesh were recognized; 98 (73%) died. One outbreak of NiV occurred in Siliguri, India, 15 kilometers north of the Bangladesh border in January and February 2001 (Chadha et al., 2006) and a second NiV outbreak was reported by newspapers in Nadia District, India also close to the border with Bangladesh, in 2007 (Mandal and Banerjee, 2007). In addition to the outbreaks, between 2001 and 2007, 17 other NiV transmission events, ranging from single sporadic human cases to clusters of 2–4 human
cases were recognized in Bangladesh (Luby et al., 2009). Thus, in contrast to the Malaysia-Singapore outbreak, which could be coherently explained by a single or perhaps a few transmissions of NiV from an infected bat to pigs, leading to a porcine epidemic which in turn led to a human epidemic (Epstein et al., 2006), in Bangladesh NiV transmission from bats to human is repeated and ongoing.
The diversity of NiV strains recovered from Bangladesh also supports multiple introductions of the virus from bats into human populations even within a single year. Among 4 NiV isolates from human NiV cases in 2004, the sequences of the nucleoprotein open reading frames of the isolates differed by 0.9% in nucleotide homology, in contrast to the sequences obtained from all of the human cases in Malaysia which were nearly identical to each other (AbuBakar et al., 2004; Chan et al., 2001; Harcourt et al., 2005).
The clinical presentation of NiV infection in Bangladesh differed from Malaysia. In Bangladesh, severe respiratory disease is more common, with 62% of cases having cough, 69% developing respiratory difficulty, and available chest radiographs showing diffuse bilateral opacities covering the majority of the lung fields (Hossain et al., 2008). By contrast, in Malaysia, 14% of patients had a nonproductive cough on presentation; only 6% of chest radiographs were abnormal and these abnormalities were mild and focal (Goh et al., 2000). The case fatality rate was higher in Bangladesh at 73%, compared with 39% from Malaysia (Goh et al., 2000; Hossain et al., 2008), but much of this difference results from the more sophisticated clinical care provided in Malaysia. One-half of Malaysian Nipah patients received mechanical ventilatory support compared to a single patient (1%) in Bangladesh (Goh et al., 2000) (unpublished data). One third of Nipah survivors in Bangladesh have moderate to severe objective neurological dysfunction 7–30 months after infection (Sejvar et al., 2007).
NiV Transmission from Bats to People
Epidemiological investigations in Bangladesh have identified three pathways of transmission of NiV from bats to people. The most frequently implicated route is ingestion of fresh date palm sap. Date palm sap is harvested from December through March, particularly in west central Bangladesh. A tap is cut into the tree trunk and sap flows slowly overnight into an open clay pot. Infrared camera studies confirm that P. giganteus bats frequently visit date palm sap trees and lick the sap during collection (Khan et al., 2008). NiV can survive for days on sugar-rich solutions such as fruit pulp (Fogarty et al., 2008). Most date palm sap is processed at high temperature to make molasses, but some is enjoyed as a fresh juice, drunk raw within a few hours of collection. In the 2005 Nipah outbreak in Tangail District, Bangladesh, the only exposure significantly associated with illness was drinking raw date palm sap (64% of case patients vs 18% of control patients; odds ratio [OR], 7.9; 95% confidence interval [CI], 1.6–38; P = .01) (Luby et al., 2006). Twenty-one of the 23 index NiV case patients recognized
in Bangladesh developed their initial symptoms during the December through March date palm sap collection season (Luby et al., 2009).
A second route of transmission for NiV from bats to people in Bangladesh is via domestic animals. Fruit bats commonly drop partially eaten saliva-laden fruit. Domestic animals in Bangladesh forage for such food. Date palm sap that is contaminated with bat feces and so is unfit for human consumption is also occasionally fed to domestic animals. The domestic animals may become infected with NiV, and shed the virus to other animals, including humans. Contact with a sick cow in Meherpur, Bangladesh in 2001 was strongly associated with Nipah infection (OR, 7.9; 95% CI, 2.2–27.7; P = .001) (Hsu et al., 2004). A pig herd visited the community two weeks before the 2003 Nipah outbreak in Naogaon and contact with the pigs was associated with illness (OR, 6.1; 95% CI, 1.3–27.8; P = .007) (Khan et al., 2008). In 2004, one family explained that they owned 2 goats that their son frequently played with. The goats became ill with fever, difficulty walking, walking in circles, and frothing at the mouth. The parents believe their son had contact with goat saliva while the goats were ill. Both goats died. Within 2 weeks of the goats’ death, the child developed encephalitis that was confirmed to be Nipah by antibody testing (unpublished data). Third, some people may come into direct contact with NiV-infected bat secretions. In the Goalando outbreak in 2004, persons who climbed trees were more likely to develop NiV infection than were control patients (OR, 8.2; 95% CI, 1.3—![]() ) (Montgomery et al., 2008).
) (Montgomery et al., 2008).
Person-to-Person Transmission
Several Bangladesh Nipah outbreaks resulted from person-to-person transmission. The clearest illustration of person-to-person NiV transmission occurred during the Faridpur outbreak in 2004 (Gurley et al., 2007a). Four persons who cared for the index patient—his mother, his son, his aunt, and a neighbor—became ill 15–27 days after the index patient first developed illness (Figure A11-2). During her hospitalization, the index patient’s aunt was cared for by a popular religious leader who lived in a nearby village and who became ill 13 days later. When the religious leader became seriously ill, many of his relatives and members of his religious community visited at his home. Twenty-two persons developed Nipah infection after contact with the religious leader. One of these followers moved to his family’s house in an adjacent village to receive care after becoming ill where he was cared for by a friend and 2 family members. These 3 caregivers and a rickshaw driver, who helped carry him to the hospital as his condition deteriorated, became ill. Ultimately, the chain of transmission involved 5 generations and affected 34 people (Gurley et al., 2007a) (Figure A11-2). Physical contact with an NiV-infected patient who later died (OR, 13.4; 95% CI, 2.0–89) was the strongest risk factor for developing NiV infection in the outbreak.
The transmission pattern in Faridpur is not unique. For example, in 2007 in Thakurgaon, 6 family members and friends who cared for an NiV-infected patient
FIGURE A11-2 Chain of person to person transmission in Nipah outbreak, Faridpur, Bangladesh, 2004.
developed Nipah infection. Case patients were more likely than control patients to have been in the same room when the index case was coughing (ICDDRB, 2007). In a review of the 122 Nipah case patients identified in Bangladesh from 2001 through 2007, 62 (51%) developed illness after close contact with another Nipah patient (Luby et al., 2009). A small minority of patients infected with NiV (ie, 9 [7%] of 122 recognized cases) transmitted NiV to 62 other persons.
Respiratory secretions appear to be particularly important for person-to-person transmission of NiV. NiV RNA is readily identified in the saliva of infected patients (Chua et al., 2001; Harcourt et al., 2005). Anthropological investigations during the Faridpur outbreak highlighted multiple opportunities for the transfer of NiV contaminated saliva from a sick patient to care providers (Blum et al., 2009). Social norms in Bangladesh require family members to maintain close physical contact during illness. The more severe the illness, the more hands-
on care is expected. Family members and friends without formal health care or infection control training provided nearly all the hands on care to Nipah patients both at home and in the hospital (Hadley et al., 2007). Care providers during the Faridpur outbreak continued to share eating utensils and drinking glasses with sick patients. Leftovers of food offered to ill Nipah patients were commonly distributed to other family members. Family members maintained their regular sleeping arrangements, which often involved sleeping in the same bed with a sick, coughing Nipah patient. There was a particularly strong desire to have close physical contact near the time of death, demonstrated by such behaviors as cradling the patients head on the family member’s lap, attempting to give liquids to the patient with a spoon or glass between bouts of coughing, or hugging and kissing the sick patient (Blum et al., 2009). In both the Faridpur outbreak in 2004 and the Thakurgaon outbreak in 2007, persons who were in a room when a Nipah patient was coughing or sneezing were at increased risk of Nipah virus infection (Gurley et al., 2007a; ICDDRB, 2007). Across all recognized outbreaks in Bangladesh from 2001 through 2007, Nipah patients with respiratory symptoms were more likely to transmit Nipah (Luby et al., 2009).
The capacity for NiV to spread in hospital settings to both staff and patients was clearly illustrated in a large outbreak affecting 66 people in Siliguri, India in 2001. The outbreak apparently originated from an unidentified patient admitted to Siliguri District Hospital who transmitted infection to 11 additional patients, all of whom were transferred to other facilities. In 2 of the facilities, subsequent transmission infected 25 staff and 8 visitors (Chadha et al., 2006). However, transmission to health care workers is rarely recognized. Among a cohort of 338 health care workers who cared for Nipah patients at 3 Malaysian hospitals and reported a combined 89 episodes of Nipah patient blood or body fluid directly contacting their bare skin, 39 splash exposures of blood or body fluid into their eyes, nose or mouth, and 12 needle stick injuries, none developed clinical illness associated with Nipah infection (Mounts et al., 2001). Health care workers in Bangladesh have much less direct physical contact with patients than in western hospitals (Hadley et al., 2007). Hands-on care is generally provided by family members and friends. No health care workers in Bangladesh who cared for identified Nipah patients have been identified with illness, although confirmed cases include 1 physician whose source of infection is unknown. A serosurvey among 105 health care workers who cared for at least 1 of 7 patients admitted with Nipah infection at one hospital in Bangladesh identified 2 health care workers with serological evidence of NiV infection; however, their antibody responses (IgG only, no IgM) and lack of symptoms suggest a previous infection, not recent nosocomial transmission (Gurley et al., 2007b).
Might person-to-person transmission be associated with particular strains of NiV that have genetic characteristics that lead to person-to-person transmission? The closely related strains in Malaysia resulted in less frequent and less severe respiratory disease than observed in Bangladesh and were not associated
with frequent person-to-person transmission. However, the pattern of the outbreaks in Bangladesh and India suggests that person-to-person transmission is more dependent on the characteristics of the occasional Nipah transmitter than a specific strain. If the NiV strain was central to person-to-person transmission, then secondary cases of NiV would be more likely to become NiV transmitters, than primary cases (because secondary cases would already have selected for strains predisposed to person-to-person transmission). However, in the review of 7 years of human Nipah infection in Bangladesh, secondary cases were no more or less likely to become Nipah transmitters than were primary cases (Luby et al., 2009). All persons who transmitted Nipah died, suggesting that late stages of infection, presumably with high virus titers, increases the risk of transmission. Even the pattern in Siliguri, the largest recognized Nipah outbreak from apparent person-to-person transmission, is consistent with the review of 7 years of human Nipah infection in Bangladesh. The unidentified index case in Siliguri District Hospital infected 11 patients, 2 of whom infected an additional 33 patients. The 13 day duration of the outbreak at Medinova Hospital suggests 2 generations of transmission likely occurred there. Taken together, this pattern suggests 4 NiV transmitters propagated human infection across 4 generations. There were 67 cases (66 recognized plus the unidentified index case), 4 (5.9%) of whom became Nipah transmitters, a proportion very close to the 7% recognized in Bangladesh. This suggests that the virus strain responsible for this largest recognized person-to-person outbreak was not exceptional. Its rate of secondary transmission was similar to other strains circulating in South Asia.
Exposures Not Associated with NiV Transmission
Outbreak investigations have both identified important routes of transmission of human NiV infection, and identified exposures not associated with transmission. NiV was recovered from the urine of Pteropus bats in Malaysia, Cambodia, and Thailand (Iehle et al., 2007; Reynes et al., 2005; Wacharapluesadee et al., 2005). In Bangladesh, P. giganteus bats live in close proximity to human populations, often roosting in trees located in rural Bangladeshi villages. Thus, bat urine, intermittently laced with NiV, contaminates the immediate physical environment in many villages in Bangladesh. Yet in each of the 8 Nipah outbreaks investigated in Bangladesh, an association between living near a bat roost and infection with Nipah was looked for but was never found. This suggests that the quantity of viable virus shed in bat urine is too low to initiate clinically apparent infection in humans.
Eating bat-bitten fruit is often suggested as a pathway of transmission for human Nipah infection. NiV was recovered from fruit dropped by Pteropus bats in Malaysia (Chua et al., 2002). It is the most commonly suggested pathway for NiV transmission from bats to domestic animals. In contrast to general environmental contamination with urine, punctured fruit contaminated with bat saliva
may favor virus survival. In Bangladesh where 43% of children under the age of 5 years meet the World Health Organization standards for chronic malnutrition (NIPORT, 2007), little food is wasted. In outbreak investigations, villagers, especially children, commonly report consuming fruit which was partially eaten by bats. However, in the 6 NiV outbreak investigations where the question was asked, case patients never reported consuming partially eaten fruit significantly more than did controls.
Unanswered Questions
Did outbreaks of human NiV infection occur in Bangladesh before the first outbreak was recognized in 2001? Almost certainly. P. giganteus are widely distributed across Bangladesh (Nowak, 1994), and wherever Pteropus bats have been tested they have antibody to henipavirus (Epstein et al., 2008; Hsu et al., 2004; Iehle et al., 2007; Reynes et al., 2005; Wacharapluesadee et al., 2005). When Pteropus bats are experimentally infected with NiV they do not become clinically ill (Middleton et al., 2007), which suggests that NiV likely coevolved with its Pteropus hosts over millennia. Bangladesh has long been densely populated, and date palm sap harvesting is an old profession using techniques and simple tools that are passed on from father to son. Moreover, people frequently die in Bangladesh of unknown causes, often outside of hospitals. Three factors that have contributed to recognition of Nipah outbreaks recently include development of diagnostic tests for Nipah infection following the Malaysian outbreak, expansion of surveillance for a range of communicable disease by the government of Bangladesh, and expansion of news media coverage in rural Bangladesh.
Unanswered questions regarding Nipah transmission include the following: (1) Why is respiratory disease and person-to-person transmission more common among human NiV infection in Bangladesh compared to Malaysia? Are certain strains of virus more likely to cause respiratory tract disease in humans, or might the different clinical syndromes in Bangladesh and Malaysia reflect differences in host susceptibility from malnutrition or other causes? (2) How stable is the genome of Nipah? The overall nucleotide homology between a prototypical Malaysian strain of NiV and a strain of NiV from Bangladesh was 91.8% ( Harcourt et al., 2005). Is there a substantial risk of mutation that would improve the efficiency of person-to-person transmission of the virus? (3) How common is unrecognized, including subclinical, infection with NiV?
Prevention Strategies
The epidemiology of NiV transmission in Bangladesh suggests two avenues to prevent human disease. The first is limiting exposure of Bangladeshi villagers to NiV contaminated fresh date palm sap. Date palm sap collection provides critical income to low-income collectors and is a seasonal national delicacy enjoyed
by millions every year. Steps to make the date palm sap consumption safer include diverting more of the production to molasses where the sap is cooked at temperatures above the level that NiV can survive and limiting bat access to date palm trees where the sap will be consumed fresh. A number of methods have been occasionally employed by date palm sap collectors to restrict bat access to date palm trees (Nahar et al., 2008). We are currently evaluating the effectiveness and scalability of these methods.
A second area for targeted intervention is reducing the exposure of caretakers to the saliva of seriously ill persons. When a Nipah outbreak is recognized, it is appropriate to implement standard precautions (Siegel et al., 2007), but recommendations to improve infection control practices more broadly in Bangladesh must consider the social and health care context in the country, where (1) the annual total per capita spending on health is $12 per person per year (Health Economics Unit MoHaFW, 2007); (2) over 99% of respiratory disease and over 99% of acute meningoencephalitis in Bangladesh is not caused by Nipah; (3) most of the people who contract Nipah are dead by the time the diagnosis is considered by local practitioners; and (4) even in the hospital setting most hands-on care is provided by family members, not health care professionals. If we recommend an unachievable level of infection control practices for persons caring for pneumonia and acute meningoencephalitis patients from rural communities in Bangladesh, we will not reduce the risk of person-to-person transmission of NiV in Bangladesh. An important research priority is to identify approaches that can be consistently implemented in these low income settings where family members are caring for patients with severe respiratory and neurological disease. For example, family members who washed their hands with soap after caring for Nipah patients were significantly less likely to become infected (Gurley et al., 2007b). If such practices were widely adopted, they would lessen the risk of person-to-person transmission of NiV and other pathogens.
Acknowledgments
We are grateful to the many contributors to Nipah outbreak investigations in Bangladesh since 2001, both those recognized as coauthors in earlier publications and the many field workers, laboratory technicians, and support staff whose willingness to promptly and thoroughly investigate outbreaks of this dangerous pathogen has been essential to our improved understanding of NiV epidemiology. We are also grateful to the community of scientists interested in NiV who have enlarged our understanding by posing pointed questions and engaged the authors in prolonged discussions on Nipah transmission. We thank Michelle Luby for her assistance with drawing Figure A11-1 and Milton Quiah for his administrative support.
Financial support. The investigations of human epidemiology of NiV infection in Bangladesh was funded by the Centers for Disease Control and Preven-
tion, the US National Institutes of Health Division of Microbiology and Infectious Diseases, International Collaborations in Infectious Disease Opportunity Pool, and the government of Bangladesh through the Improved Health for the Poor: Health, Nutrition and Population Research Project (grant MOHFW/HEALTH/AC-5/HNPR/ICDD,B/30/2003). The International Centre for Diarrheal Diseases Research, Bangladesh, acknowledges with gratitude the commitment of the Centers for Disease Control and Prevention, the National Institutes of Health, and the government of Bangladesh to the centre’s research efforts. Potential conflicts of interest. All authors: no conflicts.
References
AbuBakar S, Chang LY, Ali AR, Sharifah SH, Yusoff K, Zamrod Z. Isolation and molecular identification of Nipah virus from pigs. Emerg Infect Dis 2004; 10:2228–30.
Bates PJJ, Harrison DL. Bats of the Indian subcontinent. Kent, UK: Harrison Zoological Museum, 1997.
Blum LS, Khan R, Nahar N, Breiman RF. In-depth assessment of an outbreak of Nipah encephalitis with person-to-person transmission in Bangladesh: implications for prevention and control strategies. Am J Trop Med Hyg 2009; 80:96–102.
Chadha MS, Comer JA, Lowe L, et al. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg Infect Dis 2006; 12:235–40.
Chan YP, Chua KB, Koh CL, Lim ME, Lam SK. Complete nucleotide sequences of Nipah virus isolates from Malaysia. J Gen Virol 2001; 82(Pt 9):2151–5.
Chua KB, Bellini WJ, Rota PA, et al. Nipah virus: a recently emergent deadly paramyxovirus. Science 2000; 288:1432–5.
Chua KB, Koh CL, Hooi PS, et al. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect 2002; 4:145–51.
Chua KB, Lam SK, Goh KJ, et al. The presence of Nipah virus in respiratory secretions and urine of patients during an outbreak of Nipah virus encephalitis in Malaysia. J Infect 2001; 42:40–3.
Chua KB. Nipah virus outbreak in Malaysia. J Clin Virol 2003; 26:265–75.
Daszak P, Plowright R, Epstein JH, et al; HERG. The emergence of Nipah and Hendra virus: pathogen dynamics across a wildlife-livestock-human continuum. In: Collinge S, Ray C, ed. Disease ecology: community structure and pathogen dynamics. Oxford: Oxford University Press, 2006:186–201.
Epstein JH, Field HE, Luby S, Pulliam JR, Daszak P. Nipah virus: impact, origins, and causes of emergence. Curr Infect Dis Rep 2006; 8:59–65.
Epstein JH, Prakash VB, Smith CS, et al. Henipavirus infection in fruit bats (Pteropus giganteus), India. Emerg Infect Dis 2008; 14:1309–11.
Fogarty R, Halpin K, Hyatt AD, Daszak P, Mungall BA. Henipavirus susceptibility to environmental variables. Virus Res 2008; 132:140–4.
Goh KJ, Tan CT, Chew NK, et al. Clinical features of Nipah virus encephalitis among pig farmers in Malaysia. N Engl J Med 2000; 342:1229–35.
Gurley ES, Montgomery JM, Hossain MJ, et al. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg Infect Dis 2007a; 13:1031–7.
Gurley ES, Montgomery JM, Hossain MJ, et al. Risk of nosocomial transmission of nipah virus in a Bangladesh hospital. Infect Control Hosp Epidemiol 2007b; 28:740–2.
Hadley MB, Blum LS, Mujaddid S, et al. Why Bangladeshi nurses avoid ‘nursing’: social and structural factors on hospital wards in Bangladesh. Soc Sci Med 2007; 64:1166–77.
Harcourt BH, Lowe L, Tamin A, et al. Genetic characterization of Nipah virus, Bangladesh, 2004. Emerg Infect Dis 2005; 11:1594–7.
Health Economics Unit MoHaFW. A fact book on the Bangladesh HNP sector. Dhaka: Health Economics Unit, Ministry of Health and Family Welfare, 2007.
Hossain MJ, Gurley ES, Montgomery JM, et al. Clinical presentation of nipah virus infection in Bangladesh. Clin Infect Dis 2008; 46:977–84.
Hsu VP, Hossain MJ, Parashar UD, et al. Nipah virus encephalitis reemergence, Bangladesh. Emerg Infect Dis 2004; 10:2082–7.
ICDDRB. Outbreaks of encephalitis due to Nipah/Hendra-like viruses, western Bangladesh. Health Sci Bull 2003; 1(1):1–6.
ICDDRB. Person to person transmission of Nipah infection in Bangladesh, 2007. Health Sci Bull 2007; 5(4):1–6.
ICDDRB. Outbreaks of Nipah virus in Rajbari and Manikgonj, February. Health Sci Bull 2008; 6(1):12–3.
Iehle C, Razafitrimo G, Razainirina J, et al. Henipavirus and Tioman virus antibodies in pteropodid bats, Madagascar. Emerg Infect Dis 2007; 13:159–61.
Khan M, Nahar N, Sultana R, Hossain M, Gurley ES, Luby S. Understanding bats access to date palm sap: identifying preventative tech niques for Nipah virus transmission. New Orleans: American Society of Tropical Medicine and Hygiene, 2008:331.
Luby S, Hossain J, Gurley E, et al. Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001–2007. Emerg Infect Dis 2009; 15:1229–35.
Luby SP, Rahman M, Hossain MJ, et al. Foodborne transmission of Nipah virus, Bangladesh. Emerg Infect Dis 2006; 12:1888–94.
Mandal S, Banerjee R. Bat virus in Bengal. The Telegraph. 8 May 2007, 2007.
Middleton DJ, Morrissy CJ, van der Heide BM, et al. Experimental Nipah virus infection in pteropid bats (Pteropus poliocephalus). J Comp Pathol 2007; 136:266–72.
Middleton DJ, Westbury HA, Morrissy CJ, et al. Experimental Nipah virus infection in pigs and cats. J Comp Pathol 2002; 126:124–36.
Montgomery JM, Hossain MJ, Gurley E, et al. Risk factors for Nipah virus encephalitis in Bangladesh. Emerg Infect Dis 2008; 14:1526–32.
Mounts AW, Kaur H, Parashar UD, et al. A cohort study of health care workers to assess nosocomial transmissibility of Nipah virus, Malaysia, 1999. J Infect Dis 2001; 183:810–3.
Nahar N, Sultana R, Oliveras E, et al. Preventing Nipah virus infection: interventions to interrupt bats accessing date palm sap. New Orleans: American Society of Tropical Medicine and Hygiene, 2008:210.
NIPORT. Bangladesh demographic and health survey 2007. Dhaka, Bangladesh and Calverton, Maryland [USA]: National Institute of Population Research and Training, Mitra and Associates, 2007.
Nowak R. Walker’s bats of the world. Baltimore: Johns Hopkins University Press, 1994.
Parashar UD, Sunn LM, Ong F, et al. Case-control study of risk factors for human infection with a new zoonotic paramyxovirus, Nipah virus, during a 1998–1999 outbreak of severe encephalitis in Malaysia. J Infect Dis 2000; 181:1755–9.
Paton NI, Leo YS, Zaki SR, et al. Outbreak of Nipah-virus infection among abattoir workers in Singapore. Lancet 1999; 354:1253–6.
Reynes JM, Counor D, Ong S, et al. Nipah virus in Lyle’s flying foxes, Cambodia. Emerg Infect Dis 2005; 11:1042–7.
Sejvar JJ, Hossain J, Saha SK, et al. Long-term neurological and func tional outcome in Nipah virus infection. Ann Neurol 2007; 62:235–42.
Siegel JD, Rhinehart E, Jackson M, Chiarello L. 2007 guideline for isolation precautions: preventing transmission of infectious agents in health care settings. Am J Infect Control 2007; 35(10 Suppl 2):S65–164.
Uppal PK. Emergence of Nipah virus in Malaysia. Ann N Y Acad Sci 2000; 916:354–7.
Wacharapluesadee S, Lumlertdacha B, Boongird K, et al. Bat Nipah virus, Thailand. Emerg Infect Dis 2005; 11:1949–51.
Yob JM, Field H, Rashdi AM, et al. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg Infect Dis 2001; 7:439–41.
DATE PALM SAP LINKED TO NIPAH VIRUS OUTBREAK IN BANGLADESH, 200824
Muhammad Aziz Rahman,25,26Mohammad Jahangir Hossain,25Sharmin Sultana,26Nusrat Homaira,25,26Salah Uddin Khan,25Mahmudur Rahman,26Emily S. Gurley,25Pierre E. Rollin,27Michael K. Lo,27James A. Comer,27Luis Lowe,27Paul A. Rota,27Thomas G. Ksiazek,27,28Eben Kenah,29Yushuf Sharker,25and Stephen P. Luby25,27
Abstract
Introduction: We investigated a cluster of patients with encephalitis in the Manikgonj and Rajbari Districts of Bangladesh in February 2008 to determine the etiology and risk factors for disease.
Methods: We classified persons as confirmed Nipah cases by the presence of immuno globulin M antibodies against Nipah virus (NiV), or by the presence of NiV RNA or by isolation of NiV from cerebrospinal fluid or throat swabs who had onset of symptoms between February 6 and March 10, 2008. We classified persons as probable cases if they reported fever with convulsions or altered mental status, who resided in the outbreak areas during that period, and who died before serum samples were collected. For the case—control study, we compared both confirmed and probable Nipah case-patients to controls, who were free from illness during the reference period. We used motion-sensor-infrared cameras to observe bat’s contact of date palm sap.
______________
24 Reprinted with permission from Vector-borne and Zoonotic Diseases 12/1, published by Mary Ann Liebert, Inc., New Rochelle, New York.
25 International Centre for Diarrhoeal Disease Research, Bangladesh, Dhaka, Bangladesh.
26 Institute of Epidemiology, Disease Control and Research, Dhaka, Bangladesh.
27 Centers for Disease Control and Prevention, Atlanta, Georgia.
28 Galveston National Laboratory, Department of Pathology, University of Texas Medical Branch, Galveston, Texas.
29 Department of Biostatistics, University of Washington, Seattle.
Address correspondence to: Muhammad Aziz Rahman, Discipline of Public Health, The University of Adelaide, Level-9, 10 Pulteney St., Adelaide, SA-5000, Australia. E-mail: aziz.rahman@adelaide.edu.au.
Results: We identified four confirmed and six probable case-patients, nine (90%) of whom died. The median age of the cases was 10 years; eight were males. The outbreak occurred simultaneously in two communities that were 44km apart and separated by a river. Drinking raw date palm sap 2–12 days before illness onset was the only risk factor most strongly associated with the illness (adjusted odds ratio 25, 95% confidence intervals 3.3—N, p < 0.001). Case-patients reported no history of physical contact with bats, though community members often reported seeing bats. Infrared camera photographs showed that Pteropus bats frequently visited date palm trees in those communities where sap was collected for human consumption.
Conclusion: This is the second Nipah outbreak in Bangladesh where date palm sap has been implicated as the vehicle of transmission. Fresh date palm sap should not be drunk, unless effective steps have been taken to prevent bat access to the sap during collection.
Introduction
Nipah virus (NiV) causes encephalitis in humans and has a high fatality rate (Hossain et al., 2008; Luby et al., 2009). Species of fruit-bats in the Pteropus genus are the presumed natural reservoirs of NiV. NiV has been isolated and/ or NiV RNA has been identified in bats in Malaysia (Chua et al., 2002; Rahman et al., 2010), Cambodia (Reynes et al., 2005), and Thailand (Wacharapluesadee et al., 2005). Researchers identified antibodies against NiV in Pteropus fruit-bats in Malaysia, India, and Bangladesh (Yob et al., 2001; Hsu et al., 2004; Epstein et al., 2008). There were seven recognized Nipah outbreaks in Bangladesh from 2001 to 2007 (Luby et al., 2009) (Figure A12-1). Investigators implicated various routes of transmission in those outbreaks, including person-to-person transmission, drinking raw date palm sap, and contact with sick animals (Luby et al., 2006, 2009; Gurley et al., 2007).
Surveillance for human Nipah infection has been ongoing since 2006 in six hospitals serving communities in the northwestern districts of Bangladesh where previous Nipah outbreaks have been reported (Figure A12-1). The surveillance has been conducted by the Institute of Epidemiology, Disease Control and Research (IEDCR), in collaboration with the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR, B).
On February 26, 2008, government health workers reported that one child from Manikgonj District died with illness characterized by fever, generalized body ache, cough, difficulty breathing, and mental status changes. Another two siblings from the same household were admitted to a subdistrict healthcare facility in Manikgonj District with similar symptoms. Both of them were referred to the District Hospital for more advanced medical care; one of them died on the way to the referral hospital and the other child was taken to a private hospital in Dhaka District, where the child died on the following day. On February 27,
2008, a Nipah Surveillance physician from the Rajbari District reported another cluster of encephalitis cases. Four patients from the same village were admitted to the Rajbari District Hospital; another patient from the same area died before reaching the hospital and another patient from the neighboring village died upon reaching the hospital. All of them presented with fever, generalized weakness, cough or respiratory distress, progressive mental status changes, and unconsciousness.
IEDCR initiated an investigation of both clusters in collaboration with ICDDR,B on the day they received the reports. The objectives of the investigation were to determine the cause of the outbreak, identify risk factors for illness, and develop strategies for prevention.
Materials and Methods
Study Site and Study Population
The communities affected by the outbreak were located in Doulatpur Upazila (subdistrict) (population 155,674) of Manikgonj District and Baliakandi Upazila (population 186,562) of Rajbari District in central-western region of Bangladesh (Bangladesh Bureau of Statistics, 2010). These sites are located 44km apart and are separated by a river (Figure A12-1). Date palm sap is widely harvested, sold, and consumed in both areas.
Case Identification and Sample Collection
The investigation team sought for suspect cases who had fever and convulsions or altered mental status in the outbreak areas between February 6 and March 10, 2008 (Figure A12-2). First, we visited the hospitals where the patients were treated. We identified the suspect case-patients and interviewed them or their proxy respondents. Then we searched for additional case-patients in the affected villages. We collected blood, cerebrospinal fluid (CSF), and throat swab samples from all the living suspect case-patients. The serum and CSF samples were aliquoted locally. The samples were transported to ICDDR, B in cold pack or in liquid nitrogen for storage in —70°C freezer.
We classified persons as confirmed Nipah case-patients by the presence of immunoglobulin M (IgM) antibodies against NiV or by the presence of NiV RNA or by isolation of NiV from CSF or throat swabs. The probable cases were defined as suspect cases who died before sample collection or who had no IgM against NiV in serum collected within 8 days of onset of illness and who died before a follow-up serum sample could be obtained.
Case—Control Study
The investigation team returned to the outbreak communities to conduct a case—control study from March 5 to 10, 2008, to determine the risk factors for illness. We enrolled all confirmed and probable case-patients as cases. Individuals who lived in the same communities as the cases, and who were close in age and were free from any febrile illness with convulsions or altered mental status between February 6 and March 10, 2008, were eligible to be enrolled as controls. We identified controls by visiting the fourth closest house to the case-patient’s, confirming that no one in the house met the case definition, and identifying the household resident closest in age to the case-patient. We enrolled only one control per household. If the household resident closest in age to the case-patient declined to participate in the study; no other person in the household was sought as a control. This process was repeated at the next closest household until four controls were enrolled for each case-patient.
Data Collection
Trained interviewers collected information from cases and controls using a standardized structured questionnaire in Bengali language, based upon the questionnaires used in previous Nipah outbreak investigations in Bangladesh. We collected a detailed exposure history to previously identified risk factors for cases and respective controls for 1 month preceding the onset of illness of cases. For each case-patient who had died or was unable to respond and for each of the controls who were < 10 years of age, we identified proxy respondents. Proxy respondents included spouses, family members, friends, and neighbors who were knowledgeable about the illness or the exposures of the case-patients and controls. We also conducted informal interviews with several date palm sap collectors and local community residents about the date palm sap collection procedure, recognition of bats in the areas, and possible contamination of date palm sap by bats. We used global positioning system to determine the location of the outbreak areas.
Laboratory Analysis
Serum and CSF samples were tested for IgM and IgG antibodies against NiV using IgM-capture and indirect IgG enzyme immunoassay (Daniels et al., 2001).
CSF and throat swab specimens from five patients were tested at the U.S. Centers for Disease Control and Prevention (CDC) laboratory for molecular detection and virus isolation of NiV. Real-time RT-polymerase chain reaction (rRT-PCR) was performed using the following primers that amplified a 112- nucleotide (nt) fragment spanning from position 538 to 660 in the NiV N gene: forward primer NVBNF2B-5’-CTGGTCTCTGCAGTTAT CACCATCGA-3’, reverse primer NVBN593R 5’-ACGTACT TAGCCCATCTTCTAGTTTCA-3’, and probe NVBN542P 5’-CAGCTCCCGACACTGCCGAGGAT-3’, with the FAM dye incorporated at the 5’ end and a BHQ1 molecule at the 3’ end. PCR products were sequenced as previously described (Chadha et al., 2006), and were analyzed using Sequencher 4.10.1 software (Gene Codes).
Date Palm Sap Evaluation
The field team also collected date palm sap early in the morning from both the outbreak areas from February 27 to March 5, 2008. Two separate aliquots for a sample were collected from a tree: one in viral transport medium and another in trizol. The sap specimens were stored in a cold box maintaining temperature around 2°C—8°C and transferred to liquid nitrogen within several hours and later stored in —70°C. The sap was tested at CDC for the presence of NiV RNA by rRT-PCR; the sap was also cultured for NiV.
Infrared Camera Observation
We identified seven date palm trees where sap was collected for the cases’ consumption in Manikgonj and Rajbari District outbreak sites. To identify the possible contamination of date palm sap by bats’ secretions and to understand bat sap contamination behavior, we mounted one motion-sensor-infrared camera focusing on date palm trees’ shaved surface, sap stream, tap, and collection pot in each of the seven trees for a full night (5:00 PM to 6:00 AM).
Data Analysis
We used exact logistic regression to estimate the univariate odds ratios (ORs) with 95% confidence intervals (CIs) between exposures and case status. We stratified on the case—control pairs to account for the matched design. We assessed for confounding by constructing a multivariate exact logistic regression model. We included all exposures during multivariate analysis that had (p < 0.20) in the initial model and removed those exposures one at a time that were not significantly associated with case status. We performed all statistical analyses with STATA version 10.0.
Ethics
Interviewers obtained voluntary informed consent from all participants or proxies; for those <18 years of age, the team obtained individual assent as well as parental consent. This investigation was part of an emergency response to the outbreak, and so a complete human subjects review of all activities was not possible, but the Ethical Review Committee at ICDDR,B had previously reviewed and approved a general protocol for Nipah surveillance and response to outbreaks.
Results
The outbreak occurred in two adjoining Districts of Manikgonj and Rajbari over the same 6-day period in February 2008 (Figure A12-2). We identified a total of 10 case-patients: 4 from Manikgonj and 6 from Rajbari Districts. Nine of them died (90%); one 12-year-old child from Rajbari survived. The median age of all case-patients was 10 years, and eight (80%) were males. All of the cases presented with fever, progressive altered mental status, and loss of consciousness. The mean duration from illness onset to death was 6 days (Table A12-1).
CSF specimens were available from five (50%) case-patients and serum specimens were available from six (60%). Four patients died before the investigation team could collect any specimen. The field team was able to collect a second set of serum specimens from three case-patients (30%) within 1–6 days of first sample collection. There were four (40%) confirmed and six (60%) probable case-patients. Table A12-2 shows the laboratory results of each case-patient.
Sequencing of the complete NiV nucleoprotein (N) ORF amplified from these two isolated viruses from conventional two-step RT-PCR indicated an identical match. Complete genome sequence analysis of the two isolates confirmed that the two viruses were identical (Lo et al., 2011). The N ORF sequence shared nt sequence identity at all but seven positions with NiV isolated from India in 2007 (accession FJ513078), and at all but 10 positions with NiV isolated from Bangladesh in 2004 (accession AY988601). The amino acid sequence of N ORF differs at only one position from the 2007 Indian (R211/Q) and the 2004 Bangladesh (D188/E) isolates (Table A12-3).
We enrolled 40 controls for the 10 case-patients in the case—control study. All case-patient interviews were conducted by proxies as nine patients had died before the investigation began, and the final child was still recovering from the illness and was unable to communicate with us. We also identified proxies for 19 (48%) of the controls who were <10 years old. None of the selected cases, controls, or proxies refused to participate in the study.
In both outbreak areas, all of the case-patients drank fresh raw date palm sap 2–12 days before onset of their illness compared with 10 (25%) controls who consumed fresh date palm sap during the period of investigation (p < 0.001). Household members of case-patients were more frequently involved in date palm sap harvesting than household members of controls (30% vs. 3%, p < 0.05).
|
|
||
| Characteristics | Total, n=10; no. (%) | |
|
|
||
| Age | ||
|
Mean (SD) in years |
19 (17) |
|
|
Median (range) in years |
10 (7-55) |
|
| Male | 8 (80) | |
| Clinical features | ||
|
Fever |
10 (100) |
|
|
Altered mental status |
10 (100) |
|
|
Loss of consciousness |
10 (100) |
|
|
Convulsions |
0 (0) |
|
|
Severe weakness |
9 (90) |
|
|
Muscle pain |
6 (60) |
|
|
Headache |
5 (50) |
|
|
Cough |
5 (50) |
|
|
Difficulty in breathing |
4 (40) |
|
|
Diarrhea |
3 (30) |
|
|
Joint pain |
3 (30) |
|
|
Vomiting |
2 (20) |
|
|
New onset of seizures |
1 (10) |
|
| Died | 9 (90) | |
| Onset of illness to death (n=9) | ||
|
Mean (range) in days |
6 (1-10) |
|
| Category of cases | ||
|
Confirmed Nipah |
4 (40) |
|
|
Probable Nipah |
6 (60) |
|
|
|
||
| Case-patients | Sampling days after illness onset | CSF | Sampling days after illness onset | Serum-1 | |||||||
| No | Site | Category | IgM | IgG | Isolation | PCR | IgM | IgG | Isolatio | ||
| 1 | R | Confirmed | 9 | Neg | Neg | Neg | Neg | 5 | Pos | Neg | ND |
| 2 | R | Confirmed | 6 | Neg | Neg | Neg | Neg | 6 | Neg | Neg | ND |
| 3 | R | Confirmed | 3 | Neg | Neg | Neg | Neg | 3 | Neg | Neg | ND |
| 4 | M | Confirmed | 6 | Neg | Neg | Neg | Neg | 6 | Pos | Neg | NA |
| 5 | R | Probable | 8 | Neg | Neg | Neg | Neg | 8 | Neg | Neg | ND |
| 6 | R | Probable | NA | NA | NA | NA | NA | 4 | Neg | Neg | ND |
| 7 | R | Probable | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 8 | M | Probable | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 9 | M | Probable | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 10 | M | Probable | NA | NA | NA | NA | NA | NA | NA | NA | NA |
R, Rajbari; M, Manikgonj; IgM, immunoglobulin M; IgG, immunoglobulin G; PCR, polymerase chain reaction; Neg, negative; Pos, positive; NA, not available; ND, not done; CSF, cerebrospinal fluid.
None of the case-patients had any history of physical contact with bats, although people from the community often reported seeing bats in the tapped date palm trees during sap collection. A greater proportion of case-patients than controls reported physical contact with apparently healthy live cats (60% among cases vs. 10% among controls, p < 0.05). None of the case-patients had physical contact with sick animals, nor did they eat any sick animals. Two cases slept in the same room (20% among cases vs. 30% among controls, p > 0.05) and one case had physical contact (10% among cases vs. 0% among controls, p > 0.05) with other case-patients 2–3 days before their illness onset (Table A12-4).
In the Manikgonj cluster, three children from one family drank raw date palm sap, collected by their father, a local gachi or date palm sap collector. They drank the sap early in the morning on February 11 for the last time and subsequently two of them developed illness on February 20 and the third on February 23. The fourth child, who developed illness on February 21, was a resident of Dhaka District but visited his grandmother’s house on February 6 for 12 days. His grandmother, a neighbor of the date palm sap collector, purchased raw date palm sap from him and served it to her grandson the same day the other children drank the sap.
In the Rajbari cluster, three members from one family (mother and her two children) shared date palm sap purchased from the neighborhood date palm sap collector with two other neighborhood residents (brother-in-law and nephew of that mother) on February 18; all five subsequently developed illness. A salesman who resided nearly 5 km away from those households visited the village that morning and also drank the sap offered to him. He also died with the similar symptoms to the other four cases in Rajbari.
| Sampling days after illness onset | Serum-2 | Sampling days after illness onset | Throat swab | |||||||
| PCR | IgM | IgG | Isolation | PCR | IgM | IgG | Isolation | PCR | ||
| ND | 9 | Pos | Pos | ND | ND | 5 | ND | ND | Neg | Neg |
| ND | 7 | Pos | Neg | ND | ND | 6 | ND | ND | Neg | Pos |
| ND | NA | NA | NA | ND | ND | 3 | ND | ND | Pos | Pos |
| NA | 12 | NA | NA | ND | ND | 6 | ND | ND | Pos | Pos |
| ND | NA | NA | NA | ND | ND | 8 | ND | ND | Neg | Neg |
| ND | NA | Neg | Neg | ND | ND | NA | ND | ND | NA | NA |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
| Nipah virus isolate ID | Nucleotide position | ||||||||||||
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||
| 2 | 5 | 5 | 6 | 6 | 6 | 0 | 1 | 2 | 3 | 4 | 5 | 5 | |
| 7 | 4 | 6 | 3 | 3 | 5 | 7 | 1 | 6 | 1 | 9 | 0 | 3 | |
| 0 | 9 | 4 | 2 | 3 | 4 | 7 | 9 | 9 | 1 | 7 | 0 | 3 | |
| NIVBGD2004RAJBARI1 AY988601 | T | C | T | A | A | A | T | G | A | A | C | A | A |
| NIVBGD2008MANIKGONJ | C | A | G | • | G | G | • | A | G | G | T | • | G |
| NIVBGD2008RAJBARI | C | A | G | • | G | G | • | A | G | G | T | • | G |
| NIVIND2007NADIA FJ513078 | c | A | G | G | • | • | C | A | G | T | G | • | |
NIVBGD2004RAJBARI1 AY988601 serves as consensus sequence by which to compare the others.
"," Indicates nucleotide identity with consensus sequence.
| Exposures | Case patients with this exposure, n = 10; no. (%) | Control patients with this exposure, n = 40; no. (%) | p-Value | Odds ratio | 95% confidence intervals |
| Drank raw date palm sap | 10(100) | 10(25) | 0.000 | 38 | 5.4-∞ |
| Date sap | |||||
| Given | 6(60) | 4(10) | 0.656 | 2.2 | 0.3-19 |
| Purchased | |||||
| From local vendor | 0(0) | 1(3) | 0.429 | Undefined | Undefined |
| From any house | 1(10) | 2(5) | 0.486 | 1.0 | 0-39 |
| From anv other sources | 3(30) | 0(0) | 0.143 | 1.0 | 0-∞ |
| Collected from tree | 0(0) | 3(8) | 0.211 | 0.2 | 0-2.1 |
| Date sap harvesting | |||||
| Self | 0(0) | 3(8) | 1.000 | 1.1 | 0-10 |
| Household members | 3(30) | 1(3) | 0.022 | 15 | 1.0-860 |
| Climbed trees | |||||
| Fruit trees | 5(50) | 9(23) | 0.099 | 3.2 | 0.5-351 |
| Date palm trees | 0(0) | 4(10) | 0.549 | 0.4 | 0-4.3 |
| Physical contact with living animals | |||||
| Cow | 4(40) | 17 (43) | 1.000 | 0.9 | 0.2-4.5 |
| Cat | 6(60) | 4(10) | 0.002 | 11 | 1.9-84 |
| Physical contact with sick animals | |||||
| Cow | 0(0) | 1(3) | 1.000 | 3.9 | 0-152 |
| Chicken | 0(0) | 1(3) | 1.000 | 4.1 | 0-159 |
| Physical contact with dead animals (chicken) | 1(10) | 0(0) | 0.200 | 4.2 | 0.1-∞ |
| Ate sick animals | 0(0) | 3(8) | 1.000 | i.o | 0-11 |
| Ate dropped fruit | |||||
| Banana | 0(0) | 1(3) | 1.000 | n | 0-429 |
| Local plum (Boroi) | 5(50) | 24(60) | 0.389 | 0.5 | 0.1-3.9 |
| Papaya | 0(0) | 4(10) | 1.000 | Undefined | Undefined |
| Carambola (Kamranga) | 1(10) | 1(3) | 0.250 | 2.0 | 0.1-∞ |
| Guava | 0(0) | 13 (33) | 0.435 | 0.3 | 0-13 |
| Tamarind | 4(40) | 11 (28) | 1.000 | 1.0 | 0-66 |
| Custard apple | 1(10) | 0(0) | 0.333 | Undefined | Undefined |
| Traveled outside subdistrict | KI0) | 6(15) | 1.000 | 0.6 | 0-6.3 |
| Touched persons with fever and altered mental status who died later | 1(10) | 0(0) | 0.184 | 4.1 | 0.1-∞ |
| Been in the same room with persons with fever and altered mental status who died later | 2(20) | 3(8) | 0.258 | 2.9 | 0.2-30 |
Bolded type indicates significant results.
The median incubation period from intake of raw date palm sap to the onset of illness was 10 days (range: 9–12 days) in Manikgonj and 4 days (range: 2–7 days) in Rajbari District. All of the cases had consumed about 100 mL of date palm sap. All of the cases consumed the sap before 9 AM.
Cases were more likely to be exposed to three risk factors than controls in the initial bivariate analysis. However, in the multivariate analysis, only a single risk factor, drinking raw date palm sap, was significantly and independently associated with the illness. Nipah cases were 25 times more likely than controls to have consumed raw date palm sap (adjusted ORs 25, 95% CIs 3.3—![]() , p < 0.001) in the preceding month. Physical contact with a living cat was also associated with illness in univariate analysis (ORs 11, 95% CIs 1.9–84, p = 0.002), but was not statistically significant in the multivariate analysis (adjusted ORs 9.2, 95% CIs 0.6–675).
, p < 0.001) in the preceding month. Physical contact with a living cat was also associated with illness in univariate analysis (ORs 11, 95% CIs 1.9–84, p = 0.002), but was not statistically significant in the multivariate analysis (adjusted ORs 9.2, 95% CIs 0.6–675).
Infrared photographs showed that bats frequently visited date palm trees during sap collection. During seven nights of camera observation, 104 bats visits were photographed around the date palm tree’s sap producing area (mean: 14.9 visits per tree per night standard deviation [SD] 30.1) with 47 visits to the shaved surface (mean: 6.7 visits per tree per night SD 13.1). Bats were seen to lick date palm sap 59 times during the observation, and almost half of them (49%) were Pteropus bats. The fresh date palm sap samples (15 samples collected from 7 trees for 8 consecutive days), collected at least 9 days after consumption by the last case of this outbreak, were negative for NiV RNA by PCR and virus isolation.
Discussion
This outbreak, involving the death of nine people in two communities separated by a river in Manikgonj and Rajbari Districts over a 6-day period in February 2008, was almost certainly caused by NiV infection. The presenting clinical signs and symptoms of the case-patients were fever, central nervous system involvement, and rapid progression to death, which are consistent with other Nipah outbreaks in Bangladesh (Hossain et al., 2008). Four of the case-patients from both communities had laboratory evidence of Nipah infection; tight clustering in space and time of all case-patients, including those who were not laboratory confirmed and drinking of raw date palm sap on the same day from the same pot, which is a known risk factor for NiV infection, supports the hypothesis that the probable cases also had Nipah infection. This is the second Nipah outbreak where date palm sap has been implicated as the exposure most strongly associated with the illness.
Pteropus bats are the presumed reservoir of NiV (Chua et al., 2002; Rahman et al., 2010). They shed the virus in both saliva and urine (Reynes et al., 2005; Middleton et al., 2007); Pteropus bats were observed to be licking the raw date palm sap collected in the outbreak areas. Indeed, in contrast to an earlier infrared camera study in Bangladesh, where only 5% of the bats that contacted date palm
sap were Pteropus bats (Khan et al., 2011), when we evaluated the trees those were the sources of date palm sap consumed by cases in this outbreak, 49% of the bats that visited the tress and contacted date palm sap were Pteropus bats. There is evidence of survival of NiV in mango flesh, mango juice, pawpaw juice, and lychee juice for up to 3 days, depending upon the pH of the juice (Fogarty et al., 2008). Moreover, the half-life of NiV in bats’ urine, with pH adjusted to 7 at a temperature up to 22°C, is 18 h (Fogarty et al., 2008). The pH of date palm sap is 7.2 (Aidoo et al., 2006), suggesting that survival of NiV in date palm sap may be similar. In Bangladesh, winter is the peak collection period of date palm sap and this Nipah outbreak corresponds with the seasonality of all previously reported Nipah outbreaks in Bangladesh. In winter, the temperature remains between 15°C and 28°C (Bangladesh Meteorological Department, 2008) and this low temperature might extend the survival of NiV in bat secretions or in sap (Fogarty et al., 2008). As date palm sap is usually collected early in the morning (5 AM—7 AM) and all of the cases in this outbreak drank the raw date palm sap before 9 AM, NiV apparently survived until that time.
The date palm sap samples that were evaluated for the presence of NiV were collected 24 days after the onset of illness of the last identified case in the outbreak. The median incubation period of NiV is 9 days (Hossain et al., 2008). The absence of NiV in the sap 2 weeks after the putative transmission event suggests that date palm sap is only intermittently contaminated, a pattern of contamination that is consistent with the observed intermittent outbreaks in Bangladesh.
The distance between the two areas (44 km) was within the 50 km foraging ranges for the Pteropus bat (Kunz and Jones, 2000). The genetic sequences of the isolated viruses from the two sites were identical in contrast to substantial diversity in NiV isolates noted previously from different outbreaks in Bangladesh (Harcourt et al., 2005). While we do not know if the same bat or the same colony of bats contaminated date palm sap at these two sites, the near simultaneous occurrence of these uncommon outbreaks by an identical strain of NiV and the similar pattern of transmission suggests that they resulted from the same underlying process.
Although the association between cats and Nipah infection was not statistically significant in the multivariate model, cats are susceptible to infection with NiV and when infected can shed virus in their saliva (Middleton et al., 2002). Other domestic animals, including pigs and cattle, were associated with Nipah illness in earlier outbreaks in Bangladesh (Luby et al., 2009). The role of domestic animals in transmission of NiV is an important area for continued research.
Limitations include that we could not test samples from four cases as they died before specimens could be collected, which might be misclassified as cases in our study, but all of these cases were previously healthy people whose symptoms were consistent with confirmed Nipah cases. Moreover, we could not collect follow-up samples from the patients, who were negative in the samples collected 3, 4, and 8 days of illness onset. However, in previously investigated outbreaks, IgM against NiV was present in the follow-up samples collected 2 or more weeks
after illness onset among 56% of the Nipah cases, who did not have detectable IgM against NiV detected from earlier specimens (Hossain et al., 2008). So, it is likely that the probable cases in this outbreak also had Nipah illness. During the outbreak periods, the Government of Bangladesh conducted local awareness raising activities to notify the community about Nipah-like symptoms and to avoid drinking raw date palm sap, eating partially eaten fruits, and having contact with bats. This might have sensitized our study population regarding their response, but awareness regarding the risk factors possibly motivated study subjects and their proxies to recall their exposure history more elaborately, rather than encouraging them to hide those exposures. So, it is unlikely that the awareness raising activities affected the results of our study.
There is evidence of recurrent fatal outbreaks from 2001 through 2007 by the transmission of NiV from its fruit-bat reservoirs to humans in Bangladesh (Luby et al., 2009). The present investigation also suggests that date palm sap is an important pathway for this transmission. Drinking fresh date palm sap was the most strongly associated risk factor among the exposures investigated for this outbreak of human NiV infection in this study. The outbreak ended following local warning against drinking fresh date palm sap from the Government of Bangladesh. To prevent this illness, date palm sap should not be drunk fresh unless effective steps have been taken to prevent bat access to the sap during collection. We are working with local date palm sap collectors to develop socially acceptable low cost technologies to prevent bats’ access to the date palm sap producing parts of the tree. Studies in Bangladesh involving local date palm sap collectors suggest that using a bamboo-skirt to cover the shaved part of the date palm tree and sap collection pot might be a practical, affordable method to prevent bats’ access to the date palm sap (Nahar et al., 2008; Khan et al., 2011). Drinking raw date palm sap is a long-practiced tradition in Bangladesh, so public health recommendations to avoid drinking fresh date palm sap are unlikely to be universally followed. Research to identify culturally acceptable approaches to produce safe date palm sap may provide an additional lifesaving prevention strategy.
Acknowledgments
This research activity was funded by the Government of Bangladesh and the U.S. Centers for Disease Control and Prevention. ICDDR, B acknowledges with gratitude the commitment of Government of Bangladesh and the U.S. Centers for Disease Control and Prevention to ICDDR,B’s research efforts. We extend thanks to Dr. Imtiaz Ashraf Chowdhury and Dr. Sayma Afroze of IEDCR. Thanks also to Dr. Abu Shahid from Nipah surveillance team at Rajbari District, Dr. Khondker Mahbuba Jamil from IEDCR for laboratory support, Dr. Shahed Sazzad and Mr. Dawlat from ICDDR,B for data collection, and Ms. Dorothy Southern for her support in preparing the article. We also thank the two communities for participating in our study.
Disclosure Statement
No competing financial interests exist for any of the authors.
Disclaimer Statement
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
References
Aidoo, KE, Nout, MJR, Sarkar, PK. Occurrence and function of yeasts in Asian indigenous fermented foods. FEMS Yeast Res 2006; 6:30–39.
Bangladesh Bureau of Statistics. Hosehold and population information. 2010. Availble at http://203.112.218.65/RptPopCen.aspx?page=/PageReportLists.aspx?PARENTKEY=41 (accessed on January 27, 2011).
Bangladesh Meteorological Department. Climate of Bangladesh. 2008. Available at www.bmd.gov.bd/Document/climateofbangladesh.doc (accessed on January 10, 2009).
Chadha, MS, Comer, JA, Lowe, L, Rota, PA, et al. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg Infect Dis 2006; 12:235–240.
Chua, KB, Koh, CL, Hooi, PS, Wee, KF, et al. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect 2002; 4:145–151.
Daniels, P, Ksiazek, T, Eaton, BT. Laboratory diagnosis of Nipah and Hendra virus infections. Microbes Infect 2001; 3:289–295.
Epstein, JH, Prakash, V, Smith, CS, Daszak, P, et al. Henipavirus infection in fruit bats (Pteropus giganteus), India. Emerg Infect Dis 2008; 14:1309–1311.
Fogarty, R, Halpin, K, Hyatt, AD, Daszak, P, et al. Henipavirus susceptibility to environmental variables. Virus Res 2008; 132: 140–144.
Gurley, ES, Montgomery, JM, Hossain, MJ, Bell, M, et al. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg Infect Dis 2007; 13:1031–1037.
Harcourt, BH, Lowe, L, Tamin, A, Liu, X, et al. Genetic characterization of Nipah virus, Bangladesh, 2004. Emerg Infect Dis 2005; 11:1594–1597.
Hossain, MJ, Gurley, ES, Montgomery, JM, Bell, M, et al. Clinical presentation of Nipah virus infection in Bangladesh. Clin Infect Dis 2008; 46:977–984.
Hsu, VP, Hossain, MJ, Parashar, UD, Ali, MM, et al. Nipah virus encephalitis reemergence, Bangladesh. Emerg Infect Dis 2004; 10:2082–2087.
Khan, SU, Hossain, MJ, Gurley, ES, Nahar, N, et al. Use of infrared camera to understand bat’s access to date palm sap: implications for preventing Nipah virus transmission. Ecohealth 2011 Jan 5 [Epub ahead of print]. DOI:10.1007/s10393-010-0366-2.
Kunz, TH, Jones, DP. Pteropus vampyrus. Mamm Species 2000; 642:1–6.
Lo, MK, Sazzad, HMS, Gurley, ES, Hossain, MJ, et al. Characterization of Nipah Virus from outbreaks in Bangladesh, 2008–2010. 2011. In: Abstract Book of Annual Meeting of the American Society for Virology. Minneapolis, MN; 2011, p. 126.
Luby, SP, Hossain, MJ, Gurley, ES, Ahmed, B-N, et al. Recurrent zoonotic transmission of Nipah virus into Humans, Bangladesh, 2001–2007. Emerg Infect Dis 2009; 15:1229–1235.
Luby, SP, Rahman, M, Hossain, MJ, Blum, LS, et al. Foodborne transmission of Nipah virus, Bangladesh. Emerg Infect Dis 2006; 12:1888–1894.
Middleton, DJ, Morrissy, CJ, van der Heide, BM, Russell, GM, et al. Experimental Nipah virus infection in pteropid bats (Pteropus poliocephalus). J Comp Pathol 2007; 136:266–272.
Middleton, DJ, Westbury, HA, Morrissy, CJ, van der Heide, BM, et al. Experimental Nipah virus infection in pigs and cats. J Comp Pathol 2002; 126:124–136.
Nahar, N, Sultana, R, Gurley, ES, Hossain, MJ, et al. Date palm sap collection: Exploring opportunities to prevent Nipah transmission. Ecohealth 2010; 7:196–203.
Rahman, SA, Hassan, SS, Olival, KJ, Mohamed, M, et al. Characterization of Nipah virus from naturally infected Pteropus vampyrus bats, Malaysia. Emerg Infect Dis 2010; 16:1990–1993.
Reynes, J-M, Counor, D, Ong, S, Faure, C, et al. Nipah virus in Lyle’s flying foxes, Cambodia. Emerg Infect Dis 2005; 11:1042–1047.
Wacharapluesadee, S, Lumlertdacha, B, Boongird, K, Wanghongsa, S, et al. Bat Nipah virus, Thailand. Emerg Infect Dis 2005; 11:1949–1951.
Yob, JM, Field, H, Rashdi, AM, Morrissy, C, et al. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg Infect Dis 2001; 7:439–441.
FOOD-BORNE PATHOGEN CONTROL PROGRAMS
Cargill is an international producer and marketer of food, agricultural, financial, and industrial products and services. Founded in 1865, our privately held company employs 139,000 people in 65 countries. We help customers succeed through collaboration and innovation and are committed to sharing our global knowledge and experience to help meet economic, environmental, and social challenges.
In fiscal year 2011, Cargill had US$119.5 billion in sales and other revenues. Earnings from continuing operations were US$2.69 billion. The company also realized US$1.55 billion in income from discontinued operations.
Cargill has a very focused purpose to be the global leader in nourishing people. That purpose takes into account health and nutrition, as well as food safety and food security. We have a mission to create distinctive value, and our approach is to be trustworthy, creative, and enterprising.
Thousands of customers turn to Cargill for innovative solutions across our four major market segments.
Agriculture: We buy, process, and distribute grain, oilseeds, and other commodities to makers of food and animal nutrition products. We also provide crop and livestock producers with products and services.
Food: We provide food and beverage manufacturers, food service companies, and retailers with high-quality ingredients, meat and poultry products, and health-promoting ingredients and ingredient systems.
______________
30 Cargill, Inc.
Financial: We provide our agricultural, food, financial, and energy customers around the world with risk management and financial solutions.
Industrial: Cargill serves industrial users of energy, salt, starch, and steel products. We also develop and market sustainable products made from agricultural feedstocks.
As an agricultural and food company, food safety is fundamental to Cargill’s ongoing business. Our goal is to provide high-quality, safe food every time, everywhere. We recognize that our work in this important area is never done. Every day we work to earn the trust of our customers and consumers, beginning with the safety of the products we produce and extending to improving food safety around the world.
Our definition of food safety is simple—protecting people and animals from illness or injury from handling or consuming our food products. Our efforts to ensure this—all along the vast supply chain, from production to consumption—are much more complex. Because we touch the global food supply chain in so many ways and in so many places, we take a broad, comprehensive science- and risk-based approach to ensure the safety and integrity of all of our products. This comprehensive approach is designed to address biological, chemical, and physical hazards.
Because we recognize that food safety practices, legislation, and regulatory oversight vary between and even within nations, we have adopted one global systems approach to which we hold ourselves accountable across all of our business, in all of our geographies.
It’s everyone’s responsibility, and we take a very holistic approach from the farm all the way to the plate. We embrace the concept of One Health.
I want to share this as a roadmap. I’m going to break this down as we go through here, but I think this is a very good example of what One Health is all about. We’ve worked on this with a number of other colleagues in the food industry and through Michigan State University to create this road map for the components around global food safety (Figure A13-1).
The journey starts out with international governance up in the upper left-hand corner. Then there’s a track that goes across the top around how governments can adopt the principles, guidelines, and recommendations coming out of Codex Alimentarius (CODEX), the OIE (World Organization for Animal Health), and the International Plant Protection Commission (IPPC) as a basis for the regulatory oversight program. These organizations are the international standard setting setting bodies prescribed by the World Trade Organization’s (WTO’s) Sanitary and Phytosanitary Agreement (SPS).
The bottom track outlines how industry has taken those same principles, guidelines, and recommendations and, through an ISO framework, transformed them into food safety systems that can be implemented and then audited against to ensure that the systems have been appropriately deployed. These systems can
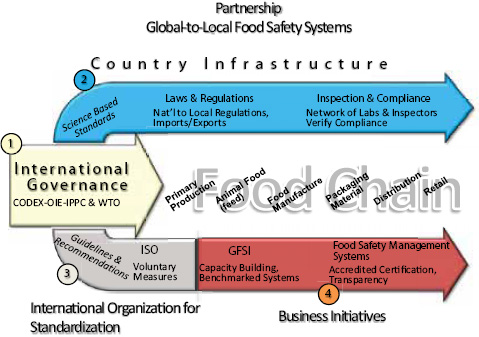
FIGURE A13-1 Roadmap for the components of global food safety.
SOURCE: Cargill.
cover the entire food network going from the farm on the left all the way through to the consumer on the right. It’s a shared responsibility, shared accountability thought process through the whole thing.
We will discuss public—private partnerships as we go through the map.
Countries are dependent on each other for food. We know that we don’t produce enough food in the areas where a lot of people live; therefore, there is going to be movement of food. This isn’t anything new. Food historically has been a basis of trade, and it used to be traded for other goods and services over time. This continues on today.
National governments established the WTO and the SPS agreements, and use CODEX, OIE, and IPPC for the process for setting international food safety standards. Out of these organizations you have science-based standards that have been internationally vetted, discussed, and adopted. Out of this comes guidelines and recommendations that can be utilized by both the public and the private sector in global food safety.
Going across the top of the road map, we actually started to create these slides as we got into discussions with various governments on regulatory reform. We’ve used this with the Chinese government (Administration of Quality Supervision, Inspection and Quarantine, Ministry of Health) in discussions about how
they can effectively implement food safety systems from a regulatory oversight standpoint.
We’ve also used it over the past year with the Food and Drug Administration (FDA) as it has looked at implementing the Food Safety Modernization Act to provide an idea of what the industry’s thinking and how we’re taking these international standards and adopting them in our food safety systems across the global food supply network.
From a government standpoint we all know that strong systems will protect customers and consumers, and also facilitate trade. A number of countries already used CODEX as a basis for a number of their regulations. Many of them reference ISO as voluntary measures, and as suggestions for the industry in terms of adoption. Regarding government inspections and compliance, if a regulatory agency is verifying compliance and evaluating a firm’s preventative measures, and the focus is on the elements that come out of international governance, then you will have industry and government looking at the same criteria and thus aligned as to what it is that is important as it relates to the safety of our food system.
On the private-sector side, the rationale is to build on science-based standards coming out of CODEX, OIE, and IPPC. A strategic partnership exists between ISO and WTO to facilitate market requirements. They’re working together to make sure that there’s a framework available for the private sector to adopt these principles.
ISO does not regulate, legislate, certify, or accredit. We have 163 countries and national standards that collaborate with ISO on the development of these voluntary measures. There is a lot of input and a lot of ownership through this process. They are voluntary measures, but there are measures for the accreditation process, for the certification process, for auditing, for auditor competency, and then also for food safety management systems. All together it gives industry the basis for consistent, harmonized food safety management systems.
The process standardizes implementation; it gives us harmonization, alignment, and consistency across the food chain from origination through consumption. In some cases there may be a market requirement, or it may be referred to in regulations and legislation. For the industry it’s a good framework—using the guidelines, recommendations, and principles out of CODEX, OIE, and IPPC and putting them into a framework that can be adopted by facilities in their food safety systems.
How do we do that? Within the industry there has been a lot of discussion about food safety being a competitive issue. Let’s go back to the mid-1990s when the beef industry got together as it was struggling with E. coli O157:H7 and how to deal with the situation. The industry got together and made a decision that food safety would not be a competitive issue in the ground beef business.
So all of a sudden companies were coming together and sharing insight, best practices, and data. Together we’ve driven the O157 presence down with the help of the ground beef industry and other stakeholders focusing in on what were the
important elements of a food safety system and aligning on how to address the challenge.
The work that we’ve done as industry is readily available. Everybody has access to that information and those processes. We all know the tools that are out there for the entire industry to achieve the goals of improved ground beef safety.
We’ve also been working through an organization called the Global Food Safety Initiative (GFSI). GFSI is a multistakeholder group that benchmarks food safety systems. We just came out with guidance document 6 earlier this year. It is based on the principles of good hygiene and hazard analysis and critical control point (HACCP) from CODEX.
The guidance document has requirements for food safety systems and their delivery. It also has a component around capacity building that allows these principles to be implemented in emerging markets where the capacity might not be there. There’s a process that takes countries or individual facilities in countries through a stepwise progression so that they can achieve this certification process.
We think food safety management systems are really the way to go in terms of having a robust program, and an accredited certification gives us third-party assurances that we’re doing the right thing. We strive to create transparency and confidence in the supply chain. This has to be done through a partnership. We believe it’s effective and efficient. We believe that it protects consumers around the globe. This has been implemented within Cargill (Figure A13-2).
This document is in every one of our facilities around the world. Everybody is aware of it. Everybody knows it and understands it. In most places around the world you’re going to have both the business unit leader and the plant manager also signing this document. It’s a true reflection of both top-down and bottom-up commitment to the policy.
We have based our policy and procedures on CODEX. It’s a focus on food safety management systems. We have general requirements that are required to be documented. The next section describes management responsibility. Every business unit leader and every plant manager has a responsibility that they must achieve in order to be compliant with the policy.
We also have a resource management section. We have a section describing planning and realization of safe products. These may sound like strange section titles to you, but they’re taken from ISO, coming right out of CODEX. These are very consistent all the way through. In fact, we just this past year renumbered our policy and procedures manual to be in line with CODEX, so it’s quite clear.
In looking at planning and realization of safe products, the key is prerequisite programs, steps you must take to enable a hazard analysis, doing the hazard analysis, and then putting in your operational prerequisite programs and establishing your HACCP plan.
You must update your plan on a regular basis; we require an annual reassessment of the plan. We also have a verification component. Traceability is included in this, and then a control of any nonconforming products. It’s very consistent in

FIGURE A13-2 Cargill food safety policy.
SOURCE: Cargill.
the approach. The next essential section includes validation and verification as well as continuous improvement of the management system.
You’ve got to be able to validate that what you’ve put in place is effective. Then you’ve got to verify that you’re doing what you said you were going to do over time. These all become important components. It’s important for us to remainoutcome based so that we can drive continuous improvement.
When new technology becomes available, new interventions become available; we want to be able to take advantage of those and not be constrained by a regulatory construct that is prescriptive and telling us how to do it. Let’s focus on outcomes. Let’s agree on what those performance standards need to be. Let’s agree what the outcome needs to be, and then let industry move forward and innovate and continuously improve and share that information across the supply chain.
There are a lot of prerequisite programs. I’m going to discuss management of purchased materials and measures for the prevention of cross-contamination as we go through here. These are prerequisite programs coming out of CODEX and some Cargill-specific prerequisite programs included. We have a lot of them. We spent a lot of time on this. This is really the soul of what we do each and every day in our food-producing plants.
I’ll outline management of purchased materials. This really gets into our supplier and external manufacturer program. We have more than 1,000 plants. We have hundreds of thousands of suppliers and at least 400-500 external manufacturers who produce product on Cargill’s behalf. They’re expected to have the same systems in place that we do.
We have a program around the selection and management of those suppliers, incoming raw materials, and then our supplier and external manufacturer qualification and management scheme. This is a very simple diagram in the way we approach this with our suppliers (Figure A13-3). We believe this will mitigate food safety and quality risks, keeping people and animals safe. That’s the core of what we do each and every day.
We do a risk assessment on the product and on the plant. We look at their competency development. We look at the relationship management. One of the most important components of this is know your supplier. We follow the “trust but verify” model. You must know who you’re working with as a supplier. We have accountability; they have accountability. We’re going to collaborate to make sure that we’re assessing food safety appropriately and that there’s
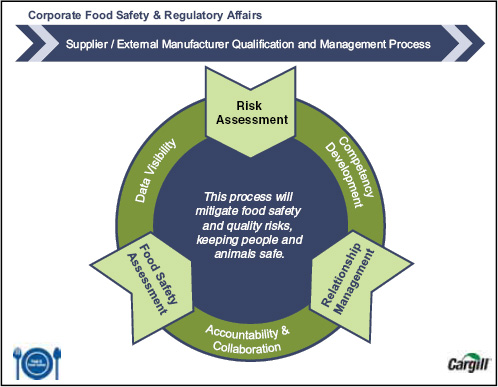
FIGURE A13-3 Corporate food safety and regulatory affairs.
SOURCE: Cargill.
transparency in the information that’s being generated. This is at the core of what we do.
We’ve put together a very simple risk assessment model that we use with all of our suppliers. There’s a material risk that you can see that goes from low to high, and we have a quantitative score that we go through in looking at what that risk is associated with that product. But then we also assess the capability of the supplier to manage that risk.
If you’ve got a high-risk material, but you’ve got a company that’s controlling that risk, they’re going to go into that medium category. If I’ve got a high-risk material and a high-risk supplier, they’re in the priority list. About 6 percent of our suppliers fall into that range. We’re working intensely with them, and they’re under increased scrutiny, obviously, as we go through the process. We’re rolling this out across the entire company over the course of the next 3 years, and it’s quite an undertaking.
Microbiological cross-contamination is extremely important for us. We focus on environmental monitoring for most of our facilities. We also have a specific Salmonella control program and a Listeria monocytogenes control program in those facilities where those hazards are identified as being reasonably likely to occur.
The environmental monitoring program goes into play where you have a risk that’s reasonably likely to occur and could find its way into product (Figure A13-4). We’ve put together a decision tree that we’ve taken through every one of our facilities so that everybody is operating against the same criteria.
We look at the difference between shelf-stable and not-shelf-stable products. But then there are very specific processes that everybody goes through to assess the risk associated with their product and their facility. That will take you down to whether or not you need an environmental monitoring program in place.
We’ve found this to be very useful in really driving people to think critically about the products that they produce, the facilities that they manage, and the products—they have to understand where they’re going and who’s consuming them. This has been a really valuable tool for us to get people really thinking critically about the business that they’re managing.
In summary, I believe that we do have a path forward here. I think the One Health approach makes a lot of sense. We live it every day. We’re originating grain and producing ready-to-eat food. To me, One Health is what we do every day.
We believe that we have a structure and a mechanism for effective global partnerships in place. We work closely not only with our supply chain and our competitors in the industry, but also with our customers and with the regulatory agencies. And we do a lot of work with the Centers for Disease Control and Prevention (CDC)—sharing information with CDC, talking with them, understanding epidemiology and microbiology.
Working with academia, consumer groups, government, and industry is the way forward. We’ve all tried to do it alone. The private sector has tried to do it alone. Government has tried to do it alone. It doesn’t work. We’ve got to work
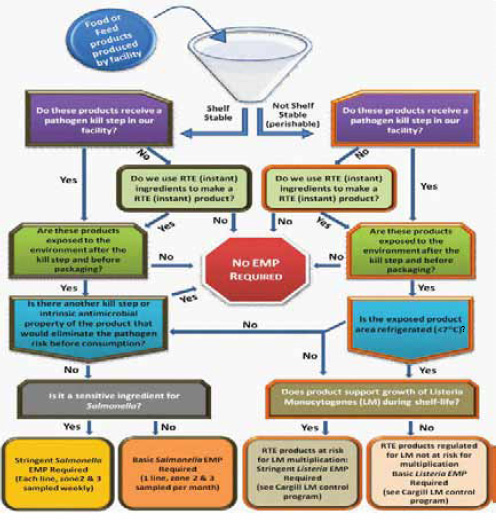
FIGURE A13-4 Cargill environmental monitoring decision tree.
SOURCE: Cargill.
together. We’ve got to get on the same page. We’ve got to get aligned around some of these issues.
We believe that resources must be deployed based on risk. You must have a science base and a risk base to apply resources. We’re all operating with reduced resources.
We’re trying to do more with less, so it becomes even more important that we’re focused on the science, we’re focused on the risk, and we’re applying resources against the areas of greatest need. Focus has to be on prevention. It has to be on preventing issues from happening in order to maintain confidence in the food supply and to have a shared goal of safe, affordable food.
Food security plays into this in a major way, and the more preventative measures we can have in place around the world, the more assurance we’re going to have of an abundant, safe food supply. It builds confidence in food safety, enhances global trade. It enhances food security. It enhances people’s enjoyment of their nutrition.
Lastly, I have to finish with this statement. Business shoulders the responsibility for safe food. I know a lot of times government thinks it has the responsibility. It doesn’t. We do. It’s our product. It’s our brand. They’re our customers. We want to work together, and we want to work collaboratively. But at the end of the day, we’re the ones who have the responsibility, and we accept that.
EMERGING FOODBORNE PATHOGENS AND PROBLEMS: EXPANDING PREVENTION EFFORTS BEFORE SLAUGHTER OR HARVEST
Casey Barton Behravesh, Ian T. Williams, Robert V. Tauxe31
Introduction
Infections caused by microbes that contaminate the food supply are a frequent reminder of the complex food web that links us with animal, plant, and microbial populations around the world. In the United States, an estimated 46 million foodborne infections occur each year, along with 250,000 hospitalizations and 3,000 deaths (Scallan et al., 2011a, 2011b). While all are at risk, the consequences are the most severe in the vulnerable populations of the very young, the elderly, and those with compromised immune systems. Of the many pathogens that can contaminate food, some, like norovirus and Salmonella serotype Typhi, are sustained in human reservoirs and contaminate the food supply via the excreta of infected humans. Many others are sustained in animal reservoirs and contaminate our food supply because they are present in the flesh, milk, or eggs in the living animal, or because they are in the excreta of infected animals that subsequently contaminate the foods we eat. Some pathogens persist in the environment, or in multiple hosts, and can contaminate the foods we eat via pathways that reflect the variety of ecosystems that make up our food supply.
Food safety depends on understanding these pathways well enough to prevent them. In the United States, substantial progress during the 20th century in animal disease control efforts has greatly reduced the foodborne infections re-
______________
31 Division of Foodborne, Waterborne and Environmental Diseases, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, GA.
lated to zoonotic diseases such as brucellosis and bovine tuberculosis (Tauxe and Esteban, 2006). At the same time, an increasing number of microbes have been recognized that can cause serious illness in humans, but rarely cause illness in the animals that carry them. The presence of these microbes is thus not apparent to the rancher or farmer, and the animal appears entirely healthy on inspection at slaughter; addressing these microbes requires a different prevention paradigm based on reducing levels of microbial contamination throughout the food chain. This effort starts on the farm or ranch where animals are raised, with attention to fodder, water, and biosecurity there. An early success was the virtual elimination of the parasite Trichinella from the nation’s swineherds, and the prevention of pork-related trichinosis in people, through attention to the fodder fed to pigs (Schantz, 1983). Recent outbreaks show that plants can also be contaminated with human pathogens on the farm, through manures, water, and wild animal incursions (Lynch et al., 2009). The need to reduce and prevent contamination continues through harvest and slaughter, subsequent processing, and the food preparation steps in the final kitchen (Figure A14-1). Indeed, reducing the num-
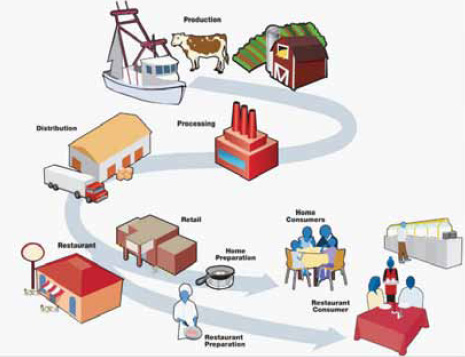
FIGURE A14-1 The food production chain from the farm to the table.
SOURCE: Adapted from Tauxe (2006) Table 3-1, page 73. http://www.cdc.gov/outbreaknet/investigations/figure_food_production.html (accessed April 10, 2012).
ber of foodborne infections by making food safer is the result of efforts by many partners in the food safety system.
Concentrated animal production has parallels with human urbanization, like the challenge of providing water, food, and fecal disposal for thousands of individuals every day. Just as the spread of many infections in human cities depends critically on treating the drinking water, and collecting sewage and keeping it out of the food and water supplies, and immunizing ourselves against many infections, so will the health of animals raised in virtual cities depend on attention to the safety of their water and food supplies, coupled with selective immunization.
New Pathogens and Problems
New food-borne pathogens emerge when previously unrecognized pathogens are identified and are linked to foodborne transmission from the beginning, or when foodborne transmission is documented for pathogens that are already well known. The list of foodborne agents that have emerged in the past three decades includes bacteria, viruses, parasites, biotoxins, and a prion (Table A14-1). They often emerge from animal reservoirs; 70 percent are sustained in animal populations and affect humans only incidentally. Many were identified in the course of outbreak investigations, when both their pathogenicity and association with food could be determined. Some common bacterial foodborne pathogens are adapted to particular reservoirs, making targeted control strategies feasible (Table A14-2).
TABLE A14-1 Major Pathogens Identified as Foodborne Since 1970
|
|
||
| Bacterial | Parasitic | |
| Arcobacter butzleri* | Cryptosporidium* | |
| Campylobacter jejuni* | Cyclospora cayetanensis | |
| Campylobacter fetus* | Sarcocystis* | |
| Cronobacter sakazakii | Trypanosoma cruzi* | |
| E. coli O157:H7* | ||
| E. coli, non-O157 STEC* | Viral | |
| E. coli, enteroaggregative/STEC | Astrovirus | |
| E. coli, other diarrheagenic | Caliciviridae (norovirus and sapovirus) | |
| Listeria monocytogenes* | Hepatitis E* | |
| Vibrio cholerae O139, toxigenic* | Nipah virus* | |
| Vibrio vulnificus* | Rotavirus | |
| Vibrio parahaemolyticus* | ||
| Yersinia enterocolitica* | Fungal | |
| Yersinia pseudotuberculosis* | Aspergillus flavus aflatoxin | |
| Algal | Prion Agent | |
| Pseudo-nitzschia pungens* (domoic acid-producing) |
new Variant Creutzfeld Jacob Disease prion* | |
|
|
||
* Reservoir or major transmission pathway through animals.
SOURCE: Adapted from Tauxe (2006), Table 3-1, p. 73.
TABLE A14-2 Major Food-Animal Reservoirs for Human Foodborne Bacterial Pathogens
|
|
|
| • | Campylobacter—poultry, other birds, cattle |
| • | Shiga toxin-producing E. coli O157:H7—cattle and other ruminants |
| • | Salmonella—poultry, cattle, swine, reptiles, and others — Salmonella serotype Enteritidis—poultry — MDR Salmonella serotype Newport—dairy and beef cattle — Salmonella serotype Choleraesuis—swine |
| • | Vibrio—molluscan shellfish |
| • | Yersinia enterocolitica—swine |
|
|
|
For example, Campylobacter are adapted to birds and particularly to poultry, among which they are commensal intestinal flora, and contaminate poultry meat by cross-contamination at slaughter. Some strains can also colonize cattle and are transmitted via raw cows’ milk. Shiga toxin—producing E. coli O157:H7 can colonize the peri-rectal glands of ruminants and transfer from hides and feces to meat during the slaughter process. Strains of Salmonella serotype Enteritidis that spread around the world in the 1980s colonize the peri-ovarian tissues of the hen’s reproductive tract, where they come in contact with the egg yolk as it forms, and contaminates the internal contents of normal-appearing eggs. If the egg is fertilized, these Salmonella then colonize the reproductive tissues of the chick embryo and reach the next generation, while in the unfertilized table egg, Salmonella can multiply in the yolk and infect the eater of a less than fully cooked egg.
The reservoirs where these pathogens persist, and the pathways by which they reach humans, are revealed in outbreak investigations. By epidemiological methods, the illnesses in an outbreak can often be associated with consuming a particular food, the food vehicle of infection. Between 2003-2008, the food vehicles identified in 1,565 outbreaks reported to the Centers for Disease Control and Prevention (CDC) with specific food vehicles are a broad spectrum of animal- and plant-derived foods (Figure A14-2). The list of implicated foods is regularly expanded as new ones are identified in outbreak investigations. Between 2006 and early 2012, 15 new specific food types were identified as food vehicles in outbreaks affecting the United States (Table A14-3). It is curious that while many of the pathogens have animal reservoirs, many new food vehicles are plant derived. This includes plant-derived processed foods, like peanut butter, peanut paste, and spinach powder; spices such as black and white pepper; tree nuts; and fresh produce items.
The identification of new food safety problems has been accelerated by important improvements in surveillance and response. These new surveillance tools capture information about infections in humans as well as in animals and contamination of foods, providing important information that is integrated across sectors.

SOURCE: CDC, unpublished data.
|
|
|
| • | Bagged spinach |
| • | Pasteurized carrot juice |
| • | Peanut butter |
| • | Broccoli powder on a children’s snack food |
| • | Dry dog food |
| • | Frozen pot pies |
| • | Canned chili sauce |
| • | Hot peppers |
| • | White and black pepper |
| • | Raw cookie dough (likely flour) |
| • | Hazelnuts |
| • | Fenugreek sprouts |
| • | Papayas |
| • | Pine nuts |
| • | Raw frozen scraped ground tuna |
|
|
|
SOURCE: CDC, unpublished data.
In the United States, public health surveillance that tracks the frequency of human infections with specific pathogens has expanded greatly since 1996 to capture different kinds of information that is needed for making public health decisions. FoodNet, the network for active surveillance of infections often transmitted through foods in 10 sentinel sites around the country, led by the CDC and supported by the Food Safety and Inspection Service of the U.S. Department of Agriculture (USDA) and the Food and Drug Administration (FDA), provides accurate tracking of what is diagnosed in human clinical laboratories, overcoming local variation in reporting requirements (Scallan, 2007). FoodNet data are fundamental to estimating the total number of illnesses that occur and to tracking trends over time. By 2010, FoodNet data showed that the incidence of E. coli O157 infections had declined by 44 percent since the baseline period of 1996-1998, that of Campylobacter by 27 percent, and that of Listeria infections by 38 percent, while those caused by Salmonella had not decreased at all (Figure A14-3) (CDC, 2011e). The substantial progress made in reducing E. coli O157, Campylobacter, and Listeria infections between 1996 and 2003 was largely the result of improvements in sanitation at slaughter and meat processing for meat and poultry. There has been little progress in more recent years.
A second surveillance enhancement is PulseNet, the national molecular subtyping network for bacterial foodborne pathogens, which has enhanced detection of outbreaks (Swaminathan et al., 2001). In each state, clinical laboratories send strains of Shiga toxin—producing E. coli (STEC), Salmonella, and Listeria
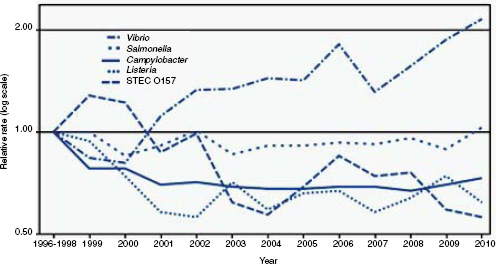
SOURCE: CDC (2011e).
monocytogenes isolated from ill persons to the public health laboratories where they are subtyped by molecular methods and added to a national database. To find clusters of infections that may be related, the database is scanned looking for surges in particular subtypes. Each state can review its own data and national data, and the CDC looks for multistate increases. The same laboratory methods are applied by the USDA to Salmonella and E. coli isolated from animals and meats, and by FDA to isolates from other foods, so that PulseNet can explore possible connections between animal reservoirs and foods, in addition to clusters of human infections. Epidemiological investigation of such clusters may reveal an exposure that the cases all have in common, such as eating a particular food, contact with an unusual pet, or travel to a particular place. The growth of this surveillance system, which now adds patterns of 50,000 isolates a year to the national database, and the increasingly sophisticated epidemiological approach to the clusters identified have led to a dramatic increase in the number of multistate outbreaks detected and investigated.
A third surveillance enhancement tracks the frequency of antimicrobial resistance in enteric bacterial pathogens (CDC, 2011c; Holmes and Chiller, 2004). The National Antimicrobial Resistance Monitoring System (NARMS) for enteric bacteria also depends on the submission of isolates to public health laboratories. For NARMS, a systematic subset of those isolates is tested at the CDC to determine their resistance to a panel of antimicrobial agents. In parallel, isolates from retail meats are tested at FDA, and isolates from animal carcasses are tested in USDA laboratories. These three arms of NARMS provide ongoing monitoring of the progression of resistance to specific agents that are used in agriculture and human health and have been important to promoting prudent use and regulation.
Finally, the National Outbreak Reporting System (NORS) collects reports of outbreaks of foodborne and waterborne illnesses that are investigated by public health departments, providing the summary information about frequency by pathogen and vehicle type (CDC, 2011d). In 2009, this system was expanded to include outbreaks of gastroenteritis caused by contact with animals and by person-to-person transmission. NORS thus captures information about a range of transmission events likely to lead to gastrointestinal disease.
Improving Preharvest Prevention: The Animal Sector
Long-term prevention of foodborne disease depends on actions of many partners in the food production chain, stretching from farm to table. Some critical prevention measures include quality assurance programs at egg farms; safe agricultural practices for produce farmers; inspection systems at meat processing plants; use of pasteurization, canning, cooking, irradiation, and other steps to kill pathogens in food processing; and food safety education for consumers and staff in the food industry. Much of the progress has been focused on safer processing of animals and plants after they are harvested, with less emphasis on the preven-
tion that can be achieved before harvest or slaughter. Making food safer in the future will depend on reducing preharvest contamination.
Outbreak investigations show that contamination events often start with problems in production, that is, while growing the plants we harvest or raising food animals. Many factors throughout all stages of the food production and distribution system can affect food safety. For meat products, what happens on farms, in feedlots, during transport and lairage before slaughter, as well as during slaughter and further processing can have a major effect on human health (Miller and Griffin, 2012). Domesticated food animals can also serve as a source of contamination of nearby produce-growing fields and can lead to human infection through direct contact at petting farms and mail order hatcheries. Preventing such infections also means reducing the carriage and spread of human pathogens among live animals.
Bacterial and other microbial pathogens in animal feces can contaminate the environment in which animals are raised, where they roam, and where they are kept while awaiting slaughter. Because animal hides and intestinal contents may have pathogens, efforts at slaughter are focused on cleaning the hides, removing them with care, and preventing the contamination of meat with intestinal contents. Poultry farms with large populations of birds are a setting where infectious agents can spread rapidly. When birds are slaughtered, hot water dips help remove feathers but can spread intestinal contents to subsequent carcasses. Campylobacter jejuni/coli, a common cause of illness in the Unites States, contaminates at least 40 percent of chicken breasts at retail (FDA, 2011). As a result, poultry is a major source of Campylobacter infections in humans. People become infected by consuming inadequately cooked poultry or other foods that become cross-contaminated via contact with poultry. Even infants riding in shopping carts containing raw poultry are at increased risk (Patrick et al., 2010).
Control measures for Campylobacter focused on slaughter sanitation; chlorinating water baths and chiller tanks have been associated with a decrease in Campylobacter infections in the United States in the late 1990s, although there has been no progress since 2002 (Figure A14-3). In New Zealand, similar control measures implemented at slaughter led to a dramatic 50 percent decline in campylobacteriosis in 2008 and a parallel decline in Guillain-Barre syndrome cases (Baker et al., 2012; Sears et al., 2011). In Scandinavia, a new control strategy is “test and freeze,” developed first in Iceland and then adopted in Norway and Denmark, in which flocks are tested preslaughter for the presence of Campylobacter (Tustin et al., 2011). Birds from flocks that have Campylobacter must be frozen after slaughter, which reduces the level of Campylobacter contamination, while birds from poultry farms without Campylobacter can be sold fresh at a premium price. This provides farmers with an economic incentive to reduce Campylobacter contamination and may be a model for how to incentivize preharvest food safety measures. To make further progress, short of irradiating poultry, control measures will need to include a preharvest focus ( Wagenaar et
al., 2008). Such measures may include chlorinating the drinking water, probiotics, and vaccination (Wagenaar et al., 2008). In Denmark, field evaluations show that putting insect screens on henhouses can lead to a 70 percent decrease in Campylobacter flock prevalence (Hald et al., 2007).
Escherichia coli O157:H7 infection has emerged as an important cause of human illness ranging from simple diarrhea, to hemorrhagic colitis, to hemolytic-uremic syndrome, characterized by hemolytic anemia, thrombocytopenia, and renal injury (Griffin and Tauxe, 1991; Rangel et al., 2005). It was first recognized as a human pathogen in foodborne outbreaks associated with ground beef in 1982 (Riley et al., 1983). In 1993, after a large multistate E. coli O157 outbreak was linked to undercooked ground beef patties sold from a fast-food restaurant chain, E. coli O157 became broadly recognized as an important human pathogen (Bell et al., 1994). In 1994, officials at the USDA declared E. coli O157:H7 an adulterant of ground beef, so that finding these bacteria in ground beef resulted in its mandatory recall, and then implemented a new inspection procedure for beef carcasses based on hazard analysis critical control point strategies. In 2002, after a large multistate outbreak and recall of ground beef, regulators and slaughter and beef grinding companies focused more intensive effort on preventing the contamination of ground beef itself, including increased focus on hide removal, testing beef trim before it reached the grinder, and holding ground beef lots until they were found not to be contaminated. These efforts helped to reduce the contamination of ground beef and in turn may have led to the decrease in laboratory-confirmed E. coli O157:H7 cases measured in the U.S. FoodNet active surveillance system (See Figure A-14-3) (CDC, 2011e). However, beef remains the most frequently identified vehicle for E. coli O157:H7 infections, followed by produce-associated outbreaks (CDC, 2010a). Reducing these infections further will depend on preharvest interventions to decrease the shedding of E. coli O157 by cattle before they come to slaughter. E. coli O157 is common among cattle, particularly in the summertime, and reducing carriage may be achieved using a suite of interventions, including vaccines (two are currently available for evaluation), probiotics, and bacteriophage treatments, and microbicidal agents such as sodium chlorate (Loneragan and Brashears, 2005). Leveraging the need for safer meat with economic incentives for lower contamination rates in cows has yet to be achieved.
Better prevention can occur with multifaceted on-farm preharvest control measures and with test and diversion strategies. One example is egg safety in the United States. Shell eggs and poultry are common sources of human Salmonella serotype Enteritidis (SE) infections, which cause 6,000 to 7,000 laboratory-confirmed illnesses annually in the United States. These strains of SE increased dramatically in the 1980s in the United States and many other countries, causing egg-associated infections because they can silently infect the ovaries of healthy hens, resulting in internally contaminated eggs (St. Louis et al., 1988). In the United States, initial control measures included use of pasteurized liquid egg product for high-risk foods and institutions, refrigeration during transporta-
tion and sale, and voluntary programs of flock-based interventions. These egg safety programs typically included obtaining SE-free chicks from hatcheries, preventing spread among flocks by biosecurity, cleaning and disinfection, and testing henhouse environments with diversion of eggs to pasteurization if SE was found; these programs were associated with significant decreases in SE infections (Mumma et al., 2004). However, by themselves they are not enough. In July 2010, a nationwide increase in SE infections was identified in the most common pulsed-field gel electrophoresis (PFGE) pattern (CDC, 2010b). In PulseNet, this common PFGE pattern accounted for 30 to 40 percent of all SE isolates. In 2010, approximately 1,900 more laboratory-confirmed illnesses with this outbreak strain were reported than were expected; adjusting for underreporting approximately 55,000 illnesses likely occurred. Epidemiological investigations focused on 29 restaurant and event clusters in 11 states; egg suppliers were identified for 15 of these clusters. A single producer in Iowa was identified as a supplier of shell eggs in 92 percent of clusters with completed tracebacks, and a second Iowa producer supplied eggs to at least one cluster. The first producer was found to sell feed to the second; both producers shared the same source of pullets. Inspection of these producers identified 13 environmental samples matching the outbreak strain and found substantial potential for egg contamination. These producers recalled more than 550 million shell eggs in August 2010. Also in 2010, a year after publication, a new regulation titled “Prevention of Salmonella Enteritidis in Shell Eggs During Production, Transportation and Storage” was implemented for producers with 50,000 or more laying hens (FDA, 2009). The shell egg rule mandates what had previously been voluntary in egg safety programs to prevent SE contamination of eggs during production; to prevent further growth of SE during transportation and storage of shell eggs; and to require recordkeeping of testing results for SE. Shell eggs remain an important vehicle for SE infection, and this new rule is an important step toward enhancing egg safety.
A similar program was launched in the United Kingdom in 1998 to reduce SE infections. In the “British Lion” program, egg producers implemented measures voluntarily; including on-farm biosecurity, cleaning and disinfecting henhouses between flocks, vaccinating hens against SE, and monitoring them for the presence of infection (see http://www.britegg.co.uk/). Eggs from those producers were stamped with a red lion rampant and were thus differentiated from other eggs, including those imported from continental Europe. As with the Scandinavian Campylobacter program, this aligned the consumers’ desire for local and safer eggs to support the cost of the flock-based control programs. Following this launch of this control program, the incidence of SE infections in the United Kingdom fell substantially (Figure A14-4).
SE infections are transmitted through chicken meat as well as through eggs. Recent sampling of retail chicken breasts as part of NARMS indicated that 2 percent are contaminated with SE (FDA, 2011). This is occurring despite efforts to improve slaughter hygiene and inspection processes. As with Campylobacter, it
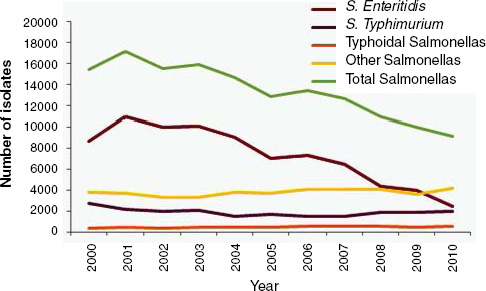
FIGURE A14-4 Number of reported cases of salmonellosis, by serotype, England and Wales, 2000-2010.
SOURCE: Health Protection Agency, http://www.hpa.org.uk/Topics/InfectiousDiseases/InfectionsAZ/Salmonella/EpidemiologicalData/salmDataHuman (accessed April 8, 2012).
is likely that making further progress with poultry-associated SE and other types of Salmonella will require on-farm preharvest interventions. Indeed, it is notable that, despite the extensive efforts to reduce flock contamination of egg-laying flocks, virtually nothing similar has been done with broiler flocks. And as with Campylobacter, there is evidence that on-farm control measures for Salmonella may be effective. For example, parent flock vaccination, combined with hatchery sanitation and on-farm biosecurity in production flocks was recently documented to greatly reduce the Salmonella contamination of poultry carcasses at slaughter (Berghaus et al., 2011; Dorea et al., 2010). This suggests that Salmonella in poultry flocks may often be vertically acquired and that a focused program to reduce contamination in parent flocks may have value, although questions remain about the impact of concurrent immunocompromising infections on immune response to vaccines in chicken flocks (Hoerr, 2010). Aligning the benefit of less contaminated poultry meat with the cost to the producer of taking such measures could lead to their adoption.
Another emerging challenge for food safety officials is multidrug-resistant Salmonella infections. In the United States, many outbreaks of multidrug-resistant Salmonella infections have been investigated in recent years. Because infections with multidrug-resistant Salmonella may be associated with an increased risk of hospitalization and antibiotic treatment failure, these illnesses
are especially concerning (Anderson et al., 2005). In 2011, an outbreak of 136 laboratory-confirmed infections with Salmonella serotype Heidelberg infections were identified in 32 states (CDC, 2011b). Epidemiological and traceback investigations linked these illnesses with consumption of ground turkey from a common production facility, resulting in a recall of 36 million pounds of ground turkey, the largest USDA Class I recall in U.S. history. Antibiotic resistance profiles of patient and environmental samples matched an identical multidrug-resistant pattern, which included several clinically relevant antibiotics. These outbreaks of multidrug-resistant salmonellosis highlight the importance of preharvest food safety programs to reduce the need for antibiotic usage in animals, and for considering further measures to reduce contamination with multidrug-resistant Salmonella.
Traditional animal disease control measures, such as those for trichinosis and brucellosis, have had important impacts on the reduction of infections. Presently, concern exists for agents that may be less pathogenic in animals but can cause serious human disease. Often these animals appear asymptomatic, but they can be shedding bacteria and other agents that cause disease in humans. One example of a recurring public health issue involving human illness linked to contact with asymptomatic animals is the problem of human Salmonella infections linked with live poultry (chicks, ducklings, chickens, ducks, turkeys, and geese) (Loharikar et al., 2012). Since 1990, at least 35 such outbreaks of human Salmonella infections have been reported in the United States (CDC, 2011a). Various Salmonella serotypes have been associated with these outbreaks, and specific outbreak strains have been linked to single mail-order hatcheries over several years (CDC, 2006a). These illnesses are especially severe among young children who account for the majority of infections. Chicks and ducklings appear healthy and clean, but their bodies and areas where they live and roam can be contaminated with Salmonella, leading to human illness. For example, since 2004, Salmonella serotype Montevideo infections having the same genetic fingerprint profile have been reported annually and linked to a single mail-order hatchery. Starting in late 2007, this hatchery implemented numerous interventions to reduce Salmonella transmission in its birds, including biosecurity measures and introduction of an autogenous vaccine specific to the outbreak strain. As of 2011, these measures reduced, but did not eliminate, corresponding human Salmonella infections associated with live poultry from the implicated mail-order hatchery.
Human contact with animals of many types and in various settings carries a risk of infectious illnesses. Petting zoos and similar venues that feature animals like goats, sheep, cattle, pigs, and poultry can be particularly risky for high-risk individuals including children, elderly, and those with weakened immune systems, especially when handwashing facilities are also inadequate. Since the 1990s, more than 150 outbreaks linked to animals displayed in public settings have been reported in the United States (CDC, 2011a). Cultures of specimens from patients, animals, and animal environments have yielded the outbreak strain
in numerous investigations. These animals usually appear healthy but can be shedding zoonotic pathogens that also contaminate the areas where the animals are displayed, leading to infections in people who do not directly contact an animal (Friedman et al., 1998). Strategies that reduce contamination of live animals used for food could also help prevent transmission by direct contact.
Improving Preharvest Prevention: The Plant Sector
Large outbreaks of human infections linked to fresh produce consumed after minimal processing have been more frequently identified in recent decades (Lynch et al., 2009; Sivapalasingam et al., 2004). There is little that consumers can do to protect themselves because these foods are not cooked, washing them has little effect on contamination, and the contamination itself is undetectable, so it is particularly important to prevent such contamination from happening in the first place.
Outbreak investigations have revealed direct links between produce and animal reservoirs. Several recent produce-associated outbreaks have followed wildlife intrusion into growing fields or fecal contamination from nearly animal production facilities that likely led to produce contamination. In 2006, a multistate outbreak of approximately 200 illnesses with E. coli O157:H7 infection from 26 states was linked to the consumption of fresh spinach (CDC, 2006b). An environmental investigation identified E. coli O157:H7 isolates with a PFGE pattern indistinguishable from the outbreak strain in samples obtained from river water, cattle manure, and wild pig feces in and around a field used to grow one brand of spinach from the implicated lot (Wendel et al., 2009). The investigation team also found evidence that wild pigs had been in the spinach fields (FDA, 2006). In August 2006, FDA launched a lettuce safety initiative to address recurring outbreaks of E. coli O157 infections. After this outbreak, the initiative was expanded to include all leafy greens.
An instructive outbreak of produce-related illness linked to wildlife intrusion was identified in Alaska in 2008, when there was a sharp increase in the incidence of Campylobacter infections around Anchorage (Gardner et al., 2011). Raw peas had been suspected as the source of a small cluster in 2005, and a larger increase in 2008 was rapidly shown to be associated with eating raw peas, from one local farm, which was adjacent to a nature preserve for the Sandhill Crane, Grus canadensis. Peas were harvested mechanically and washed in a tank without added chlorine. After harvest, shelled peas were bagged and labeled with directions for blanching, though they were often repacked in bags without this advice, and eaten without blanching. Cranes were observed feeding on peas in the growing fields at the time of harvest, and molecular subtyping studies confirmed that some Campylobacter bacteria isolated from patients were indistinguishable from strains isolated from peas, and from crane feces. Harvest was halted that year, and resumed the following year with scarecranes in the fields, chlorination
in the wash water, and clearly visible advice on the packages to blanch the peas before eating. To date, the outbreak has not recurred. This investigation shows that wild birds may be an underrecognized source of produce contamination, and that some basic prevention measures may make it safer. Animal intrusions have also been suspected as the likely source of contamination of apples in cider orchards by cattle or deer with E. coli O157 and Cryptosporidium (Besser et al., 1993; CDC, 1997), strawberries by deer with E. coli O157 (Laidler and Keene, 2012), and lettuces by wild animals with Yersinia pseudotuberculosis in Finland (Nuorti et al., 2004).
Water contaminated with animal feces and then used to irrigate plants has also been a route connecting plant production with animal reservoirs. In 2006, an outbreak of approximately 80 persons with E. coli O157:H7 infection was linked to lettuce served at locations of a Mexican-style fast food restaurant chain in Iowa and Minnesota (CDPH, 2006). An investigation identified dairy farms near lettuce fields in California that provided lettuce to the restaurants where ill persons had eaten. Environmental samples from the dairy farm and water in soil samples in close proximity to the growing fields identified E. coli O157 indistinguishable from the outbreak strain. The irrigation system was connected to the dairy wastewater blending and distribution system, with inadequate backflow protection devices, presenting a possible route for contaminated water to be used on fields adjacent to the lettuce-growing fields associated with this outbreak. These findings indicated that the nearby dairy farm was the likely source of this outbreak. Contaminated water used for irrigation or for processing has been suspected as the likely source of contamination of outbreaks of E. coli O157 infections traced to lettuce (Hilborn et al., 1999), and tomatoes, mangoes, and cantaloupes with Salmonella (Bowen et al., 2006; Greene et al., 2008; Hedberg et al., 1999; Sivapalasingam et al., 2003).
Sprout-associated outbreaks represent a special scenario, in which the presence of even a few bacterial cells on seeds can be amplified to a large number as a result of the sprouting process itself (Taormina et al., 1999). As seeds are a raw agricultural commodity rather than a food, they may not be expected to be free of pathogens, and their transformation into a food (the sprouts themselves) actually increases the risk, unless special measures are taken to decontaminate the seeds before sprouting and to regularly test the sprouting environment for contamination.
Observations of the biology of human pathogens on plants suggest that interactions between pathogens and produce may sometimes lead to internalization of the pathogen into edible parts of the plant, where it cannot be washed off or eliminated by surface treatments (Berger et al., 2010; Erickson, 2012). This internalization can occur via different mechanisms. Human bacterial enteric pathogens can enter cut or bruised surfaces of leaves and fruit and then multiply in the interior. Air spaces in many fruits contract with a sudden decease in temperature, and this contraction can draw in water and pathogens from the outside of a fruit,
as shown for apples, mangoes, and tomatoes (Burnett et al., 2000; Penteado et al., 2004; Rushing et al., 1996). The interactions may be active as well as passive. In the case of sprouts, grown hydroponically without accompanying soil flora, E. coli O157 and Salmonella present in seeds can enter via the young sprouts’ root hairs and are rapidly found thoughout the entire plant (Itoh et al., 1998; Jaquette et al., 1996). The interactions may be complex. For example, in the dark, Salmonella distribute randomly over the surface of fresh lettuce leaves, but when light stimulates photosynthesis, they concentrate at the stomatal openings that are the respiratory pores on the leaf, as though drawn to products of photosynthetic metabolism (Kroupitski et al., 2009). Some pathogens may be able to manipulate the stomata directly, with a type 3 secretion system that targets the guard cells that ordinarily hold stomata closed in the presence of Gram-negative flagellated bacteria (Saldaña et al., 2011).
These observations raise the question of whether some enteric bacterial pathogens have a life cycle with plant as well as animal hosts. An enteric organism that colonizes herbivores and that also can enter and persist in the plants the herbivores eat gains ready access to the next generation of herbivores. Transfer events from herbivore to plant and plant to herbivore are frequent in prairie or pasture. If enteric pathogens cycle between animal and plant hosts, then the omnivorous human can encounter them on both sides of the cycle. This suggests that, just as food animals may need safer water, fodder, and environments, so food plants may be safer with further attention to the water, soil amendments, and environments used to grow them.
Emerging Foodborne Infections Around the World
Major changes in the global food trade in the past several decades have led to a transformation in the patterns of food production (Florkowski, 2008). The food we eat is sourced from around the globe and distributed over larger distances than ever before. This global trade provides opportunities for exporting countries to earn foreign exchange and drives increases in the standard of living in developing countries. Not only have supply chains become longer, but also the global trade in food has become more specialized. Higher-income countries export grains and processed food to low- and middle-income countries, which in turn export labor-intensive horticultural and fishery products to higher-income countries. Finally, there has been a move toward integration and consolidation of agriculture and food industries, and large corporations have ownership and control across all stages of food production and distribution.
Changes in the globalization of the food trade have important implications for food safety (Tauxe et al., 2010). More imported foods and food ingredients means we depend on food safety systems in other countries. Centralized production of foods means when a problem occurs, it can lead to a widespread outbreak. In this setting, a contaminated food can rapidly cause a geographically
widespread or “dispersed scenario” type of foodborne disease outbreak. In these outbreaks, there are a small numbers of cases in many jurisdictions, typically detected by lab-based subtype surveillance, leading to a multistate or country investigation, and they are usually a result of an industrial contamination event with broad implications. Effective investigations of these types of outbreaks are key to reducing the burden of foodborne disease as we identify food vehicles and factors that lead to outbreaks.
A recent example of a dispersed scenario outbreak was an outbreak of Salmonella serotype Montevideo infections in the United States associated with salami products from one company made with contaminated imported black and red pepper (Julian et al., 2010). A total of 272 cases were identified from 44 states and the District of Columbia during 2009 and 2010. In a multistate case-control study, consumption of salami was associated with illness. The outbreak strain was identified in salami products, one company facility environmental sample, and sealed containers of black and red pepper used to produce the salami products. Pepper tracebacks revealed that the pepper originated from Asian countries, although the locus of contamination was not determined.
Multicontinental outbreaks have been recognized when the same subtype of a pathogen is recognized as a simultaneous source of infection in widely separated populations as a result of global trade in foods and feeds. Detecting such events depends on using the same subtyping strategies in many countries, and on collaboration and information sharing when possible links are recognized, and it is likely that many are missed. For exam ple, a global pandemic of Salmonella serotype Agona in the early 1970s was the result of contaminated anchovy meal shipped from Peru and used in chicken feed around the world. This was first recognized as a restaurant-associated outbreak in Arkansas traced back to one poultry farm and was subsequently linked to sudden increases in Salmonella Agona infections in many countries (Clark et al., 1973). In 1995, simultaneous outbreaks of sprout-associated Salmonella serotype Stanley infections in Arizona, Michigan, and Finland were all linked to seeds from one shipper in the Netherlands, who obtained and blended seeds from many other countries ( Mahon et al., 1997). In 1998, a savory snack produced in Israel caused Salmonella serotype Typhimurium infections in the United Kingdom and North America and was subsequently shown to be a common source of infections in Israel itself; contamination had apparently occurred at the factory where the snack was made (Killalea et al., 1996; Shohat et al., 1996). In 2001, contaminated peanuts from China caused outbreaks of Salmonella serotype Stanley infections in Canada, the British Isles, and Australia (Kirk et al., 2004). In 2010, semi-dried tomatoes from Turkey were recognized as the source of similar hepatitis A virus infections in Australia, France, and the Netherlands (Donnan et al., 2012; Petrignani et al., 2010). These events are surely the tip of the proverbial iceberg and are likely to become more frequently recognized as subtype-based surveillance networks increase worldwide (Swaminathan et al., 2006). They illustrate how a foodborne
pathogen arising in one part of the world can be rapidly disseminated if it is introduced into the global trade networks.
New and unusual foodborne infections continue to arise around the world, as the expanding ecologies of human food production provide niches for the emergence of foodborne pathogens. Two striking examples are described elsewhere in this symposium: Nipah virus encephalitis in Bangladesh, and Shiga toxin—producing E. coli O104 infections in Germany (See Burger, 2011, and Luby et al., 2011, in this volume). In Brazil, Chagas disease, a parasitic infection caused by Trypanosoma cruzi, transmitted by the feces of the triatomid bug, and carried by the opossum, is a classic vectorborne infection, long associated with primitive rural thatched housing. An urban foodborne outbreak occurred in 1986, linked to consumption of fresh sugar cane juice, apparently contaminated by triatomids or opossum feces present in the cane as it was crushed to extract the juice (Shikanai-Yasuda et al., 1991). Since then, such outbreaks have been more frequently recognized, particularly with production of fresh juice of the açai berry, a jungle fruit that is now being grown in orchards to satisfy consumer demand (Nobrega et al., 2009). The transmission may depend on the intersection of this production with Amazonian ecology ( Valente et al., 2009). Such outbreaks may be due to the direct contamination of freshly processed juice with bugs; illness has not been reported from commercial pasteurized product.
In western China, between 2006 and 2009, public health officials investigated several foodborne outbreaks of infection with toxigenic Vibrio cholerae O139, a variant strain of the dominant V. cholerae O1 that first appeared in India in 1995 and spread through Southeast Asia (Li et al., 2008; Tang et al., 2010; Xia et al., 2010). In these outbreaks, the food vehicle has been the soft-shelled turtle, steamed and served at banquet celebrations. These turtles are brought from sources in other parts of China or Asia, where they are produced commercially. A survey of such turtles in Hunan markets found that 7 of 437 had toxigenic V. cholerae O1 or O139 (Xie et al., 2009). The traditional virtues of the long-lived turtle may make it an attractive delicacy for banquets. As with the crab-associated cholera in the United States, future control for this problem may depend on better understanding of circumstances where the turtles are harvested, as well as on better cooking practices (Lowry et al., 1989).
In Israel, where Tilapia species have been raised in fish farms for more than 30 years, a new pathogen emerged in the 1990s, after marketing practices changed. The previous practice had been to market the fish frozen, but in 1996, some began to sell them alive. In 1996-1997, 62 cases of severe Vibrio vulnificus biotype 3 infections were reported, among persons handling the live fish (Bisharat et al., 1999). These infections were food-associated, although not caused by eating the fish itself. They typically started as local wound infection in the person buying the fish, after a penetrating injury from the many spines on the dorsal fin, and rapidly progressed to bacteremia; 41 required surgical wound debridement. Unlike infections with Vibrio vulnificus biotype 1, which cause severe wound
infections in persons who have poor immune systems, and primary bacteremia in oyster eaters with serious liver disease, the patients in Israel were previously healthy. The implicated fish were also healthy and came from a variety of farms. Biotype 3 is a novel recombination of biotype 1 and biotype 2, a pathogen of eels (Bisharat et al., 2005). In 1998, the marketing policy was changed back to selling fish frozen, although cases still occur among those who handle the live fish (Zaidenstein et al., 2008). While it remains unclear where the recombination first occurred, the event is an example of how a pathogen can expand in an agricultural niche and reach the consumer if the circumstances are right.
In Taiwan, since 2000, extremely resistant strains of Salmonella serotype Choleraesuis have caused serious infections in humans and have also been detected in local swineherds (Chiu et al., 2002). This highly invasive serotype has a predilection in humans for endothelial tissues. Most hospitalized patients were admitted with primary bacteremia, sepsis, aortitis, and aortic valve infections (Jean et al., 2006). Human strains have high levels of resistance to ampicillin, chloramphenicol, trimethoprim-sulfamethoxazole, and ciprofloxacin (Chang et al., 2005). More recently, some have been resistant to extended-spectrum cephalosporins as well, due to a transmissible plasmid with blaCMY2 that has recently been found in four other common Salmonella serotypes in addition to Choleraesuis (Su et al., 2011). Indistinguishable strains have been reported from pigs, for which Choleraesuis is a host-adapted pathogen, and it was noted that 40 percent of pig farmers used fluoroquinolones to treat their herds (Hsueh et al., 2004). It seems likely that many of the human infections come from the porcine source, and full control is likely to require changes in agriculture practices to control the infection in pigs.
The challenge of global emerging foodborne infections underlines our interdependence on the public health and food safety systems around the world. Several collaborative programs are actively improving basic public health capacity, promoting standard laboratory identification and subtyping methods, and providing rapid communication. For example, the Training Programs in Epidemiology and Public Health Interventions Network, and the European Program for Intervention Epidemiology Training provide long-term training and practical experience in more than 56 countries (Anonymous, 2006; Moren et al., 1996). The World Health Organization (WHO) Global Foodborne Infections Network (formerly known as WHO Global Salm-Surv) has held 73 short-term multinational multidisciplinary courses to train microbiologists and epidemiologists together from public health, food, and animal medicine sectors (WHO, 2010). PulseNet International has training laboratories in all regions of the world, members in more than 75 counties who have been trained in standardized methods for molecular subtyping, and a global platform for evaluating and introducing new standard methods as they are developed (CDC, 2010c; Swaminathan et al., 2006). The WHO INFOSAN communication channel can link food safety authorities in all countries, so that information about newly identified hazards can be rapidly
disseminated (WHO, 2007). Managing international foodborne outbreaks relies on robust investigations in the countries where disease occurs, and in the countries where the implicated food is grown or manufactured (Tauxe et al., 2008). In many of these investigations, close collaboration with the exporting country authorities and with the food industry can lead to better prevention strategies for the long term.
Conclusion
The complex and changing biological web of the human food supply means that we can expect new pathogens to emerge and novel food vehicles to be identified. Many of these will start in animal reservoirs and may reach us through both animal- and plant-derived foods. Much of the recent progress that has occurred in food safety has been the result of focused efforts to reduce contamination after harvest, for example with better sanitation and process control for meat and poultry at slaughter and in subsequent processing, and better control of processed foods to reduce contamination with Listeria. Contamination can start well before harvest or slaughter, and interventions that focus on the live animal or plant are needed to make further progress in making food safer. Such intervention will depend on understanding the biology of pathogens in the field, their life cycles, and the points at which contamination can be prevented or interrupted. Detecting the new problems will depend on robust capacity for public health surveillance and investigation and on multidisciplinary understanding of the ecologies that sustain them. With our global food supply, problems that arise in one part of the world can spread rapidly, if they enter the global food trade. Improving the safety of the food supply thus depends on stronger public health capacity around the world, better understanding of new challenges wherever they are identified, and translating that understanding into effective prevention from farm to table.
References
Anderson, A. D., M. A. Nelson, N. L. Baker, S. Rossiter, and F. Angulo. 2005. Public health consequences of use of antimicrobial agents in agriculture. In Food safety assurance and veterinary public health, edited by F. J. M. Smulders and J. D. Collins. Wangeningen, NL: Wageningen Academic.
Anonymous. 2006. Global epidemiology. Proceedings of the third TEPHINET Conference, Beijing, China, November 8-12, 2004. Morbidity and Mortality Weekly Report 55(Suppl.).
Baker, M. G., A. Kvalsvig, J. Zhang, R. Lake, A. Sears, and N. Wilson. 2012. Declining Guillain-Barre syndrome after campylobacteriosis control, New Zealand, 1988-2010. Emerging Infectious Diseases 18(2):226-233.
Bell, B. P., M. Goldoft, P. M. Griffin, M. A. Davis, D. C. Gordon, P. I. Tarr, C. A. Bartleson, J. H. Lewis, T. J. Barrett, J. G. Wells, R. Baron, and J. Kobayashi. 1994. A multistate outbreak of Escherichia coli O157:H7-associated bloody diarrhea and hemolytic uremic syndrome from hamburgers: The Washington experience. Journal of the American Medical Association 272:1349-1353.
Berger, C. N., S. V. Sodha, R. K. Shaw, P. M. Griffin, D. Pink, P. Hand, and G. Frankel. 2010. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environmental Microbiology 12(9):2385-2397.
Berghaus, R. D., S. G. Thayer, J. J. Maurer, and C. L. Hofacre. 2011. Effect of vaccinating breeder chickens with a killed Salmonella vaccine on Salmonella prevalences and loads in breeder and broiler chicken flocks. Journal of Food Protection 74:727-734.
Besser, R. E., S. M. Lett, J. T. Weber, M. P. Doyle, T. J. Barrett, J. G. Wells, and P. M. Griffin. 1993. An outbreak of diarrhea and hemolytic uremic syndrome from Escherichia coli O157:H7 in fresh-pressed apple cider. Journal of the American Medical Association 269(17):2217-2220.
Bisharat, N., A. Agmon, R. Finkelstein, R. Raz, G. Ben-Dror, L. Lerner, S. Soboh, R. Colodner, D. N. Cameron, D. L. Wykstra, D. Swerdlow, and J. J. Farmer III; Israel Vibrio Study Group. 1999. Clinical, epidemiological, and microbiological features of Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Lancet 354:1421-1424.
Bisharat, N., D. I. Cohen, R. M. Harding, D. Falush, D. W. Crook, T. Peto, and M. C. Maiden. 2005. Hybrid Vibrio vulnificus. Emerging Infectious Diseases 11(1):30-35.
Bowen, A., A. Fry, G. Richards, and L. Beuchat. 2006. Infections associated with cantaloupe consumption: A public health concern. Epidemiology and Infection 134(4):675-685.
Burnett, S. L., J. Chen, and L. R. Beuchat. 2000. Attachment of Escherichia coli O157:H7 to the surfaces and internal structures of apples as detected by confocal scanning laser microscopy. Applied and Environmental Microbiology 66(11):4679-4687.
CDC (Centers for Disease Control and Prevention). 1997. Outbreaks of Escherichia coli O157:H7 infection and cryptosporidiosis associated with drinking unpasteurized apple cider—Connecticut and New York, October 1996. Morbidity and Mortality Weekly Report 46(1):4-8.
———. 2006a. Three outbreaks of salmonellosis associated with baby poultry from three hatcheries—United States, 2006. Morbidity and Mortality Weekly Report 56(12):273-276.
———. 2006b. Update on multi-state outbreak of E. coli O157:H7 infections from fresh spinach, Octo ber 6, 2006. http://www.cdc.gov/foodborne/ecolispinach/100606.htm (accessed April 8, 2012).
———. 2010a. Foodborne Disease Outbreak Surveillance. http://www.cdc.gov/outbreaknet/surveillance_data.html (accessed April 8, 2012). Atlanta, GA: CDC.
———. 2010b. Investigation update: Multistate outbreak of human Salmonella Enteritidis infections associated with shell eggs. http://www.cdc.gov/salmonella/enteritidis/ (accessed April 8, 2012). Atlanta, GA: CDC.
———. 2010c. PulseNet International. http://www.pulsenetinternational.org (accessed April 8, 2012). Atlanta, GA: CDC.
———. 2010d. WHO Global Foodborne Infection Network. http://www.who.int/gfn/en (accessed April 8, 2012). Geneva: WHO.
———. 2011a. Compendium of measures to prevent disease associated with animals in public settings, 2011. MMWR Recommendations and Reports 60(4):1-28.
———. 2011b. Multistate outbreak of human Salmonella Heidelberg infections linked to ground turkey. http://www.cdc.gov/salmonella/heidelberg/index.html (accessed April 7, 2012).
———. 2011c. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Final Report, 2010. http://www.cdc.gov/narms/pdf/2010-annual-report-narms.pdf (accessed April 7, 2012). Atlanta, GA: U.S. Department of Health and Human Services.
———. 2011d. Surveillance for foodborne disease outbreaks—United States, 2008. Morbidity and Mortality Weekly Report 60(35):1197-1202.
———. 2011e. Vital signs: Incidence and trends of infection with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 1996–2010. Morbidity and Mortality Weekly Report 60(22):749-755.
———. 2012. Notes from the field: Multistate outbreak of Salmonella Altona and Johannesburg infections linked to chicks and ducklings from a mail-order hatchery—United States, February—October 2011. Morbidity and Mortality Weekly Report 61(11):195.
CDPH (California Department of Public Health). 2006. E. coli O157:H7 outbreak associated with iceberg lettuce at Taco John’s December 2006. http://www.cdph.ca.gov/pubsforms/Documents/fdb%20eru%20IceLet%20TacoJohn022008.pdf (accessed March 24, 2012).
Chang, C. C., Y. H. Lin, C. F. Chang, K. S. Yeh, C. H. Chiu, C. Chu, M. S. Chien, Y. M. Hsu, L. S. Tsai, and C. S. Chiou. 2005. Epidemiologic relationship between fluoroquinolone-resistant Salmonella enterica serovar Choleraesuis strains isolated from humans and pigs in Taiwan (1997 to 2002). Journal of Clinical Microbiology 43(6):2798-2804.
Chiu, C. H., T. L. Wu, L. H. Su, C. Chu, J. H. Chia, A. J. Kuo, M. S. Chien, and T. Y. Lin. 2002. The emergence in Taiwan of fluoroquinolone resistance in Salmonella enterica serotype Choleraesuis. New England Journal of Medicine 346(6):413-419.
Clark, G. M., A. F. Kauffman, E. J. Gangarosa, and M. A. Thompson. 1973. Epidemiology of an international outbreak of Salmonella agona. Lancet 2:490-493.
Donnan, E. J., J. E. Fielding, J. E. Gregory, K. Lalor, S. Rowe, P. Goldsmith, M. Antoniou, K. E. Fullerton, K. Knope, J. G. Copland, D. S. Bowden, S. L. Tracy, G. G. Hogg, A. Tan, J. Adamopoulos, J. Gaston, and H. Vally. 2012. A multistate outbreak of hepatitis A associated with semidried tomatoes in Australia, 2009. Clinical Infectious Diseases 54(6):775-781.
Dorea, F. C., D. J. Cole, C. Hofacre, K. Zamperini, D. Mathis, M. P. Doyle, M. D. Lee, and J. J. Maurer. 2010. Effect of Salmonella vaccination of breeder chickens on contamination of broiler chicken carcasses in integrated poultry operations. Applied and Environmental Microbiology 76:7820-7825.
Erickson, M. C. 2012. Internalization of fresh produce by foodborne pathogens. Annual Review of Food Science and Technology 3:283-310.
FDA (Food and Drug Administration). 2006. Ensuring food safety: Tracking and resolving the E. coli spinach outbreak. Rockville, MD: Food and Drug Administration. http://www.fda.gov/NewsEv-ents/Testimony/ucm110926.htm (accessed April 8, 2012).
———. 2009. Final Rule: Prevention of Salmonella Enteritidis in shell eggs during production, storage, and transportation, 21 CFR Parts 16 and 118. Federal Register 74(130):33029-33101. http://www.gpo.gov/fdsys/pkg/FR-2009-07-09/pdf/E9-16119.pdf (accessed April 8, 2012).
———. 2011. 2010 Retail meat report—National Antimicrobial Resistance Monitoring System. Rockville, MD: Food and Drug Administration.
Florkowski, W. J. 2008. Status and projections for foods imported into the United States. In Imported foods: Microbiological issues and challenges, edited by M. Doyle and M. Erickson. Washington, DC: ASM Press. Chap. 1.
Friedman, C., C. Torigan, P. Shillam, E. Hoffman, D. Heltzel, J. Beebe, G. Malcolm, W. DeWitt, L. Huwagner, and P. Griffin. 1998. An outbreak of salmonellosis among children attending a reptile exhibit at a zoo. Journal of Pediatrics 132:802-807.
Gardner, T. J., C. Fitzgerald, C. Xavier, R. Klein, J. Pruckler, S. Stroika, and J. B. McLaughlin. 2011. Outbreak of campylobacteriosis associated with consumption of raw peas. Clinical Infectious Diseases 53(1):26-32.
Greene, S. K., E. R. Daly, E. A. Talbot, L. J. Demma, S. Holzbauer, N. J. Patel, T. A. Hill, M. O. Walderhaug, R. M. Hoekstra, M. F. Lynch, and J. A. Painter. 2008. Recurrent multistate outbreak of Salmonella Newport associated with tomatoes from contaminated fields, 2005. Epidemiology and Infection 136(2):157-165.
Griffin, P. M., and R. V. Tauxe. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiologic Reviews 13:60-98.
Hald, B., H. M. Sommer, and H. Skovgard. 2007. Use of fly screens to reduce Campylobacter spp. introduction in broiler houses. Emerging Infectious Diseases 13(12):1951-1953.
Hedberg, C. W., F. J. Angulo, K. E. White, C. W. Langkop, W. L. Schell, M. G. Stobierski, J. M. Schuchat, J. M. Besser, S. Dietrich, L. Helsel, P. M. Griffin, J. W. McFarland, M. T. Osterholm, and Investigation Team. 1999. Outbreaks of salmonellosis associated with eating uncooked tomatoes: Implications for public health. Epidemiology and Infections 122:385-393.
Hilborn, E. D., J. H. Mermin, P. A. Mshar, J. L. Hadler, A. Voetsch, C. Wojtkunski, M. Swartz, R. Mshar, M. Lambert-Fair, J. A. Farrar, K. Glynn, and L. Slutsker. 1999. A multistate outbreak of Escherichia coli O157:H7 infections associated with consumption of mesclun lettuce. Journal of the American Medical Association 159:1758-1764.
Hoerr, F. J. 2010. Clinical aspects of immunosuppression in poultry. Avian Disease 45:2-15.
Holmes, C. N., and T. C. Chiller. 2004. National Antibiotic Resistance Monitoring System for enteric bacteria. Emerging Infectious Diseases 10(11):2061.
Hsueh, P. R., L. J. Teng, S. P. Tseng, C. F. Chang, J. H. Wan, J. J. Yan, C. M. Lee, Y. C. Chuang, W. K. Huang, D. Yang, J. M. Shyr, K. W. Yu, L. S. Wang, J. J. Lu, W. C. Ko, J. J. Wu, F. Y. Chang, Y. C. Yang, Y. J. Lau, Y. C. Liu, C. Y. Liu, S. W. Ho, and K. T. Luh. 2004. Ciprofloxacin-resistant Salmonella enterica Typhimurium and Choleraesuis from pigs to humans, Taiwan. Emerging Infectious Diseases 10(1):60-68.
Itoh, Y., Y. Sugita-Konishi, F. Kasuga, M. Iwaki, Y. Hara-Kudo, N. Saito, Y. Noguchi, H. Konuma, and S. Kumagai. 1998. Enterohemorrhagic Escherichia coli O157:H7 present in radish sprouts. Applied and Environmental Microbiology 64(4):1532-1535.
Jaquette, C. B., L. R. Beuchat, and B. E. Mahon. 1996. Efficacy of chlorine and heat treatment in killing Salmonella Stanley inoculated onto alfalfa seeds and growth and survival of the pathogen during sprouting and storage. Applied and Environmental Microbiology 62(7):2212-2215.
Jean, S. S., J. Y. Wang, and P. R. Hsueh. 2006. Bacteremia caused by Salmonella enterica serotype Choleraesuis in Taiwan. Journal of Microbiology, Immunology, and Infection 39(5):358-365.
Julian, E., K. MacDonald, N. Marsden-Haug, L. Saathoff-Huber, R. Bonavolante, D. Otero, J. Nosari, C. Austin, D. Von Stein, A. Garvey, G. Kline, C. Lord, R. Groepper, B. Kissler, M. Parish, D. Elder, V. Howard-King, J. Pringle, J. Besser, S. Brown, K. Cooper, S. Sodha, I. Williams, C. Barton Beravesh, and L. Bettencourt Gieraltowski. 2010. Salmonella Montevideo infections associated with salami products made with contaminated imported black and red pepper—United States, July 2009-April 2010. Morbidity and Mortality Weekly Report 59(50):1647-1650.
Killalea, D., L. Ward, D. Roberts, J. de Louvois, F. Sufi, J. Stuart, P. Wall, M. Susman, M. Schwieger, P. Sanderson, I. Fisher, P. Mead, O. Gill, C. Bartlett, and B. Rowe. 1996. International epidemio-logical and microbiological study of outbreak of Salmonella agona infection from a ready to eat savoury snack—I: England and Wales and the United States. British Medical Journal (Clinical Research Edition) 313:1105-1107.
Kirk, M. D., C. L. Little, M. Lem, M. Fyfe, D. Genobile, A. Tan, J. Threlfall, A. Paccagnella, D. Lightfoot, H. Lyi, L. McIntyre, L. Ward, D. J. Brown, S. Surnam, and I. S. T. Fisher. 2004. An outbreak due to peanuts in their shell caused by Salmonella enterica serotypes Stanley and Newport—sharing molecular information to solve international outbreaks. Epidemiology and Infection 132:571-577.
Kroupitski, Y., D. Golberg, E. Belausov, R. Pinto, D. Swartzberg, D. Granot, and S. Sela. 2009. Internalization of Salmonella enterica in leaves is induced by light and involves chemotaxis and penetration through open stomata. Applied and Environmental Microbiology 75(19):6076-6086.
Laidler, M., and W. Keene. 2012. An outbreak of E. coli O157:H7 infections linked to commercial strawberries contaminated by deer, poster 139. In 2012 international conference on emerging infectious diseases. Atlanta, GA: ASM Press.
Li, H. X., X. M. Wang, Z. G. Cui, H. J. Zhou, S. L. Xia, and B. Kan. 2008. Analysis of the source of a cholera outbreak caused by O139 Vibrio cholerae. Disease Surveillance 23(4):218-220.
Loharikar, A. B. E., C. Schwensohn, S. Weninger, J. Wagendorf, J. Scheftel, A. Garvey, K. Warren, E. Villamil, J. A. Rudroff, K. Kurkjian, S. Levine, K. Colby, B. Morrison, A. May, S. Anderson, E. Daly, N. Marsden-Haug, M. M. Erdman, T. Gomez, A. Rhorer, J. Castleman, J. K. Adams, L. Theobald, P. Lafon, E. Trees, J. Mitchell, M. J. Sotir, and C. B. Behravesh. 2012. Four multistate outbreaks of human Salmonella infections associated with live poultry contact, United States, 2009. Zoonoses and Public Health. doi: 10.1111/j.1863-2378.2012.01461.x.
Loneragan, G. H., and M. M. Brashears. 2005. Pre-harvest interventions to reduce carriage of E. coli O157 by harvest-ready feedlot cattle. Meat Science 71:72-78.
Lowry, P. W., A. T. Pavia, L. M. McFarland, B. H. Peltier, T. J. Barrett, H. B. Bradford, J. M. Quan, J. Lynch, J. B. Mathison, R. A. Gunn, and P. A. Blake. 1989. Cholera in Louisiana: Widening spectrum of seafood vehicles. Archives of Internal Medicine 149:2079-2084.
Lynch, M. F., R. V. Tauxe, and C. W. Hedberg. 2009. The growing burden of foodborne outbreaks due to contaminated fresh produce: Risks and opportunities. Epidemiology and Infection 137(3):307-315.
Mahon, B., A. Ponka, W. Hall, K. Komatsu, S. Dietrich, A. Siitone, G. Cage, P. Hayes, M. Lambert-Fair, N. Bean, P. Griffin, and L. Slutsker. 1997. An international outbreak of Salmonella infections caused by alfalfa sprouts grown from contaminated seeds. Journal of Infectious Diseases 175:876-882.
Miller, J. M., and P. M. Griffin. 2012. One Health through eyes of clinical and public health microbiology. Microbe 7(1):23-27.
Moren, A., M. Rowland, F. Van Loock, and J. Giesecke. 1996. The European Programme for Intervention Epidemiology Training. Eurosurveillance 1:30-31.
Mumma, G. A., P. M. Griffin, M. I. Meltzer, C. R. Braden, and R. V. Tauxe. 2004. Egg quality assurance programs and egg-associated Salmonella Enteritidis infections, United States. Emerging Infectious Diseases 10(10):1782-1789.
Nobrega, A. A., M. H. Garcia, E. Tatto, M. T. Obara, E. Costa, J. Sobel, and W. N. Araujo. 2009. Oral transmission of Chagas disease by consumption of acai palm fruit, Brazil. Emerging Infectious Diseases 15(4):653-655.
Nuorti, J. P., T. Niskanen, S. Hallanvuo, J. Mikkola, E. Kela, M. Hatakka, M. Fredericksson-Ahomaa, Lyytikäinen, A. Siitonen, H. Korkeala, and P. Ruutu. 2004. A widespread outbreak of Yersinia pseudotuberculosis O:3 infections from iceberg lettuce. Journal of Infectious Diseases 189:766-774.
Patrick, M. E., B. E. Mahon, S. M. Zansky, S. Hurd, and E. Scallan. 2010. Riding in shopping carts and exposure to raw meat and poultry products: Prevalence of, and factors associated with, this risk factor for Salmonella and Campylobacter infection in children younger than 3 years. Journal of Food Protection 73(6):1097-1100.
Penteado, A. L., B. S. Eblen, and A. J. Miller. 2004. Evidence of Salmonella internalization into fresh mangos during simulated postharvest insect disinfestation procedures. Journal of Food Protection 67:181-184.
Petrignani, M., M. Harms, L. Verhoef, R. van Hunen, C. Swaan, J. van Steenbergen, I. Boxman, R. Peran, I. Sala, H. J. Ober, H. Vennema, M. Koopmans, and W. van Pelt. 2010. Update: A food-borne outbreak of hepatitis A in the Netherlands related to semi-dried tomatoes in oil, January—February 2010. Eurosurveillance 15(20). Available at: http://www.eurosurveillance.org/images/dynamic/EE/V15N20/art19572.pdf (accessed August 7, 2012).
Rangel, J. M., P. H. Sparling, C. Crowe, P. M. Griffin, and D. L. Swerdlow. 2005. Epidemiology of Escherichia coli O157: H7 outbreaks United States, 1982-2002. Emerging Infectious Diseases 11(4):603-609.
Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. New England Journal of Medicine 308:681-685.
Rushing, J. W., F. J. Angulo, and L. R. Beuchat. 1996. Implementation of a HACCP program in a commercial fresh-market tomato packinghouse: A model for the industry. Dairy, Food and Environmental Sanitation 16(9):549-553.
Saldaña, Z., E. Sanchez, J. Xicohtencatl-Cortes, J. L. Puente, and J. A. Giron. 2011. Surface structures involved in plant stomata and leaf colonization by shiga-toxigenic Escherichia coli O157:H7. Frontiers in Microbiology 2:119.
Scallan, E. 2007. Activities, achievements, and lessons learned during the first 10 years of the Foodborne Diseases Active Surveillance Network: 1996-2005. Clinical Infectious Diseases 44(5):718-725.
Scallan, E., P. M. Griffin, F. J. Angulo, R. V. Tauxe, and R. M. Hoekstra. 2011a. Foodborne illness acquired in the United States—unspecified agents. Emerging Infectious Diseases 17(1):16-22.
Scallan, E., R. M. Hoekstra, F. J. Angulo, R. V. Tauxe, M. A. Widdowson, S. L. Roy, J. L. Jones, and P. M. Griffin. 2011b. Foodborne illness acquired in the United States—major pathogens. Emerging Infectious Diseases 17(1):7-15.
Schantz, P. M. 1983. Trichinosis in the United States—1947-1981. Food Technology 83-86.
Sears, A., M. G. Baker, N. Wilson, J. Marshall, P. Muellner, D. M. Campbell, R. J. Lake, and N. P. French. 2011. Marked campylobacteriosis decline after interventions aimed at poultry, New Zealand. Emerging Infectious Diseases 17(6):1007-1015.
Shikanai-Yasuda, M. A., C. B. Marcondes, L. A. Guedes, G. A. Siqueira, A. A. Barone, J. C. P. Dias, V. Amato Neto, J. E. Tolezano, B. A. Peres, E. R. Arruda Jr., M. H. Lopes, M. Shiroma, and E. Chapadeiro. 1991. Possible oral transmission of acute Chagas’ disease in Brazil. Revista do Instituto de Medicina Tropical de São Paulo 33(5):351-357.
Shohat, T., M. S. Green, D. Merom, O. N. Gill, A. Reisfeld, A. Matas, D. Blau, N. Gal, and P. E. Slater. 1996. International epidemiological and microbiological study of outbreak of Salmonella agona infection from a ready to eat savoury snack—II: Israel. British Medical Journal (Clinical Research Edition) 313:1107-1109.
Sivapalasingam, S., E. Barrett, A. Kimura, M. S. Van Duyne, W. De Witt, M. Ying, A. Frisch, Q. Phan, E. Gould, P. Shillam, V. Reddy, T. Cooper, M. Hoekstra, C. Higgins, J. P. Sanders, R. V. Tauxe, and L. Slutsker. 2003. A multistate outbreak of Salmonella enterica serotype Newport infections linked to mango consumption: Impact of a water-dip disinfestation technology. Clinical Infectious Diseases 37:1585-1590.
Sivapalasingam, S., C. R. Friedman, L. Cohen, and R. V. Tauxe. 2004. Fresh produce: A growing cause of outbreaks of foodborne illness in the United States; 1973 through 1997. Journal of Food Protection 67(10):2342-2353.
St. Louis, M. E., D. L. Morse, M. E. Potter, T. M. DeMelfi, J. J. Guzewich, R. V. Tauxe, and P. A. Blake. 1988. The emergence of grade A eggs as a major source of Salmonella Enteritidis infections: Implications for the control of salmonellosis. Journal of the American Medical Association 259:2103-2107.
Su, L. H., W. S. Teng, C. L. Chen, H. Y. Lee, H. C. Li, T. L. Wu, and C. H. Chiu. 2011. Increasing ceftriaxone resistance in Salmonellae, Taiwan. Emerging Infectious Diseases 17(6):1086-1090.
Swaminathan, B., T. J. Barrett, S. B. Hunter, R. V. Tauxe, and CDC PulseNet Task Force. 2001. PulseNet, the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerging Infectious Diseases 7(3):382-389.
Swaminathan, B., P. Gerner-Smidt, L. K. Ng, S. Lukinmaa, K. M. Kam, S. Rolando, E. Perez Gutierrez, and N. Binsztein. 2006. Building PulseNet International: An interconnected system of laboratory networks to facilitate timely public health recognition and response to foodborne disease outbreaks and emerging foodborne diseases. Foodborne Pathogens and Diseases 3:36-50.
Tang, X. F., L. G. Liu, H. L. Ma, B. P. Zhu, C. X. Hao, X. Y. Wu, N. Fei, X. P. Zhu, and L. J. Zhang. 2010. Outbreak of cholera associated with consumption of soft-shelled turtles, Sichuan Province, China, 2009. Chinese Journal of Epidemiology 31(9):1050-1052.
Taormina, P. J., L. R. Beuchat, and L. Slutsker. 1999. Infections associated with eating seed sprouts: An international concern. Emerging Infectious Diseases 5:626-634.
Tauxe, R. V. 2006. The burden of illness associated with foodborne threats to health, and the challenge of prevention. In Addressing foodborne threats to health: Policies, practices and global coordination. Workshop summary. Washington, DC: The National Academies Press.
Tauxe, R. V., and E. Esteban. 2006. Advances in food safety to prevent foodborne diseases in the United States. In Silent victories: The history and practice of public health in twentieth-century America, edited by J. Ward and C. Warren. Oxford: Oxford University Press. Chap. 2.
Tauxe, R. V., S. J. O’Brien, and M. Kirk. 2008. Outbreaks of foodborne diseases related to the international food trade. In Imported foods: Microbial issues and challenges, edited by M. P. Doyle and M. C. Erickson. Washington, DC: ASM Press. Pp. 69-112.
Tauxe, R. V., M. P. Doyle, T. Kuchenmüller, J. Schlundt, and C. E. Stein. 2010. Evolving public health approaches to the global challenge of foodborne infections. International Journal of Food Microbiology 139(Suppl. 1):S16-S28.
Tustin, J., K. Laberge, P. Michel, J. Reiersen, S. Dadadottir, H. Briem, H. Hardardottir, K. Kristinsson, E. Gunnarsson, V. Fridriksdottir, and F. Georgsson. 2011. A national epidemic of campylobacteriosis in Iceland, lessons learned. Zoonoses and Public Health 58(6):440-447.
Valente, S. A. S., V. C. Valente, A. Y. N. Pinto, M. J. B. Miranda, M. P. dos Santos, C. O. S. Miranda, P. Cuervo, and O. Fernandes. 2009. Analysis of an acute Chagas disease outbreak in the Brazilian Amazon: Human cases, triatomines, reservoir mammals, and parasites. Transactions of the Royal Society of Tropical Medicine and Hygiene 103:291-297.
Wagenaar, J. A., W. Jacobs-Rietsma, M. Hofshagen, and D. Newell. 2008. Poultry colonization with Campylobacter and its control at the primary production level. In Campylobacter, 3rd ed., edited by I. Nachamkin, C. Szymanski, and M. Blaser. Washington, DC: ASM Press. Pp. 667-668.
Wendel, A. M., U. Sharapov, J. Grant, J. R. Archer, T. Monson, C. Koschmann, and J. P. Davis. 2009. Multistate outbreak of Escherichia coli O157:H7 infection associated with consumption of packaged spinach, August-September 2006: The Wisconsin investigation. Clinical Infectious Diseases 48(8):1079-1086.
WHO (World Health Organization). 2007. The International Food Safety Authorities Network (INFOSAN). http://www.who.int/foodsafety/fs_management/infosan/en/ (accessed April 8, 2012).
———. 2010. WHO Global Foodborne Infection Network. http://www.who.int/gfn/en (accessed August 7, 2012).
Xia, B. Z., Q. Li, J. Long, J. Xia, and H. Zhao. 2010. Analysis of a foodborne outbreak of cholera. Journal of Tropical Medicine (Chinese) 10(1):88-89.
Xie, Q., X. J. Zeng, and W. Zheng. 2009. Survey of Vibrio cholerae-carrying rate of short-shelled turtles sold in markets of Chenzou City. China Tropical Medicine 9(1):135-136.
Zaidenstein, R., C. Sadik, L. Lerner, L. Valinsky, J. Kopelowitz, R. Yishai, V. Agmon, M. Parsons, C. Bopp, and M. Weinberger. 2008. Clinical characteristics and molecular subtyping of Vibrio vulnificus illnesses, Israel. Emerging Infectious Diseases 14(12):1875-1882.
ANTIBIOTIC RESISTANCE—LINKING
HUMAN AND ANIMAL HEALTH
This paper will address the transmission of antibiotic-resistant microorganisms between animals and humans in a One Health perspective. It will give a general introduction to the epidemiology of antibiotic resistance in zoonotic pathogens and then focus on some national and international programs for integrated surveillance and control of antimicrobial resistance at the human—animal interface, with particular emphasis on programs implemented in the authors’ home country, Denmark.
______________
32 Provost, Chief Academic Officer, Technical University of Denmark, Anker Engelunds Vej 1, 2800 Lyngby, Denmark.
Overview
The epidemiology of antimicrobial-resistant microorganisms at the human—animal interface involves complex and largely unpredictable systems that include transmission routes of resistant bacteria as well as resistance genes and the impact of antimicrobial selective pressures in several reservoirs (animals, humans, and the environment) (Figure A15-1).
Thus the One Health approach is useful when it comes to addressing zoonotic transmission of pathogens that are resistant to antimicrobials, because we need to engage a wide range of stakeholders including farmers, veterinarians, food safety professionals, medical doctors, as well as environment and wildlife experts in monitoring and control activities.
The feature that particularly differentiates antimicrobial resistance from other food safety—related problems is the role of the chemical driver, the antimicrobials, which selects for the resistant bacteria that subsequently can spread between animals and humans.
Antimicrobials are used widely to prevent or treat disease in food animals. The major part of the usage is for prevention of disease, and their use has become an integral part of modern industrialized food-animal production, to the extent where nearly all feed for growing animals is supplemented with antimicrobials in various doses, ranging from so-called “subtherapeutic concentrations” to full therapeutic doses. It is estimated that the volumes of antimicrobials used in food animals exceeds the use in humans worldwide, and nearly all the classes of antimicrobials that are used for humans are also being used in food animals, including the newest classes of drugs such as third- and fourth-generation cephalosporins, fluoroquinolones, glycopeptides, and streptogramins (Aarestrup et al., 2008).
The massive use of antimicrobial agents in agriculture has supported the intensification of modern food-animal production since the early 1960s by facilitating earlier weaning, higher animal densities, and the use of cheaper feed sources, among others, and has most likely contributed to increased outputs and lower prices of meat. However, the gains have come at a cost, which is being borne, in part, by other stakeholders, in particular public health. Furthermore, the production gains achieved by indiscriminate antimicrobial usage in the 1960s production systems may to a large extent be achievable by other means in modern and more environmentally sustainable food-animal production systems, where higher emphasis is placed on animal welfare, a smaller environmental footprint, and disease prevention through hygiene and intelligent herd management.
The amounts and patterns of antimicrobials used in food animals is the major determinant for the propagation of resistant bacteria in the animal reservoir. Thus, the levels and patterns of resistance observed in food animals to a wide extent reflect the patterns of drug usage; however, other determinants also play a part, such as spread of bacterial clones between animals, in particular vertical spread between the generations (e.g., the spread of resistant Salmonella in the poultry
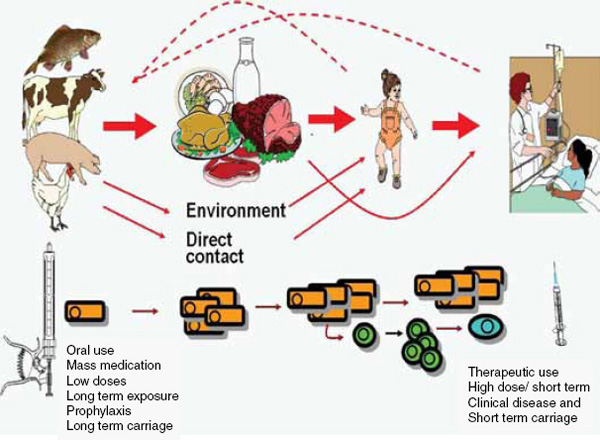
FIGURE A15-1 The epidemiology of antimicrobial resistance at the human-animal interface is invariably complex. It involves a multitude of potential transmission routes and vehicles, antimicrobial selective pressures and other ecological drivers, as well as horizontal transmission of resistance genes between bacterial species and genera.
and swine breeding pyramids), and successful adaptation of clones resistant to the animal reservoir (e.g., MRSA CC398) (Aarestrup et al., 2008).
Transmission of resistance from animals to humans can take place through a variety of routes (Figure A15-1), where the food-borne route probably is the most important (most infections with enteric bacterial pathogens, such as Salmonella enterica, Campylobacter coli/jejuni, and Yersinia enterocolitica, probably occur through this route in industrialized countries), whereas, for other resistant pathogens, direct contact between animal and humans may be the major route of transmission (e.g., MRSA CC398). Bacteria as well as antibiotic residues from food-animal production are spread widely in the environment, mainly with manure, where it affects bacteria in the environment as well as in wild fauna. Thus, the environment and wild fauna can become reservoirs of resistance and a source of reintroduction of resistant bacteria into the food-animal and human reservoirs.
The public health consequences of zoonotic antibiotic resistance are invariably difficult to assess for a number of reasons: the epidemiology is highly complex because it involves complex production and distribution systems of animals and food, it involves the spread of bacterial clones as well as resistance genes, and, finally, the impact on public health includes several end points that are difficult to determine, such as infections that would otherwise not have occurred, increased morbidity and mortality, and higher costs of treatment of disease. In the most comprehensive assessment of the problem to date, an expert group gathered by the World Health Organization (WHO), the Food and Agriculture Organization (FAO), and the World Animal Health Organization (OIE) in 2003 concluded
there is clear evidence of adverse human health consequences due to resistant organisms resulting from non-human usage of antimicrobials. These consequences include infections that would not have otherwise occurred, increased frequency of treatment failures (in some cases death) and increased severity of infections, as documented for instance by fluoroquinolone resistant human Salmonella infections. Evidence shows that the amount and pattern of non-human usage of antimicrobials impact on the occurrence of resistant bacteria in animals and on food commodities and thereby human exposure to these resistant bacteria. The foodborne route is the major transmission pathway for resistant bacteria and resistance genes from food animals to humans, but other routes of transmission exist. There is much less data available on the public health impact of anti microbial usage in aquaculture, horticulture and companion animals.” (FAO et al., 2003)
Investigating the zoonotic antimicrobial resistance problem in its full complexity requires monitoring of antimicrobial usage and resistance in all relevant reservoirs and stages in the transmission route, and coherent analysis of the data (i.e., “integrated monitoring”). For the purpose of intervention, there are multiple potential points of control that may be used, depending on the specific nature of
the problem. Identifying and intervening at the most efficient points of control requires a comprehensive assessment of the risk based on integrated monitoring, as well as good collaboration between all the stakeholders involved.
Already in the early 1960s, findings of resistant Salmonella in food animals and humans, and studies that showed that they could pass their resistance traits on to other enteric bacteria, gave rise to major concern in the United Kingdom. This led to the formation of the “Swann Committee,” which recommended
that only antibiotics which “have little or no application as therapeutic agents in man or animals and will not impair the efficacy of a prescribed therapeutic drug or drugs through the development of resistant strains of organisms” should be usable for growth promotion. (Swann et al., 1969)
This was put into the UK legislation and subsequently the European Union legislation. The United States and the rest of the world, however, did not follow the same path.
In the mid-1990s the detection of vancomycin-resistant Enterococcus faecium as well as quinolone-resistant Salmonella and Campylobacter in food animals and evidence of their spread to humans elevated the scientific and public concerns to new levels. This prompted a series of international expert consultations and meetings under the auspices of the WHO and/or the OIE, and it also led to implementation of specific interventions to contain antimicrobial resistance in the food-production chain in many countries, most importantly the complete termination of the use of antimicrobial growth promoters in Europe (FAO et al., 2004; WHO, 1997).
Recently a number of antimicrobial-resistant pathogens have emerged in the food-production chain: extended beta-lactamase producing Salmonella and Escherichia coli, transmissible quinolone resistance (qnr) in Salmonella and E. coli and animal-associated methicillin-resistant Staphylococcus aureus (MRSA), which can transmit to, and cause infections in, humans. These emergences can all be associated with the use of antimicrobial agents in food animals, and they have led to renewed attention to the use of certain types of antimicrobials in food animals that are considered critically important for human health (Aarestrup et al., 2008; Xia et al., 2010).
Residues of antimicrobial agents that may occur in animal-derived products appear to be of a lesser concern for public health than the resistant bacteria. A WHO expert committee concluded in 2003 that residues of antimicrobials in foods, under present regulatory regimes, represent a significantly less important human health risk than the risk related to antimicrobial-resistant bacteria in food (FAO et al., 2003).
Use of antibiotic resistance genes as marker genes in genetically modified plants, which may serve as feed for animals or food for humans, has also raised concerns in this context. Recently, the European Food Safety Authority (EFSA) conducted a risk assessment based on the current state of the science and con-
cluded the following: “Notwithstanding these uncertainties, the current state of knowledge indicates that adverse effects on human health and the environment resulting from the transfer of these two antibiotic resistance genes from GM plants to bacteria, associated with use of GM plants, are unlikely” (EFSA, 2009).
Increased overlap between humans and wildlife populations may increase the risk for novel disease emergence in wildlife in a recent study by Wheeler et al. (2012). Antibiotic resistance was used as a molecular marker for the intensity of human—wildlife microbial connectivity in the Galápagos Islands. Antibiotic-resistant bacteria were found in reptile feces from tourism sites, whereas no resistance was detected at protected beaches on more isolated islands, indicating that human contact may be the source of resistant enteric bacteria (E. coli and Salmonella) in Galápagos wildlife (Wheeler et al., 2012).
Recognizing the continued emergence of new bacterial pathogens, in animals, that are resistant to antimicrobials considered critically important for human therapy, there is good reason to further strengthen global efforts to prevent and control the emergence and spread of resistance from animals to humans. The One Health concept and its focus on the interdependencies and links between the three health systems of animals, humans, and the environment, respectively, are extremely well suited for this purpose.
The remaining part of this article describes some examples of national and international interventions to contain antimicrobial resistance in the food-production chain, with main emphasis on interventions employed in the authors’ home country, Denmark, which happen to be some of the most advanced and also best documented interventions in this regard.
National and International Attempts to Monitor and Control Transmission of Antimicrobial Resistance Between Animals and Humans
A large number of national and international rules and regulations are involved in the regulation and control of food-borne antimicrobial resistance.
Legal Framework at the International Level
The Codex Alimentarius Commission (CAC), under the WHO and the FAO, has issued recommendations that should be implemented by all countries as a code of practice to minimize and contain antimicrobial resistance (CAC, 2005). This code of practice gives recommendation for the responsibilities of regulatory authorities, the veterinary pharmaceutical industry, veterinarians, and wholesale and retail distributors and producers.
As examples, regulatory authorities should ensure that antimicrobial agents are prescription only (thus, not used for growth promotion), only drugs that are efficacious and with well-established dosages should be approved, surveil-
lance programmes for monitoring drug use and resistance should be established, research should be encouraged, and all unused drug should be collected and destroyed. It is stated that veterinarians should only prescribe drugs for animals under their care and ensure that the drugs used are aimed at clinical disease. In addition, the professional organization should develop clinical practice guidelines on responsible use. In addition to these international recommendations a large number of different national legislation regulates the use of antimicrobial agents and the control of antimicrobial-resistant bacteria.
Below are some examples of control options and their effect on resistance.
Possible Risk Management Options
Currently from a practical and legal point of view, control options are usually divided into pre-harvest (e.g., on farm) and post-harvest (e.g., slaughterhouse and food). However, a more logical way to look at this problem would be to either avoid selection and/or stop the spread of resistant bacteria. Thus, the control of antimicrobial resistance can be controlled either through management of the selective pressure leading to resistance or through interventions aimed at limiting the spread of the selected resistance.
Continuous and updated information is essential to guide risk management and to determine the effect of possible interventions. Thus, continuous monitoring of the occurrence of food-borne pathogens, antimicrobial resistance, and drug use as well as research studies determining the effects of interventions and the associations between different reservoirs, the spread of bacterial clones and genes, and risk factors for the development and spread of resistance are essential for efficient risk management.
Monitoring of Antimicrobial Resistance Information on the occurrence of antimicrobial resistance is needed at the local, national, and international levels to guide policy and detect changes that require intervention strategies. Such monitoring programmes should be continuous and standardized, enabling comparison between countries as well as over time. The main aspects to be considered in establishing a monitoring system include animal or food groups to be sampled, the number of samples to take and the strategy for collection, bacterial species to be included, methods for susceptibility testing, antimicrobials to test, break points to use, quality control, data to be reported, analysis and interpretation of data, and reporting (Bager et al., 1999). The Danish Integrated Antimicrobial Resistance Monitoring Programme (DANMAP) established in 1995 was the first integrated program in Europe (Figure A15-2). Recently a proposal for a common protocol for antimicrobial resistance monitoring was proposed for Europe (EFSA, 2008). It can be hoped that this can form the basis for a future establishment of a common global standard.
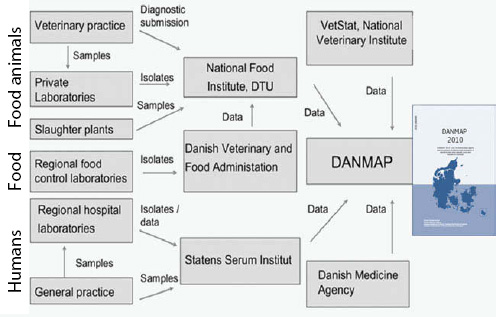
SOURCE: DANMAP (2010).
Monitoring of Antimicrobial Drug Use Data on drug usage is essential for the development of national and international policies for containment of antimicrobial resistance. In Denmark, a programme called Vetstat was implemented in August 2000 and has since collected data from veterinarians, pharmacies, and feed mills. The programme monitors the use of all prescription medicine in production animals, including sera and vaccines, as well as the use of coccidiostatics (Stege et al., 2003). Data are collected at the farm level and include information concerning animal species, age of animal, disease, farm identification number, veterinarians’ number, drug identification number, amount of medicine, and date for use of medicine. Today Vetstat enables the authorities to assess usage patterns at the level of the individual herd and individual prescriber. Furthermore, many veterinarians use Vetstat daily as a tool in relation to their service for their clients (farmers). Because all data are converted to defined animal daily doses (ADDs) it is possible to compare the use of antibiotics on one farm with a similar average for the whole country.
In 2010 the Danish Veterinary and Food Authority (DVFA) introduced the “Yellow Card Initiative” based on Vetstat (DVFA, 2012). Each year, DVFA will issue threshold limits for antimicrobial consumption in pigs (other animal species may follow later). The limits for pigs in 2010 were as follows:
1. Weaners (7-30 kg): 28 ADD per 100 weaners per day.
2. Young pigs, including young females (over 30 kg), excluding sows, gilts, and boars: 8 ADD per 100 pigs per day.
3. Sows, gilts, and boars: 5.2 ADD per 100 pigs per day.
If the average antimicrobial consumption in a holding within a 9-month period exceeds one or more of the threshold limits, DVFA may issue an order or injunction (the yellow card) compelling the owner of the holding, in collaboration with the veterinary practitioner, to reduce the antimicrobial consumption in the holding below the threshold limits within 9 months.
The total use of antimicrobials in swine has been reduced by 21 percent in Denmark, following the introduction of the Yellow Card Initiative, when comparing national data on usage for the years 2009 and 2011, respectively (DVFA, 2012).
Recently a first attempt to collect comparable veterinary antibiotic usage data for the European countries was carried out (Figure A15-3) (Grave et al., 2010). The rather large differences between the different countries can be explained by differences in types of animal production systems, different veterinary antibiotic policies and practices, or differences in disease occurrence. A recent study reported that a conservative estimate of the comparable figure for the United States was considerably higher, approximately 300 mg/kg (Aarestrup et al., 2010).
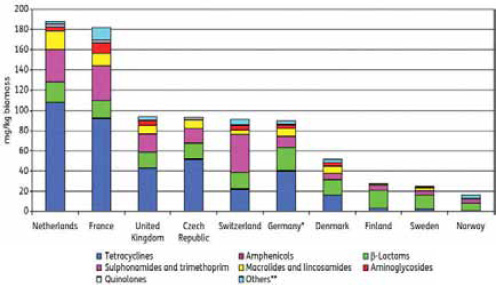
SOURCE: Grave et al. (2010). The comparable figure for the United States is estimated to be approximately 300 mg/kg according to Aarestrup et al., 2010.
Limiting the Selective Pressure
Prescription One of the basic principles in the Codex Alimentarius codes of practice to minimize and contain antimicrobial resistance is that all antimicrobial agents should be on prescription, and the right to prescribe drugs should rest with the veterinarians or other animal health professionals with an appropriate education. Prescribing and dispensing should be separated to avoid conflicts of interest.
Drug Approval All drugs intended for human or animal use undergo an approval process before licensing, which differs somewhat between countries even though some general guidelines are used. The traditional risks that are considered in the approval process include proof of efficacy against the target pathogen, target animal safety, environmental safety, and human health safety with a focus on toxicological effects (residues). Human hazards related to the transfer of antimicrobial resistance are of more recent concern and have so far only had limited focus in the approval process. In 2003 the U.S. Food and Drug Administration (FDA) published a guidance document for a qualitative risk assessment to be performed prior to the approval of an antimicrobial agent for animal use (FDA, 2003). This guideline outlines an evidence-based approach to preventing antimicrobial resistance from emerging in humans as a consequence of using antimicrobial agents in animals. This guidance requires a ranking into high, medium, and low of the following factors: (1) probability that resistant bacteria are present in target animals as a consequence of drug use, (2) probability for humans to ingest the bacteria in question, and (3) probability that human exposure results in an adverse effect. These three factors are then joined together in an overall risk estimate ranked as high, medium, or low. In combining the three factors, the most value is put on the consequence estimate. Thus, in essence, antimicrobial agents considered “critically important” will be ranked as having a high risk no matter what the probability for selection or transfer. Thus, already in the approval process consideration as to whether antimicrobials are of critical importance for human health can be taken into account. As an example, in Australia fluoroquinolone use was never approved for use in food animals. Fluoroquinolone-resistant strains are either at very low levels or nonexistent in food animals. The rates of resistance are also very low in human isolates in comparison to other countries (e.g., community onset bloodstream infection resistance rate in E. coli of 2 percent) (Kennedy et al., 2008).
It is also possible in the approval process to implement certain restrictions. Thus, it could be possible to approve drugs for a limited number of indications, without accepted extralabel or off-label usage or for some modes of administration only (e.g., only parenteral use). This has recently been done by FDA in the United States, which in July 2008 issued an order coming into effect by October 2008 prohibiting the extralabel use of cephalosporin antimicrobial drugs in food-producing animals (FDA, 2008).
Treatment formularies and prescriber guidelines In Denmark a veterinary treatment formulary was published by the National Food Institute in 1997 ( Pedersen et al., 1999). This formulary was mainly targeted toward concerns for human health, but it also took into account the prevalence of antimicrobial resistance among bacteria causing infections in animals. In the formulary, antimicrobials for every disease and associated pathogen(s) are listed and scored (1-3) within the following four categories: efficacy, resistance among the pathogen causing infection in animals, national criteria for human importance in Denmark, and WHO criteria for Critically Important Antimicrobials (WHO, 2005).
It is difficult to evaluate the specific effects of the guidelines. However, considering that one of the main recommendations in Denmark has been to limit the use of macrolides and cephalosporins and that the use of these classes of antimicrobials for pigs, which constitutes 80 percent of the usage for animals in Denmark, has increased by 30 and 33 percent, respectively, between 2005 and 2006 (DANMAP, 2006), the effect seems to be minor.
Restrictions on the use of certain antimicrobial classes It is also possible to implement national or international restrictions on certain antimicrobial classes. As mentioned, in Australia fluoroquinolones are not registered for use in food animals. In Denmark fluoroquinolones were approved for use in production animals in 1993, and in the following years an emergence of resistance was observed. In the year 1999 the farmers voluntarily stopped the use of in-feed fluoroquinolones, and in 2002 the veterinarians’ use and prescription of fluoroquinolones to food- producing animals were further restricted by the authorities. Thus, fluoroquinolones can only be used in food-producing animals if a current laboratory test of resistance patterns shows that no other antimicrobial will be effective in treatment of the disease in question and this has been reported to the regional veterinary officer. Furthermore, fluoroquinolones can only be administered by injection and by the veterinarian only. This reduced the total usage of fluoroquinolones in animals in Denmark from 183 kg in 2001 to 49 kg in 2006 (Figure A15-4).
Limiting the prescribers’ profit on the sale of antimicrobial agents In many countries a considerable part of the veterinarians’ income comes from the direct sale of antibiotics to the farmers. This could tempt some veterinarians to overprescribe antibiotics because of the financial benefit. An example from Denmark showed that limiting the possibility of veterinarians to profit from the sale of drugs led to a reduction in total usage. In 1995 the Danish government issued a new legislation reducing and fixing the veterinarians’ profit from the direct sale of antibiotics to a maximum of 5 percent. Furthermore, a veterinarian can only sell antibiotics to a farmer during a visit and for a maximum of 5 days of use. The rest has to be bought at a pharmacy. This resulted in a reduction of 40 percent in total use of
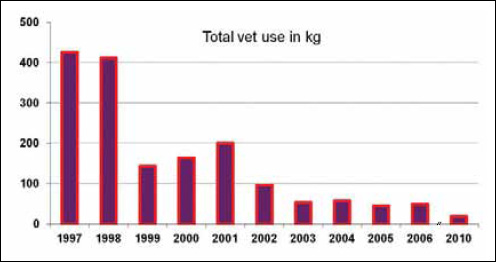
therapeutic agents and a reduction in tetracycline use from almost 37 tonnes in 1994 to 9 tonnes in 1995 (Grave and Wegener, 2003).
Price and taxation In human medicine several studies have shown an association between expenses and the prescription of a specific drug. It is a reasonable assumption that the cost of the drug is a considerable factor for the farmer’s decision on when and how to use antimicrobials over other disease control and prevention options. In Denmark, a tax was imposed on antimicrobial growth promoters in 1998. The purpose of the tax was to remove the postulated financial benefit from using the antimicrobial growth promoters. From 1998, and until the ban in 2000, a sharp reduction on overall use of antimicrobial growth promoters occurred, but this could also be explained by other factors such as public and media attention, implementation of industry codes of practice, etc. More scientific studies addressing the effects of taxation as a risk management tool are needed.
Voluntary withdrawals or banning of drugs In the United Kingdom the use of tetracyclines and penicillin as growth promoters was banned following the recommendation the Swann report.
In 1995 the Danish Ministry of Agriculture, Fisheries and Food decided to ban the use of the growth promoter avoparcin because of its cross-resistance to vancomycin, a critically important antimicrobial for human use. In 1997, the European Union (EU) banned the use of avoparcin. In 1998 Denmark banned
the use of virginiamycin because of cross-resistance to the critical important Quinupristin-Dalfopristin used in humans. In 1998, the Danish animal production industry voluntarily stopped the use of growth promoters; only swine up to 35 kg bodyweight were still treated with growth promoters until January 2000. In 1999 the EU banned tylosin, spiramycin, virginiamycin, and bacitracin, and the remaining growth promoters were banned in the EU from January 2006. The gradual banning of growth promoters in Denmark resulted in a 50 percent reduction of the usage of antimicrobial agents in animal production from 1997 to 1998, and consequential reductions in the levels of antimicrobial resistance in a range of different bacterial species in food animals (Figures A15-5 and A15-6) (Aarestrup et al., 2008).
In the first quarter of 2005 there was a voluntary withdrawal in Québec chicken hatcheries of the extralabel use of ceftiofur. After the withdrawal, a significant decrease in ceftiofur resistance was seen in Salmonella Heidelberg isolates from retail chicken and humans, as well as in E. coli from retail chickens (Figure A15-7) (Dutil et al., 2010).
The examples above show that reduction in the use of antimicrobial agents can have a positive effect on the occurrence of antimicrobial resistance. The
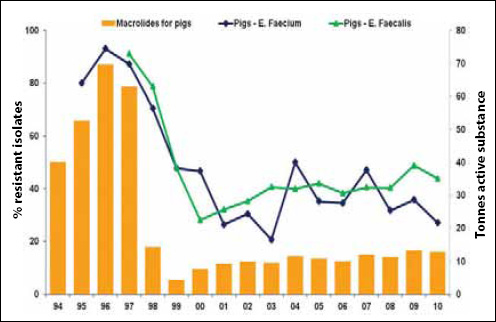
SOURCE: DANMAP (2010).
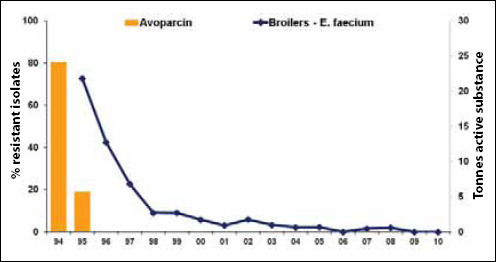
SOURCE: DANMAP (2010).
disadvantage of relying on voluntary withdrawals is that there are no controls that prevent the same groups from later reintroducing these antibiotics and the consequential rapid rise in resistance rates that will result. In fact the chicken hatcheries in Québec have already begun using ceftiofur again.
Preventive veterinary medicinal strategies Disease prevention is an integrated part of food-animal production, and Specific Pathogen Free swine and poultry production systems use this option actively. Preventing disease is considered an essential factor in reducing antimicrobial usage. Strangely only few published scientific studies seem to have addressed this specific point. The most likely explanation for the lack of scientific confirmation thereof is the lack of combined data on management systems, drug use, disease incidence, and antimicrobial resistance.
In a study from Norway the effect of introducing vaccines for prevention of disease in farmed salmon was investigated. The introduction of vaccines led to a substantial reduction in the use of antimicrobials in Norwegian aquaculture (Figure A15-8) (Markestad and Grave, 1997).
It is important to note that whenever antibiotics have been removed as routine food additives for growth promotion and disease prevention purposes there has been no or little evidence that this has resulted in any decrease in animal health or food production.
Controlling Spread of Resistant Bacteria Improved hygiene and infection control is a well-established and essential part of controlling infectious diseases.
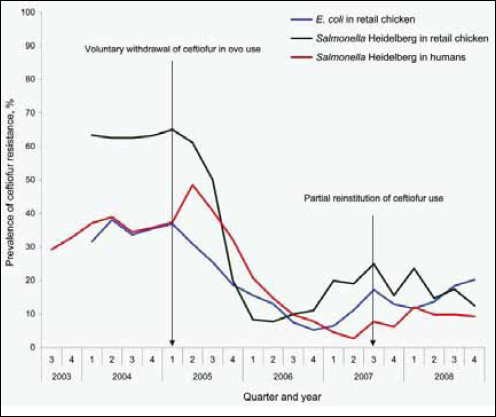
Improving the general hygiene in all stages of production and thereby reducing the microbial load on food products will also reduce the antimicrobial resistance load. However, a number of additional options aimed directly at reducing antimicrobial resistance are available for authorities and other stakeholders.
Setting thresholds for the acceptable level of pathogenic bacteria in foodstuffs is a well-established risk management praxis. Thresholds exist for a wide range of bacterial species or subtypes in foodstuffs (e.g., Listeria monocytogenes and E. coli O157:H7). Thus, establishing thresholds for bacteria resistant to certain antimicrobials is a valid, however rarely used, option to be considered. In Denmark, a specific control programme aimed at S. Typhimurium DT104 was implemented in 1998 (Wegener, 2006). As a part of this programme a zero tolerance for DT104 in food was established. The programme has led to a reduction of DT104 in domestically produced and imported foodstuffs.
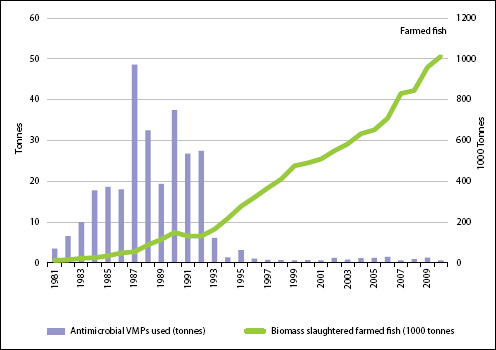
SOURCE: NORM/NORM-VET (2010).
While several food safety standards for traded food products exist, there seems to be a bigger problem in relation to the trade of live animals. Requirements in relation to epizootic diseases do exist, but none seem to be in place in relation to zoonotic bacteria or in particular antimicrobial resistance. Thus, today breeding animals with resistant Salmonella and other pathogens can be traded freely between countries, constituting an efficient route of global dissemination of resistant bacteria.
Conclusions
Integrated surveillance systems are essential to monitor the emergence and spread of antimicrobial resistance along the food production chain. Such systems require
• systematic sampling, harmonized laboratory methods, and good data management;
• detailed denominator data about the origin of the samples;
• subtyping of bacterial isolates, and molecular characterization of resistance genes;
• detailed antimicrobial usage data; and,
• flawless collaboration and coordination, including sharing and comparing data.
Based on existing surveillance systems it is fair to conclude the following:
• There is a close relationship between the patterns of antimicrobial usage and the observed patterns of antimicrobial resistance in food animals; however, other factors such as co-selection and clonal spread also play a part.
• There is a close relationship between levels and patterns of antimicrobial resistance in the food supply and antimicrobial resistance in human food-borne infections, bearing in mind that some food is imported and other foods are consumed while travelling abroad, and that all sources need to be accounted for.
There is a great need to reduce the overall use of antimicrobials in agri- and aquaculture worldwide, and the experiences from different countries suggest that major reductions can be achieved without significant negative effects on animal health or productivity, and for the long-term benefit of public, environmental, and animal health.
A number of effective upstream interventions to reduce resistance have been documented, including banning nontherapeutic uses in food animals, enforcing prescription-only policies, removing financial incentives for prescribing therapeutic drugs, restricting the use of drugs considered critically important for human health, monitoring usage at the farm level and providing advice to high-end users, and establishing thresholds for resistant pathogens in food.
Reducing antimicrobial usage requires collaboration between experts, regulatory authorities, and producers, and integrated monitoring of the effects of interventions is essential. This may be facilitated by establishing a coordinating body, for example, an antibiotic council, including all relevant stakeholders.
References
Aarestrup, F. M., H. C. Wegener, and P. Collignon. 2008. Resistance in bacteria of the food chain: Epidemiology and control strategies. Expert Review of Anti-Infective Therapy 6:733-750.
Aarestrup, F. M., V. F. Jensen, H. D. Emborg, E. Jacobsen, and H. C. Wegener. 2010. Changes in the use of antimicrobials and the effects on productivity of swine farms in Denmark. American Journal of Veterinary Research 71:726-733.
Bager, F., F. M. Aarestrup, N. E. Jensen, M. Madsen, A. Meyling, and H. C. Wegener. 1999. Design of a system for monitoring antimicrobial resistance in pathogenic, zoonotic and indicator bacteria from food animals. Acta Veterinaria Scandinavica Supplement 92:77-86.
CAC (Codex Alimentarius Commission). 2005. Code of practice to minimize and contain antimicrobial resistance. CRC/RCP 61-2005. Codex Alimentarius Commission.
DANMAP (Danish Integrated Antimicrobial Resistance Monitoring and Research Programme). 2006. Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, foods and humans in Denmark, Danish Integrated Antimicrobial Resistance Monitoring and Research Programme. Technical University of Denmark. http://www.danmap.org.
Dutil, L., R. Irwin, R. Finley, L. K. Ng, B. Avery, P. Boerlin, A. M. Bourgault, L. Cole, D. Daignault, A. Desruisseau, W. Demczuk, L. Hoang, G. B. Horsman, J. Ismail, F. Jamieson, A. Maki, A. Pacagnella, and D. R. Pillai. 2010. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerging Infectious Diseases 16:48-54.
DVFA (Danish Veterinary and Food Administration). 2012. Danish Veterinary and Food Administration. The Yellow Card Initiative. http://www.foedevarestyrelsen.dk/english/Animal/AnimalHealth/Pages/The-Yellow-Card-Initiative-on-Antibiotics.aspx (accessed April 2012).
EFSA (European Food Safety Authority). 2008. European Food Safety Authority—Working Group on Developing Harmonised Schemes for Monitoring Antimicrobial Resistance in Zoonotic Agents. Harmonised monitoring of antimicrobial resistance in Salmonella and Campylobacter isolates from food animals in the European Union. Clinical Microbiology and Infection 14(6):522-533.
EFSA. 2009. Statement of EFSA on the consolidated presentation of opinions on the use of antibiotic resistance genes as marker genes in genetically modified plants. EFSA Journal 1108:1-8.
FAO (Food and Agriculture Organization), OIE (World Animal Health Organization), and WHO (World Health Organization). 2003. Joint FAO/OIE/WHO Expert Workshop on Non-Human Antimicrobial Usage and Antimicrobial Resistance: Scientific assessment, Geneva, December 1-5, 2003.
———. 2004. Second Joint FAO/OIE/WHO Workshop on Non-human Antimicrobial Usage and Antimicrobial Resistance: Management options. Oslo, March 15-18, 2004. Geneva: WHO.
FDA (Food and Drug Administration). 2003. Guidance 152. Guidance for industry. Evaluating the safety of antimicrobial new animal drugs with regard to their microbial effects on bacteria of human health concern. Docket No. 98D-1146. Bethesda, MD: FDA.
———. 2008. New animal drugs; cephalosporin drugs; extralabel animal drug use; order of prohibition. Docket No. FDA-2008-N-0326. 21 CFR Part 530. Bethesda, MD: FDA.
Grave, K., and H. C. Wegener. 2006. Comment on: Veterinarians’ profit on drug dispensing. Preventive Veterinary Medicine 77:306-308.
Grave, K., J. Torren-Edo, and D. Mackay. 2010. Comparison of the sales of veterinary antibacterial agents between 10 European countries. Journal of Antimicrobial Chemotherapy 65:2037-2040.
Kennedy, K. J., J. L. Roberts, and P. J. Collignon. 2008. Escherichia coli bacteraemia in Canberra: Incidence and clinical features. Medical Journal of Australia 188:209-213.
Markestad, A., and K. Grave. 1997. Reduction of antibacterial drug use in Norwegian fish farming due to vaccination. Developments in Biological Standardization 90:365-369.
Pedersen, K. B., F. M. Aarestrup, N. E. Jensen, F. Bager, L. B. Jensen, S. E. Jorsal, T. K. Nielsen, H. C. Hansen, A. Meyling, and H. C. Wegener. 1999. The need for a veterinary antibiotic policy. The Veterinary Record 145:50-53.
Stege, H., F. Bager, E. Jacobsen, and A. Thougaard. 2003. VETSTAT—the Danish system for surveillance of the veterinary use of drugs for production animals. Preventive Veterinary Medicine 57:105-115.
Swann, M. M., and Joint Committee on the Use of Antibiotics in Animal Husbandry and Veterinary Medicine. 1969. Report of the Joint Committee on the Use of Antibiotics in Animal Husbandry and Veterinary Medicine. London: Her Majesty’s Stationery Office.
Wegener, H. C. 2006. Risk management for the limitation of antibiotic resistance—experience of Denmark. Journal of Medical Microbiology 296(Suppl. 41):11-13.
Wheeler, E., P. Y. Hong, L. C. Bedon, and R. I. Mackie. 2012. Carriage of antibiotic-resistant enteric bacteria varies among sites in Galapagos reptiles. Journal of Wildlife Diseases 48:56-67.
WHO. 1997. The medical impact of the use of antimicrobials in food animals: Report of a WHO meeting. Berlin, October 13-17, 1997. Geneva: WHO.
———. 2005. Critically important antibacterial agents for human medicine for risk management strategies of non-human use: Report of a WHO working group consultation. Canberra, Australia, February 15-18, 2005. Geneva: WHO.
———. 2011. Tackling antibiotic resistance from a food safety perspective in Europe. Copenhagen: WHO Regional Office for Europe.
Xia, L. N., L. Li, C. M. Wu, Y. Q. Liu, X. Q. Tao, L. Dai, Y. H. Qi, L. M. Lu, and J. Z. Shen. 2010. A survey of plasmid-mediated fluoroquinolone resistance genes from Escherichia coli isolates and their dissemination in Shandong, China. Foodborne Pathogens and Disease 7:207-215.
ORIGINS OF MAJOR HUMAN INFECTIOUS DISEASES33
Nathan D. Wolfe,34Claire Panosian Dunavan,35and Jared Diamond36
Many of the major human infectious diseases, including some now confined to humans and absent from animals, are ‘new’ ones that arose only after the origins of agriculture. Where did they come from? Why are they overwhelmingly of Old World origins? Here we show that answers to these questions are different for tropical and temperate diseases; for instance, in the relative importance of domestic animals and wild primates as sources. We identify five intermediate stages through which a pathogen exclusively infecting animals may become transformed into a pathogen exclusively infecting humans. We propose an initiative to resolve disputed origins of major diseases, and a global early warning system to monitor pathogens infecting individuals exposed to wild animals.
Human hunter/gatherer populations currently suffer, and presumably have suffered for millions of years, from infectious diseases similar or identical to diseases of other wild primate populations. However, the most important infectious diseases of modern food-producing human populations also include diseases that could have emerged only within the past 11,000 years, following the rise of agriculture (Diamond, 1997; Dobson and Carper, 1996). We infer this because, as discussed below, these diseases can only be sustained in large dense human populations that did not exist anywhere in the world before agriculture. What were the sources of our major infectious diseases, including these ‘new’ ones?
______________
33 Provided courtesy of Nature Publishing Group.
34 Department of Epidemiology, School of Public Health, University of California, Los Angeles 90095-1772, USA.
35 Division of Infectious Diseases, David Geffen School of Medicine, University of California, Los Angeles 90095-1688, USA.
36 Departments of Geography and of Environmental Health Sciences, University of California, Los Angeles 90095-1524, USA.
Why do so many animal pathogens, including virulent viruses like Ebola and Marburg, periodically infect human hosts but then fail to establish themselves in human populations?
A tentative earlier formulation noted that major infectious diseases of temperate zones seem to have arisen overwhelmingly in the Old World (Africa, Asia and Europe), often from diseases of Old World domestic animals. Hence one goal of this article is to reappraise that conclusion in the light of studies of the past decade. Another goal is to extend the analysis to origins of tropical diseases ( Diamond and Panosian, 2006). We shall show that they also arose mainly in the Old World, but for different reasons, and mostly not from diseases of domestic animals. These results provide a framework for addressing unanswered questions about the evolution of human infectious diseases—questions not only of practical importance to physicians, and to all the rest of us as potential victims, but also of intellectual interest to historians and evolutionary biologists. Historians increasingly recognize that infectious diseases have had major effects on the course of history; for example, on the European conquest of Native Americans and Pacific Islanders, the inability of Europeans to conquer the Old World tropics for many centuries, the failure of Napoleon’s invasion of Russia, and the failure of the French attempt to complete construction of a Panama Canal (Crosby, 1986; McNeill, 1976; Ramenofsky, 1987). Evolutionary biologists realize that infectious diseases, as a leading cause of human morbidity and mortality, have exerted important selective forces on our genomes (Anderson and May, 1991; Dobson and Carper, 1996).
We begin by defining five stages in the evolutionary transformation of an animal pathogen into a specialized pathogen of humans, and by considering why so many pathogens fail to make the transition from one stage to the next. We then assemble a database of 15 temperate and 10 tropical diseases of high evolutionary and/or historical impact, and we compare their characteristics and origins. Our concluding section lays out some unresolved questions and suggests two expanded research priorities. We restrict our discussion to unicellular microbial pathogens. We exclude macroparasites (in the sense of Anderson and May, 1991), as well as normally benign commensals that cause serious illness only in weakened hosts. The extensive Supplementary Information provides details and references on our 25 diseases, robustness tests of our conclusions, factors affecting transitions between disease stages, and modern practices altering the risk of emergence of new diseases.
Evolutionary Stages
Box A16-1 delineates five intergrading stages (Figure A16-1) through which a pathogen exclusively infecting animals (Stage 1) may become transformed into a pathogen exclusively infecting humans (Stage 5). Supplementary Table S1 assigns each of the 25 major diseases discussed (Supplementary Note S1) to one of these five stages.
BOX A16-1
Five Stages Leading to Endemic Human Diseases
We delineate five stages in the transformation of an animal pathogen into a specialized pathogen of humans (Figure A16-1). There is no inevitable progression of microbes from Stage 1 to Stage 5: at each stage many microbes remain stuck, and the agents of nearly half of the 25 important diseases we selected for analysis (Supplementary Table S1) have not reached Stage 5.
• Stage 1. A microbe that is present in animals but that has not been detected in humans under natural conditions (that is, excluding modern technologies that can inadvertently transfer microbes, such as blood transfusion, organ transplants, or hypodermic needles). Examples: most malarial plasmodia, which tend to be specific to one host species or to a closely related group of host species.
• Stage 2. A pathogen of animals that, under natural conditions, has been transmitted from animals to humans (‘primary infection’) but has not been transmitted between humans (‘secondary infection’). Examples: anthrax and tularemia bacilli, and Nipah, rabies and West Nile viruses.
• Stage 3. Animal pathogens that can undergo only a few cycles of secondary transmission between humans, so that occasional human outbreaks triggered by a primary infection soon die out. Examples: Ebola, Marburg and monkeypox viruses.
• Stage 4. A disease that exists in animals, and that has a natural (sylvatic) cycle of infecting humans by primary transmission from the animal host, but that also undergoes long sequences of secondary transmission between humans without the involvement of animal hosts. We arbitrarily divide Stage 4 into three substages distinguished by the relative importance of primary and secondary transmission:Stage 4a. Sylvatic cycle much more important than direct human-to-human spread. Examples: Chagas’ disease and (more frequent secondary transmission approaching Stage 4b) yellow fever. Stage 4b. Both sylvatic and direct transmission are important. Example: dengue fever in forested areas of West Africa and Southeast Asia. Stage 4c. The greatest spread is between humans. Examples: influenza A, cholera, typhus and West African sleeping sickness.
• Stage 5. A pathogen exclusive to humans. Examples: the agents causing falciparum malaria, measles, mumps, rubella, smallpox and syphilis. In principle, these pathogens could have become confined to humans in either of two ways: an ancestral pathogen already present in the common ancestor of chimpanzees and humans could have co-speciated long ago, when the chimpanzee and human lineages diverged around five million years ago; or else an animal pathogen could have colonized humans more recently and evolved into a specialized human pathogen. Co-speciation accounts well for the distribution of simian foamy viruses of non-human primates, which are lacking and presumably lost in humans: each virus is restricted to one primate species, but related viruses occur in related primate species ( Switzer et al., 2005). While both interpretations are still debated for falciparum malaria, the latter interpretation of recent origins is widely preferred for most other human Stage 5 diseases of Supplementary Table S1.
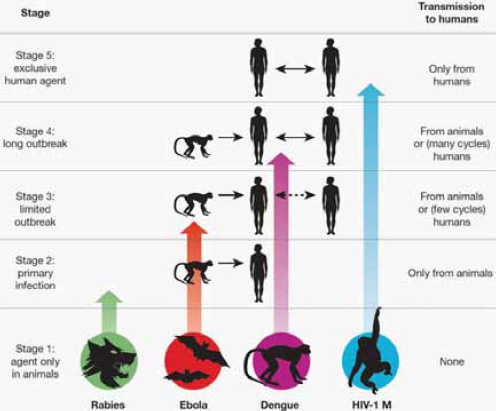
FIGURE A16-1 Illustration of the five stages through which pathogens of animals evolve to cause diseases confined to humans. (See Box A16-1 for details.) The four agents depicted have reached different stages in the process, ranging from rabies (still acquired only from animals) to HIV-1 (now acquired only from humans).
A large literature discusses the conditions required for a Stage 5 epidemic to persist (Anderson and May, 1991; Dobson and Carper, 1996). Briefly, if the disease infects only humans and lacks an animal or environmental reservoir, each infected human introduced into a large population of susceptible individuals must on average give rise during his/her contagious lifespan to an infection in at least one other individual. Persistence depends on factors such as the duration of a host’s infectivity; the rate of infection of new hosts; rate of development of host protective immunity; and host population density, size and structure permitting the pathogen’s regional persistence despite temporary local extinctions.
Less well understood are two of the critical transitions between stages, discussed in Box A16-2. One is the transition from Stage 1 to Stage 2, when a pathogen initially confined to animals first infects humans. The other is the tran-
sition from Stage 2 to Stages 3 and 4 (see also Supplementary Note S2), when a pathogen of animal origin that is nevertheless transmissible to humans evolves the ability to sustain many cycles of human-to-human transmission, rather than just a few cycles before the outbreak dies out (as seen in modern Ebola outbreaks).
BOX A16-2
Transitions Between Stages
Transition from Stage 1 to Stage 2. Most animal pathogens are not transmitted to humans, that is, they do not even pass from Stage 1 to Stage 2. This problem of cross-species infection has been discussed previously (Antia et al., 2003; May etal., 2001; Moyaetal., 2004; Taylor et al., 2001). Briefly, the probability-per-unit-time (p) of infection of an individual of a new (that is, new recipient) host species increases with the abundance of the existing (that is, existing donor) host, with the fraction of the existing host population infected, with the frequency of ‘encounters’(opportunities for transmission, including indirect ‘encounters’ via vectors) between an individual of the existing host and of the new host, and with the probability of transmission per encounter. p decreases with increasing phylogenetic distance between the existing host and new host. p also varies among microbes (for example, trypanosomes and flaviviruses infect a wide taxonomic range of hosts, while plasmodia and simian foamy viruses infect only a narrow range), and this variation is related to a microbe’s characteristics, such as its ability to generate genetic variability, or its ability to overcome host molecular barriers of potential new hosts (such as humoral and cellular defenses or lack of cell membrane receptors essential for microbe entry into host cells).
These considerations illuminate different reasons why a given animal host species may or may not become a source of many infections in humans. For instance, despite chimpanzees’ very low abundance and infrequent encounters with humans, they have donated to us numerous zoonoses (diseases that still mainly afflict animals) and one or two established human diseases (AIDS and possibly hepatitis B) because of their close phylogenetic relationship to humans. Despite their large phylogenetic distance from humans, many of our zoonoses and probably two of our established diseases (plague and typhus) have been acquired from rodents, because of their high abundance and frequent encounters with humans in dwellings. Similarly, about half of our established temperate diseases have been acquired from domestic livestock, because of high local abundance and very frequent contact. Conversely, elephants and bats are not known to have donated directly to us any established diseases and rarely donate zoonoses, because they are heavily penalized on two or three counts: large phylogenetic distance, infrequent encounters with humans, and (in the case of elephants) low abundance. One might object that Nipah, severe acute respiratory syndrome (SARS) and rabies viruses do infect humans from bats, but these apparent exceptions actually support our conclusion. While bats may indeed be the primary reservoir for Nipah and SARS, human infections by these viruses are acquired mainly from intermediate animal hosts that frequently encounter humans (respectively, domestic pigs,
and wild animals sold for food). The rare cases of rabies transmission directly to humans from bats arise because rabies changes a bat’s behaviour so that it does encounter and bite humans, which a healthy bat (other than a vampire bat) would never do.
Transition from Stage 2 to Stage 3 or 4. Although some Stage 2 and 3 pathogens, such as the anthrax and Marburg agents, are virulent and feared, they claim few victims at present. Yet if they made the transition to Stage 4 or 5, their global impact would be devastating. Why do animal pathogens that have survived the initial jump across species lines into a human host (Stages 1 to 2) usually reach a dead end there, and not evolve past Stages 3 and 4 into major diseases confined to humans (Stage 5)? Barriers between Stages 2 and 3 (consider the rabies virus) include differences between human and animal behaviour affecting transmission (for example, animals often bite humans but humans rarely bite other humans); a pathogen’s need to evolve adaptations to the new human host and possibly also to a new vector; and obstacles to a pathogen’s spread between human tissues (for example, BSE is restricted to the central nervous system and lymphoid tissue). Barriers between Stages 3 and 4 (consider Ebola virus) include those related to human population size and to transmission efficiency between humans. The emergence of novel pathogens is now being facilitated by modern developments exposing more potential human victims and/or making transmission between humans more efficient than before (Morens et al., 2004; Morse, 1995; Weiss and Michael, 2004; Wilson, 1995). These developments include blood transfusion (hepatitis C), the commercial bushmeat trade (retroviruses), industrial food production (bovine spongiform encephalitis, BSE), international travel (cholera), intravenous drug use (HIV), vaccine production (simian virus 40, SV40), and susceptible pools of elderly, antibiotic-treated, immunosuppressed patients (see Supplementary Note S2 for details).
Database and Conclusions
Database. Supplementary Table S1 lists 10 characteristics for each of 25 important ‘temperate’ (15) and ‘tropical’ (10) diseases (see Supplementary Note S3 for details of this distinction). Our aim was to select well-defined diseases causing the highest mortality and/or morbidity and hence of the highest historical and evolutionary significance (see Supplementary Note S1 for details of our selection criteria). Of the 25 diseases, we selected 17 because they are the ones assessed by Lopez et al. (2005) as imposing the heaviest world burdens today (they have the highest disability-adjusted life years (DALY) scores). Of the 17 diseases, 8 are temperate (hepatitis B, influenza A, measles, pertussis, rotavirus A, syphilis, tetanus and tuberculosis), and 9 are tropical (acquired immune deficiency syn-
drome (AIDS), Chagas’ disease, cholera, dengue haemorrhagic fever, East and West African sleeping sicknesses, falciparum and vivax malarias, and visceral leishmaniasis).
We selected eight others (temperate diphtheria, mumps, plague, rubella, smallpox, typhoid and typhus, plus tropical yellow fever) because they imposed heavy burdens in the past, although modern medicine and public health have either eradicated them (smallpox) or reduced their burden. Except for AIDS, dengue fever, and cholera, which have spread and attained global impact in modern times, most of these 25 diseases have been important for more than two centuries.
Are our conclusions robust to variations in these selection criteria? For about a dozen diseases with the highest modern or historical burdens (for example, AIDS, malaria, plague, smallpox), there can be little doubt that they must be included, but one could debate some of the next choices. Hence we drew up three alternative sets of diseases sharing a first list of 16 indisputable major diseases but differing in the next choices, and we performed all 10 analyses described below on all three sets. It turned out that, with one minor exception, the three sets yielded qualitatively the same conclusions for all 10 analyses, although differing in their levels of statistical significance (see Supplementary Note S4). Thus, our conclusions do seem to be robust.
Temperate/tropical differences. Comparisons of these temperate and tropical diseases yield the following conclusions:
• A higher proportion of the diseases is transmitted by insect vectors in the tropics (8/10) than in the temperate zones (2/15) (P < 0.005, X2-test, degrees of freedom, d.f. = 1). This difference may be partly related to the seasonal cessations or declines of temperate insect activity.
• A higher proportion (P = 0.009) of the diseases conveys long-lasting immunity (11/15) in the temperate zones than in the tropics (2/10).
• Animal reservoirs are more frequent (P < 0.005) in the tropics (8/10) than in the temperate zones (3/15). The difference is in the reverse direction (P = 0.1, NS, not significant) for environmental reservoirs (1/10 versus 6/15), but those environmental reservoirs that do exist are generally not of major significance except for soil bearing tetanus spores.
• Most of the temperate diseases (12/15) are acute rather than slow, chronic, or latent: the patient either dies or recovers within one to several weeks. Fewer (P = 0.01) of the tropical diseases are acute: 3/10 last for one or two weeks, 3/10 last for weeks to months or years, and 4/10 last for many months to decades.
• A somewhat higher proportion of the diseases (P = 0.08, NS) belongs to Stage 5 (strictly confined to humans) in the temperate zones (10/15 or 11/15) than in the tropics (3/10). The paucity of Stage 2 and Stage 3
diseases (a total of only 5 such diseases) on our list of 25 major human diseases is noteworthy, because some Stage 2 and Stage 3 pathogens (such as anthrax and Ebola) are notoriously virulent, and because theoretical reasons are often advanced (but also denied) as to why Stage 5 microbes with long histories of adaptation to humans should tend to evolve low morbidity and mortality and not cause major diseases. We discuss explanations for this outcome in Supplementary Note S5.
Most (10/15) of the temperate diseases, but none of the tropical diseases (P < 0.005), are so-called ‘crowd epidemic diseases’ (asterisked in Supplementary Table S1), defined as ones occurring locally as a brief epidemic and capable of persisting regionally only in large human populations. This difference is an immediate consequence of the differences enumerated in the preceding five paragraphs. If a disease is acute, efficiently transmitted, and quickly leaves its victim either dead or else recovering and immune to re-infection, the epidemic soon exhausts the local pool of susceptible potential victims. If in addition the disease is confined to humans and lacks significant animal and environmental reservoirs, depletion of the local pool of potential victims in a small, sparse human population results in local termination of the epidemic. If, however, the human population is large and dense, the disease can persist by spreading to infect people in adjacent areas, and then returning to the original area in a later year, when births and growth have regenerated a new crop of previously unexposed non-immune potential victims. Empirical epidemiological studies of disease persistence or disappearance in isolated human populations of various sizes have yielded estimates of the population required to sustain a crowd disease: at least several hundred thousand people in the cases of measles, rubella and pertussis (Anderson and May, 1991; Dobson and Carper, 1996). But human populations of that size did not exist anywhere in the world until the steep rise in human numbers that began around 11,000 years ago with the development of agriculture (Bellwood, 2005; Diamond, 1997). Hence the crowd epidemic diseases of the temperate zones must have evolved since then.
Of course, this does not mean that human hunter/gatherer communities lacked infectious diseases. Instead, like the sparse populations of our primate relatives, they suffered from infectious diseases with characteristics permitting them to persist in small populations, unlike crowd epidemic diseases. Those characteristics include: occurrence in animal reservoirs as well as in humans (such as yellow fever); incomplete and/or non-lasting immunity, enabling recovered patients to remain in the pool of potential victims (such as malaria); and a slow or chronic course, enabling individual patients to continue to infect new victims over years, rather than for just a week or two (such as Chagas’ disease).
Pathogen origins. (See details for each disease in Supplementary Note S10). Current information suggests that 8 of the 15 temperate diseases probably or possibly reached humans from domestic animals (diphtheria, influenza A, measles,
mumps, pertussis, rotavirus, smallpox, tuberculosis); three more probably reached us from apes (hepatitis B) or rodents (plague, typhus); and the other four (rubella, syphilis, tetanus, typhoid) came from still-unknown sources (see Supplementary Note S6). Thus, the rise of agriculture starting 11,000 years ago played multiple roles in the evolution of animal pathogens into human pathogens (Diamond, 1997; Diamond, 2002; McNeill, 1976). Those roles included both generation of the large human populations necessary for the evolution and persistence of human crowd diseases, and generation of large populations of domestic animals, with which farmers came into much closer and more frequent contact than hunter/gatherers had with wild animals. Moreover, as illustrated by influenza A, these domestic animal herds served as efficient conduits for pathogen transfers from wild animals to humans, and in the process may have evolved specialized crowd diseases of their own.
It is interesting that fewer tropical than temperate pathogens originated from domestic animals: not more than three of the ten tropical diseases of Supplementary Table S1, and possibly none (see Supplementary Note S7). Why do temperate and tropical human diseases differ so markedly in their animal origins? Many (4/10) tropical diseases (AIDS, dengue fever, vivax malaria, yellow fever) but only 1/15 temperate diseases (hepatitis B) have wild non-human primate origins (P < 0.04). This is because although non-human primates are the animals most closely related to humans and hence pose the weakest species barriers to pathogen transfer, the vast majority of primate species is tropical rather than temperate. Conversely, few tropical but many temperate diseases arose from domestic animals, and this is because domestic animals live mainly in the temperate zones, and their concentration there was formerly even more lop-sided (see Supplementary Note S8).
A final noteworthy point about animal-derived human pathogens is that virtually all arose from pathogens of other warm-blooded vertebrates, primarily mammals plus in two cases (influenza A and ultimately falciparum malaria) birds. This comes as no surprise, considering the species barrier to pathogen transfer posed by phylogenetic distance (Box A16-2). An expression of this barrier is that primates constitute only 0.5% of all vertebrate species but have contributed about 20% of our major human diseases. Expressed in another way, the number of major human diseases contributed, divided by the number of animal species in the taxonomic group contributing those diseases, is approximately 0.2 for apes, 0.017 for non-human primates other than apes, 0.003 for mammals other than primates, 0.00006 for vertebrates other than mammals, and either 0 or else 0.000003 (if cholera really came from aquatic invertebrates) for animals other than vertebrates (see Supplementary Note S9).
Geographic origins. To an overwhelming degree, the 25 major human pathogens analysed here originated in the Old World. That proved to be of great historical importance, because it facilitated the European conquest of the New World
(the Americas). Far more Native Americans resisting European colonists died of newly introduced Old World diseases than of sword and bullet wounds. Those invisible agents of New World conquest were Old World microbes to which Europeans had both some acquired immunity based on individual exposure and some genetic resistance based on population exposure over time, but to which previously unexposed Native American populations had no immunity or resistance (Crosby, 1986; Diamond, 1997; McNeill, 1976; Ramenofsky, 1987). In contrast, no comparably devastating diseases awaited Europeans in the New World, which proved to be a relatively healthy environment for Europeans until yellow fever and malaria of Old World origins arrived (McNeill, 2006).
Why was pathogen exchange between Old and New Worlds so unequal? Of the 25 major human diseases analysed, Chagas’ disease is the only one that clearly originated in the New World. For two others, syphilis and tuberculosis, the debate is unresolved: it remains uncertain in which hemisphere syphilis originated, and whether tuberculosis originated independently in both hemispheres or was brought to the Americas by Europeans. Nothing is known about the geographic origins of rotavirus, rubella, tetanus and typhus. For all of the other 18 major pathogens, Old World origins are certain or probable.
Our preceding discussion of the animal origins of human pathogens may help explain this asymmetry. More temperate diseases arose in the Old World than New World because far more animals that could furnish ancestral pathogens were domesticated in the Old World. Of the world’s 14 major species of domestic mammalian livestock, 13, including the five most abundant species with which we come into closest contact (cow, sheep, goat, pig and horse), originated in the Old World (Diamond, 1997). The sole livestock species domesticated in the New World was the llama, but it is not known to have infected us with any pathogens (Diamond, 1997; Dobson, 1996)—perhaps because its traditional geographic range was confined to the Andes, it was not milked or ridden or hitched to ploughs, and it was not cuddled or kept indoors (as are some calves, lambs and piglets). Among the reasons why far more tropical diseases (nine versus one) arose in the Old World than the New World are that the genetic distance between humans and New World monkeys is almost double that between humans and Old World monkeys, and is many times that between humans and Old World apes; and that much more evolutionary time was available for transfers from animals to humans in the Old World (about 5 million years) than in the New World (about 14,000 years).
Outlook and Future Research Directions
Many research directions on infectious disease origins merit more effort. We conclude by calling attention to two such directions: clarifying the origins of existing major diseases, and surveillance for early detection of new potentially major diseases.
Origins of established diseases. This review illustrates big gaps in our understanding of the origins of even the established major infectious diseases. Almost all the studies that we have reviewed were based on specimens collected opportunistically from domestic animals and a few easily sampled wild animal species, rather than on systematic surveys for particular classes of agents over the spectrum of domestic and wild animals. A case in point is our ignorance even about smallpox virus, the virus that has had perhaps the greatest impact on human history in the past 4,000 years. Despite some knowledge of poxviruses infecting our domestic mammals, we know little about poxvirus diversity among African rodents, from which those poxviruses of domestic mammals are thought to have evolved. We do not even know whether ‘camelpox’, the closest known relative of smallpox virus, is truly confined to camels as its name implies or is instead a rodent virus with a broad host range. There could be still-unknown poxviruses more similar to smallpox virus in yet unstudied animal reservoirs, and those unknown poxviruses could be important not only as disease threats but also as reagents for drug and vaccine development.
Equally basic questions arise for other major pathogens. While falciparum malaria, an infection imposing one of the heaviest global burdens today, seems to have originated from a bird parasite whose descendants include both the Plasmodium falciparum infecting humans and the P. reichenowii infecting chimpanzees, malaria researchers still debate whether the bird parasite was introduced to both humans and chimpanzees (Waters et al., 1991) a few thousand years ago in association with human agriculture, or instead more than five million years ago before the split of humans and chimpanzees from each other (Ayala et al., 1999). Although resolving this debate will not help us eradicate malaria, it is fascinating in its own right and could contribute to our broader understanding of disease emergence. In the case of rubella, a human crowd disease that must have emerged only in the past 11,000 years and for which some close relative may thus still exist among animals, no even remotely related virus is known; one or more may be lurking undiscovered somewhere. Does the recent identification of porcine rubulavirus and the Mapuera virus in bats as the closest known relatives of mumps virus mean that pigs infected humans, or that human mumps infected pigs, or that bats independently infected both humans and pigs? Is human tuberculosis descended from a ruminant mycobacterium that recently infected humans from domestic animals (a formerly prevalent view), or from an ancient human mycobacterium that has come to infect domestic and wild ruminants (a currently popular view)?
To fill these and other yawning gaps in our understanding of disease origins, we propose an ‘origins initiative’ aimed at identifying the origins of a dozen of the most important human infectious diseases: for example, AIDS, cholera, dengue fever, falciparum malaria, hepatitis B, influenza A, measles, plague, rotavirus, smallpox, tuberculosis and typhoid. Although more is already known about the origins of some of these agents (AIDS, influenza A and measles) than about
others (rotavirus, smallpox and tuberculosis), more comprehensive screening is still likely to yield significant new information about even the most studied agents, as illustrated by the recent demonstration that gorillas rather than chimpanzees were probably the donor species for the O-group of human immunodeficiency virus (HIV)-1 (Van Heuverswyn et al., 2006). The proposed effort would involve systematic sampling and phylogeographic analysis of related pathogens in diverse animal species: not just pigs and other species chosen for their ready availability, but a wider range of wild and domestic species whose direct contact (for example, as bushmeat) or indirect contact (for example, vector-mediated) with humans could plausibly have led to human infections. In addition to the historical and evolutionary significance of knowledge gained through such an origins initiative, it could yield other benefits such as: identifying the closest relatives of human pathogens; a better understanding of how diseases have emerged; new laboratory models for studying public health threats; and perhaps clues that could aid in predictions of future disease threats.
A global early warning system. Most major human infectious diseases have animal origins, and we continue to be bombarded by novel animal pathogens. Yet there is no ongoing systematic global effort to monitor for pathogens emerging from animals to humans. Such an effort could help us to describe the diversity of microbial agents to which our species is exposed; to characterize animal pathogens that might threaten us in the future; and perhaps to detect and control a local human emergence before it has a chance to spread globally.
In our view, monitoring should focus on people with high levels of exposure to wild animals, such as hunters, butchers of wild game, wildlife veterinarians, workers in the wildlife trade, and zoo workers. Such people regularly become infected with animal viruses, and their infections can be monitored over time and traced to other people in contact with them. One of us (N.D.W.) has been working in Cameroon to monitor microbes in people who hunt wild game, in other people in their community, and in their animal prey (Wolfe et al., 2004). The study is now expanding to other continents and to monitor domestic animals (such as dogs) that live in close proximity to humans but are exposed to wild animals through hunting and scavenging. Monitoring of people, animals, and animal die-offs (Kuiken et al., 2003) will serve as an early warning system for disease emergence, while also providing a unique archive of pathogens infecting humans and the animals to which we are exposed. Specimens from such highly exposed human populations could be screened specifically for agents known to be present in the animals they hunt (for example, retroviruses among hunters of non-human primates), as well as generically using broad screening tools such as viral microarrays (Wang et al., 2003) and random amplification polymerase chain reaction (PCR) (Jones et al., 2005). Such monitoring efforts also provide potentially invaluable repositories, which would be available for study after future outbreaks in order to reconstruct an outbreak’s origin, and as a source of relevant reagents.
References
Anderson, R. M. & May, R. M. Infectious Diseases of Humans: Dynamics and Control (Oxford Univ. Press, Oxford, UK, 1991).
Antia, R., Regoes, R. H., Koella, J. C. & Bergstrom, C. T. The role of evolution in the emergence of infectious diseases. Nature 426, 658–661 (2003).
Ayala, F. J., Escalante, A. A. & Rich, S. M. Evolution of Plasmodium and the recent origin of the world populations of Plasmodium falciparum. Parassitologia 41, 55–68 (1999).
Bellwood, P. First Farmers: the Origins of Agriculture Societies (Blackwell, Oxford, 2005).
Crosby, A. W. Ecological Imperialism: the Biological Expansion of Europe 900–1900 (Cambridge Univ. Press, Cambridge, UK, 1986).
Diamond, J. & Panosian, C. in When Disease Makes History: Epidemics and Great Historical Turning Points (ed. Hämäläinen, P.) 17–44 (Helsinki Univ. Press, 2006).
Diamond, J. Evolution, consequences, and future of plant and animal domestication. Nature 418, 34–41 (2002).
Diamond, J. Guns, Germs, and Steel: the Fates of Human Societies (Norton, New York, 1997).
Dobson, A. P. & Carper, E. R. Infectious diseases and human population history. Bioscience 46, 115–126 (1996).
Jones, M. S. et al. New DNA viruses identified in patients with acute viral infection syndrome. J. Virol. 79, 8230–8236 (2005).
Kuiken, T. et al. Pathogen surveillance in animals. Science 309, 1680–1681 (2005).
Lopez, A. D., Mathers, C. D., Ezzati, N., Jamison, D. T. & Murray, C. J. L. (eds) Global Burden of Disease and Risk Factors (Oxford Univ. Press, New York, 2006).
May, R. M., Gupta, S. & McLean, A. R. Infectious disease dynamics: what characterizes a successful invader? Phil. Trans. R. Soc. Lond. B 356, 901–910 (2001).
McNeill, J. R. in When Disease Makes History: Epidemics and Great Historical Turning Points (ed. Hämäläinen, P.) 81–111 (Helsinki Univ. Press, Helsinki, 2006).
McNeill, W. H. Plagues and Peoples (Anchor, Garden City, 1976).
Morens, D. M., Folkers, G. K. & Fauci, A. S. The challenge of emerging and re-emerging infectious diseases. Nature 430, 242–249 (2004).
Morse, S. S. Factors in the emergence of infectious diseases. Emerg. Infect. Dis. 1, 7–15 (1995).
Moya, A., Holmes, E. C. & Gonzalez-Candelas, F. The population genetics and evolutionary epidemiology of RNA viruses. Nature Rev. Microbiol. 2, 279–288 (2004).
Ramenofsky, A. Vectors of Death: the Archaeology of European Contact (New Mexico Press, Albuquerque, 1987).
Switzer, W. M. et al. Ancient co-speciation of simian foamy viruses and primates. Nature 434, 376–380 (2005).
Taylor, L. H., Latham, S. M. & Woolhouse, M. E. Risk factors for human disease emergence. Phil. Trans. R. Soc. Lond. B 356, 983–989 (2001).
Van Heuverswyn, F. et al. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature 444, 164 (2006).
Wang, D. et al. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 1, E2 (2003).
Waters, A. P., Higgins, D. G. & McCutchan, T. F. Plasmodium falciparum appears to have arisen as a result of lateral transfer between avian and human hosts. Proc. Natl Acad. Sci. USA 88, 3140–3144 (1991).
Weiss, R. A. & McMichael, A. J. Social and environmental risk factors in the emergence of infectious diseases. Nature Med. 10, S70—S76 (2004).
Wilson, M. E. Travel and the emergence of infectious diseases. Emerg. Infect. Dis. 1, 39–46 (1995).
Wolfe, N. D. et al. Naturally acquired simian retrovirus infections in central African hunters. Lancet 363, 932–937 (2004).
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgements We thank L. Krain for assistance with Supplementary Note S10; M. Antolin, D. Burke, L. Fleisher, E. Holmes, L. Real, A. Rimoin, R. Weiss and M. Woolhouse for comments; and many other colleagues for providing information. This work was supported by an NIH Director’s Pioneer Award and Fogarty International Center IRSDA Award (to N.D.W.), a W. W. Smith Foundation award (to N.D.W.), and National Geographic Society awards (to J.D. and N.D.W.).
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Correspondence should be addressed to N.W. (nwolfe@ucla.edu) or J.D. ( jdiamond@ geog.ucla.edu).
THE OUTLOOK FOR PUBLIC FOOD SAFETY
RESEARCH AND USDA SCIENCE
Thank you and good afternoon. I’d like to take this opportunity to speak with you today to talk about food safety research at the U.S. Department of Agriculture (USDA) and One Health.
As some of you might know, I served as the first Under Secretary for Food Safety at USDA from 1997 to 2001, where I oversaw U.S. government food safety policy development and USDA’s continuity of operations planning.
I’m now USDA’s Chief Scientist and Under Secretary for Research, Education and Economics, where I play a role in managing USDA’s food and agriculture research portfolio.
My own academic background is in human nutrition—so I’ve been lucky to have been able to focus on different aspects of our food and agricultural system.
But in reality, all three of these fields—food safety, nutrition, and production agriculture research—are intimately connected.
At USDA these three fields converge, and in order to ensure our food and feed systems are safe, health promoting, productive, and sustainable, we need to
______________
37 USDA Chief Scientist and Under Secretary, Research, Education, and Economics.
38 Remarks delivered at “Improving Food Safety Through One Health—Institute of Medicine/Forum on Microbial Threats,” December 14, 2011.
make sure that our research programs are planned with these goals in mind. As we used to say in Food Safety—no food is nutritious unless it is safe.
As you are well aware, 65 percent of the emerging infectious diseases of the past 60 years have come from pathogens that have jumped from animals to humans. Many of the pathogens that cause food-borne illness for thousands of Americans every year reside in animals without causing severe illness but can cause life-threatening illness in people.
Greater coordination between human and animal health professionals is becoming a prominent theme among many infectious disease professionals and is becoming the new norm for addressing emerging pathogens.
USDA is the premier organization with veterinary, food safety, nutrition, wildlife, plant, economics, and biotechnology expertise to meet the challenges of this growing coordination and communication among human, animal, and environmental infectious disease professionals.
Given this diversity of expertise, USDA identified a need for a comprehensive national strategy for One Health based upon pandemic planning for both highly pathogenic H5N1 avian influenza and 2009 pandemic H1N1 influenza viruses.
USDA formed two new department-wide interdisciplinary groups to support interdepartmental initiatives at both the policy and technical levels that enhance human, animal, and environmental health.
In a world where disease knows no boundaries, the One Health concept has evolved as the most practical and common-sense means for coordinating between the public health and animal health sectors as well as acknowledging the impact of the environment in the incubation and transfer of infectious diseases.
This comprehensive approach will improve global capabilities to detect, prevent, prepare for, and respond to emerging diseases, pandemic threats, and other issues at the human animal and ecosystem interface.
USDA is developing a greater clarity, understanding, and definition of its One Health approach.
The many cross-cutting organizational structures being created around One Health will help USDA meet the complex challenges of world hunger, food security, environmental stewardship, climate change, and emerging diseases in an ever-changing world. USDA is using the One Health working group to coordinate efforts addressing the use of antibiotics in farm animals and its impact on antibiotic resistance. USDA is also working with the Food and Drug Administration (FDA) in addressing this important societal issue.
By applying One Health principles, it is USDA’s hope to encourage a synergy of ideas, reduce program redundancy, and apply this holistic approach—ultimately—to improving global (human, animal, and environmental) health.
As many of you know, USDA, through its Food Safety and Inspection Service (FSIS), ensures the safety of meat, poultry, and processed egg products both domestically and from countries approved to export to the United States.
Prevention is the guiding principle of USDA’s food safety efforts.
USDA, FDA, and Centers for Disease Control and Prevention (CDC), along with help from consumers and the industry, have made great strides in reducing E. coli O157 illnesses. O157 illnesses have declined by nearly half over the past 14 years, and in the past 2 years, the nation’s public health objective in this area has been met. This success is due, in large part, to the Pathogen Reduction and Hazard Analysis Critical Control Point (PR_HACCP) introduced to enhance meat safety practices and is followed up with other preventive measures within USDA and the industry. HACCP by its nature is a holistic approach to an environment where food is processed.
But prevention and inspections are most effective when they are based on good scientific principles. USDA’s primary research agencies—through intramural and extramural programs—provide the science-based knowledge to inform food safety policies and regulatory decisions.
As USDA’s Chief Scientist, I believe we need to reinforce the role that science is playing in our food safety efforts. Food safety research really is a prerequisite for food safety intelligence, and our ability to keep our food system safe is circumscribed by our knowledge of threats to the health of that system.
Research is often a silent partner in food safety, working behind the scenes, before the inspections. We often speak of taking a “farm-to-table” approach to food safety—food and agricultural research is a vital third factor in this equation.
Only occasionally is the value of food and agricultural science brought into the limelight—an unfortunate outbreak of food-borne illness, for example, or recent movies like Contagion, raise awareness of ongoing research—but are quickly forgotten when the crisis passes or movie ends.
Outbreaks draw the public into the conversation. But we need to raise awareness of the food safety research that outlasts the news cycle surrounding outbreaks. For food safety, we can’t afford to take our eyes off the ball.
Our research programs are our best weapon for identifying new threats. We monitor the latest food-borne illness epidemiological data to identify emerging threats. We work closely with our research partners to develop tests and new technological approaches that work in a regulatory setting, as well as to develop intervention strategies to reduce risk throughout the food chain.
USDA’s Agricultural Research Service (ARS) conducts research on the highest priority national and international food-borne pathogens and contaminants. ARS also conducts research for its stakeholders, including its regulatory clients, FSIS and FDA.
Because of its infrastructure, ARS is able to conduct long-term research as well as to quickly respond to newly identified threats. ARS remains flexible to emerging needs, and can and does redirect programs to respond to requests from FSIS, FDA, and CDC.
The ARS food safety program has several centers across the United States dedicated to research covering important food-borne pathogens and contaminants. ARS research focuses on identifying ways to assess, control, or eliminate potentially harmful food contaminants, including those that are accidentally or
intentionally introduced and naturally occurring pathogenic bacteria, viruses, parasites, and chemical contaminants to ensure the food supply is safe and secure and that foreign and domestic regulatory requirements are met.
Based on stakeholders’ needs, ARS research focuses on several major areas, including pathogen sources and reservoirs, detection methods, and postharvest processing.
Scientists around the world recognize that the emerging human diseases of this century will continue to arise from the animal kingdom. This past decade has seen an unprecedented epizootic, or animal epidemic, of highly pathogenic avian influenza viruses, mainly affecting poultry, but also infecting several other animal species and humans. Human infections have been associated with direct or indirect contact with live or dead poultry, and animals have been infected through the consumption of infected birds or their products, so there continues to be great concern with these viruses.
USDA-funded scientists have been researching this disease since 1963 and have developed and evaluated avian influenza vaccines, helped assess public health threats, evaluated virus virulence, and helped develop protocols for inactivating flu viruses in food.
When the 2009 pandemic H1N1 influenza A virus emerged in April that year, scientists at first concluded that the virus came from pigs. Pigs can serve as one of the vessels for the “mixing” of avian influenza and human influenza that could set the stage for pandemic avian influenza. We now know that H1N1 is a “triple reassortant virus,” which means it contains genetic material from swine, avian, and human influenza viruses—a mix that may help the virus spread quickly and pass between humans and pigs and, importantly, become more virulent.
As early as 2007, USDA-funded scientists had been monitoring for strains of influenza that could spread between pig and human populations. When, in August 2007, several people exhibiting their pigs at a county fair in Ohio developed flulike symptoms, ARS scientists quickly characterized the virus and found that in pigs it was more virulent than average, instigating immediate close monitoring of the virus in swine, birds, and other species.
The following year USDA and CDC launched a collaborative effort to develop a national swine influenza virus (SIV) surveillance pilot program to better understand the epidemiology of SIV infections and to improve diagnostic tests, preventive management, and vaccines for swine and humans. This program was instrumental in implementing surveillance for the 2009 outbreak.
USDA-funded research contributes to public health by identifying emerging disease strains, assessing current vaccines against emerging strains, and developing standards for inactivating food-borne elements. These research programs are vital as emerging strains continue to evolve, including the nonseasonal H3N2 virus that has just been reported in children in the United States in 2011.
Recognizing that food safety and food security are global issues, the ARS
Food Safety and Animal Health national programs participate in both national and international collaborations through formal and informal partnerships.
Currently, ARS has numerous federal research relationships, including with FSIS, APHIS, FDA, CDC, EPA, DHS, and NIFA. ARS, APHIS, NIFA, FSIS, and FDA have annual and quarterly meetings with leadership to discuss ongoing and upcoming research needs and set priorities.
ARS has many research relationships with academia, especially where the food safety program is collocated with or near a university. ARS is also a member of the National Alliance for Food Safety and Security (NAFSS), a consortium of more than 20 State Agricultural Universities. ARS is co-chair of the Joint Committee on Research (JCR) to address food security research (under Homeland Security Presidential Directive-9) with industry and government.
Our extramural program is the National Institute of Food and Agriculture (NIFA). NIFA provides grants that support important research, education, and extension needs and can be used to conduct large population-based studies and other types of basic and applied food safety research.
USDA’s extramural grant programs at NIFA reach out annually to FDA and FSIS to share research priorities and to identify areas where joint research can benefit each agency, as well as our shared publics. In 2011, a series of joint meetings were held to determine priorities, to identify areas of potential collaboration, and to identify gaps in the current research.
In 2009, FDA and NIFA collaborated to solicit research focused on integrating food system signals with geospatial or other innovative technologies used to detect produce contamination. Food system signals are clusters of illnesses reported by government authorities, or problems identified through routine testing. Geospatial technologies include a range of tools for mapping and analyzing data derived from natural resource information, such as climate and environmental monitoring, to predict a future event.
Recently, NIFA awarded two Coordinated Agricultural Project (CAP) grants. One of the CAP grants will help to facilitate research on norovirus, which is a little-understood but difficult virus that causes food-borne and environmentally transferred illness. The other CAP grant will focus on developing intervention and risk management strategies for reducing Shiga toxin—producing E. coli contamination in pre- and postharvest environments for beef and beef products. Both CAP grants were awarded through the Agriculture and Food Research Initiative (AFRI), the flagship competitive grant program administered by NIFA.
The National Integrated Food Safety Initiative (NIFSI), another competitive grant program administered by NIFA, has awarded more than $15 million annually to support a variety of food safety priorities in applied research, education, and extension. For the past several years, the NIFSI program identified produce safety as a special emphasis area in its annual Request for Applications. Special emphasis areas are selected based on current food safety trends (illness
outbreaks), stakeholder input, and collaboration with other federal food safety agencies.
Other NIFA competitive grant programs that provide extramural grant funding for food safety include the Expanded Food and Nutrition Education Program, the Specialty Crop Research Initiative, and the Water Quality Program. Food safety program priorities for all NIFA grant programs are developed with stakeholder input from USDA’s sister federal food safety agencies, university, and industry partners and stakeholders.
The major advantage here is that when it comes to food safety there is great deal of consensus that cuts across institutional and international borders as well as public—private interests.
Collectively, USDA agencies, such as FSIS, APHIS Veterinary Services, ARS, and NIFA, are working with industry partners to ensure that hazards are identified and controlled throughout various stages of food production.
But despite the value our research brings to food safety, the continued success and growth of that system is currently being challenged on two fronts.
In 2006, the total domestic food and agriculture R&D performed was just over $11 billion, with $5 billion from the public sector and $6 billion from the private sector. The public sector tends to do the more fundamental, precompetitive, public good research that does not provide an immediate “return on investment,” while the private sector picks up the public-sector research and does the development that leads to new products and new technologies.
We know that other countries, most notably China, are ramping up their investments in agriculture research just as the United States is cutting back. Historically, over much of the life of our 150-year-old public research system, the United States has been the leader in agriculture research, which has driven the evolution of science and technology. Recently that dedication has fallen off.
This trend doesn’t bode well for our country, its health, the health of our economy, or our food safety research leadership. There is no country other than ours that holds the leadership position or the trust of the rest of the world to do this crucial research.
Our research has a proven track record of success—now more than ever, policy needs to be as scientific as our science: evidence and performance based.
Part of the issue here is that the USDA science agencies are suffering from a funding gap when compared to other U.S. government science agencies.
As many of you know, much of USDA’s capacity for doing cutting-edge research depends on both the authorizing and appropriating cycles, and in the past several weeks we’ve seen a lot of activity on both fronts. On November 17, Congress approved the annual spending bill for USDA and it was signed into law the next day. While the final version emerging from Congress was not as damaging as the House Agriculture Appropriations Committee’s proposal earlier this year, this 2012 agriculture appropriations legislation continues the steady stream of cuts to agricultural science that started with the 2011 spending bill.
On the authorizing front, with the demise of the “Supercommittee” process, it is expected now that the House and Senate Agriculture Committees will consider a reauthorization of the 2008 Farm Bill next year. As many of you may know, the Farm Bill process comes around every 5 years when existing authorities for the Department expire. Not every Farm Bill is expected to be as transformational for agricultural science as the 2008 Farm Bill, which created NIFA to be the foremost extramural agricultural research granting agency in the nation, as well as NIFA’s flagship granting program, the Agriculture and Food Research Initiative (AFRI). But every Farm Bill has a significant impact on research.
More fundamentally than funding (and this is the second primary challenge), our country simply isn’t doing enough to educate a sufficient number of students in the STEM (science, technology, engineering, and mathematics) disciplines, and particularly in the food, agriculture, and natural resource sciences, to meet future demand.
Over the past 30 years, the total number of Ph.D. recipients in agricultural fields has only remained constant, while the numbers of Ph.D.s awarded in other life science fields has grown. Because of the tight correspondence of grant funding to graduate student training, it’s not surprising that flat funding of research leads to flat education of graduate students.
Within agricultural disciplines, there has actually been a decline in the number of Ph.D.s awarded in plant, animal, and forestry sciences while the number in environmental science has risen. So our education isn’t keeping up with our scientific needs. The private sector often highlights that it does not have the workforce needed for agricultural research—meaning that, at a time where jobs are in short supply for most of the population, there are jobs going unfilled in these crucial sciences.
Training the scientists today to solve the food and agricultural challenges of tomorrow is one of the smartest investments we can make—must make—if we are serious about leading the world to a food secure future.
At every turn, in every partnership, USDA science agencies are delivering on their mission to help ensure a healthy, productive, safe, and sustainable food and agricultural system, while protecting our precious natural and human resources.
Now more than ever we cannot relent in our support for food and agricultural science, nor neglect to educate and train the future scientists who will take the advances made today to new heights.
So we need to continue to stress the vital importance of this research and emphasize the benefits it brings to society.
As awareness grows, sustained support for food and agricultural research will follow. I look forward to continue working with many of you here today to strengthen the ability of food and agricultural science to keep our food system safe and secure. Thank you.