2. Experiences with Developing Open Scientific Knowledge Discovery in Research and Applications
Case Studies
International Online Astronomy Research
Alberto Conti
-Space Telescope Science Institute-
I am the archive scientist at the Optical UV Archive at the Space Telescope Science Institute, which is the place that operates the Hubble Space Telescope for the National Aeronautics and Space Administration (NASA) and which is going to operate the successor of Hubble, the James Webb Space Telescope. As archive scientists, we have to do a lot of work to bring data to the users, but we focus mainly on three areas. We like to optimize the science for the community by not just storing the data that we get from users but also trying to deliver value-added data. We also try to develop and nurture innovation in space astronomy, particularly for Hubble and its successor. We are good at that because we have been doing it for 20 years. And we like to collaborate with the next generation of space telescopes as well as with ground-based telescopes, because that field is quite busy.
Figure 2-1 shows some key astronomical missions, which provides an idea of the kind of astronomy and astrophysics data that exist and are being developed.
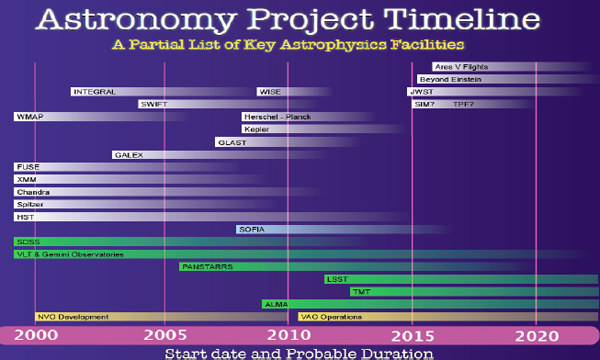
FIGURE 2-1 Astronomy project timeline.
SOURCE: Space Telescope Science Institute
The Kepler mission, for example, is a planet-finding mission. The ones on top are space based, and the bottom ones are ground based, some of which are going to produce very large amounts of data. At my institution, we collaborate directly or indirectly with most of these missions and deal in some way with the data they produce. How are we going to manage the large amounts of data from these missions?
Over the past 25 years, astronomy has been changing quite radically. Astronomers have been very good at building large telescopes to collect more data. We have been much better at building larger mirrors. But we have been a hundred times better than that at building detectors that allow us to collect extremely large amounts of data. Since the detectors roughly follow Moore’s Law, every year or so our data collection doubles, and this raises many important issues.
We realize that we are not alone in this area. We know that fields from earth science and biology to economics are dealing with massive amounts of data that must be handled. I am not particularly fond of using the word “e-science” to describe this field; I prefer to speak of “data-intensive scientific discovery,” because I think that is exactly the field that we should be moving to, because we are going to be driven by data.
However, while astronomy is similar to other fields in managing big volumes of data, the astronomy field is somehow special—not because the data are intrinsically different and special in their own right, but because they have no commercial value. They belong to every one of us, so they are an ideal test bed for trying out complex algorithms that are based on very large dimensions. Currently there are missions that have in excess of 300 dimensions, which can be very useful if a scientist wants a dataset with many dimensions to analyze.
We also have to be aware that things have been changing, for example, in the geographic information system world, where our perception of our planet has been changed by such tools as TerraServer, Microsoft’s Virtual Earth, Google Earth, and Google Maps. The way that we interact with our planet is now different than it was, and as we use all of these tools, we have no concept of what is really going on underneath, because we are focusing on the discoveries. Therefore, some of us are trying to do the same thing for data in other areas.
What is this new size paradigm? Why is so much data a problem?
In Figure 2-2, the red curve represents how much data we collect in our systems as a function of time. The big spike in the middle corresponds to the installation of new instruments for Hubble, for example. This is a trend, but it is not a problem. The real problem is the curve above it, which is the amount of data that we serve to the community. This is a large multiple of the data that we collect, so this potentially can be a problem.
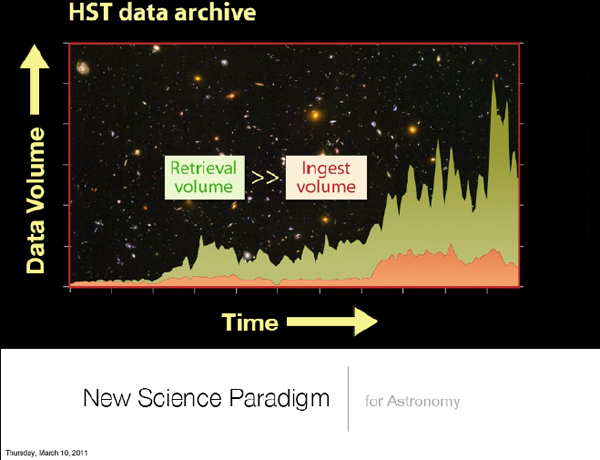
FIGURE 2-2 New science paradigm for astronomy.
SOURCE: Space Telescope Science Institute
This was a concern, at least until a few years ago, because of how we used to work with the data. As a researcher with the Space Telescope Science Institute, I can use a few protocols to access the very different data centers, but every time I interface with a different data center, I have to learn the jargon—the language of that data center. I have to learn all the idiosyncrasies of the data in this particular data center. After I learn all this, my only option is to download the data to my local machine and spend a long time filtering and analyzing them. In astronomy this limits us to small observations of very narrowly selected samples.
In 2001, scientists realized that this was not a good model, so the National Science Foundation (NSF) funded the National Virtual Observatory for Astronomy. The goal was to lower the barrier for access to all the different data centers so that a scientist did not have to worry about the complexities behind the data centers. They just had to worry about how to get data and do their science. The National Virtual Observatory was established in 2001, and for several years it was in the development phase. It moved into the operational phase with funding from NSF and NASA in 2010. By this time it was called the Virtual Astronomical Observatory.
At the same time these developments were taking place in the United States, people across the planet were building their own standards, their own protocols, and their own data centers. At some point it became evident that we could not ignore what was happening around
the world, because many of the ideas that were being proposed to deal with this deluge of data were very worthwhile and could be integrated into the thought process that goes on inside of any astronomical archive. Hence, in the middle of developing the virtual observatory, a collaboration called the International Virtual Observatory Alliance (IVOA) was begun with the goal of lowering the barrier for access at all of the data centers across the world.
Today the IVOA allows scientists to access all these different missions and to become more knowledgeable about what each mission does and how they are different from each other. It also has helped scientists identify where data are stored and what they are called. This is a major leap, but it has come at a cost, and the cost is that we have developed very ad hoc data standards for storing and sharing the data. After a long development process, we adopted protocols that turned out to not be very effective at certain times. Furthermore, after that many years of development, we do not have very effective tools, because doing the data mining is very difficult. We are still in the mode where most of the data have to be downloaded and filtered locally, which is not what we would like to do. Although we have standards and protocols, the IVOA has come under some pressure lately because it is not changing perhaps as fast as some would like.
To some people’s credit—particularly Dr. Alex Szalay of Johns Hopkins University, who is one of the founding fathers of the virtual observatory idea—it was predicted that this transition would be chaotic, and it has been chaotic. It is very hard to integrate very large data centers across the planet into a seamless process of discovery, but some people are trying to do that. The chaotic part has come to pass. Unfortunately we do not yet have a uniformly federated and distributed regime of sources across the planet that we can access in a seamless manner, so this is part of our existing problem.
In the future, I think the IVOA would still be at the core of developing such a distributed system. Some people, like me, believe that the IVOA should have a much smaller and limited role in defining the requirements and standards for this distributed system that allows people to access and process on a cloud-like system. The IVOA should try to build standards that are not ad hoc, but rather are based on industry standards. Metadata is another important frontier that we must explore so that we can characterize these data and mine them appropriately. The idea is that we want to enable new science and new discoveries.
Here is what I see as the challenges we are facing. We capture data very efficiently, but we need to reduce obstacles to organizing and analyzing the data. We are also not very good at summarizing, visualizing, and curating these large amounts of data. Another issue is that most of the time the algorithms that produce the papers that we see are never associated with the data. We should consider this as a problem. A scientist should be able to reproduce exactly the research result once he or she finds the data.
Semantic technology is key to being able to deal with many of these obstacles. We have to be prepared, because this will introduce a fundamental change in the way astronomical research is conducted. Another issue is that we do not have available to us an intensive data-mining infrastructure, and if we are to build one ourselves, that will be very expensive. Many of these efforts in astronomy are not funded at a level that would allow us to build our own. A solution for handling very large datasets is emerging. Such solutions are not pervasive in astronomy, which is also a problem. As for hosting, if we host our large datasets on the cloud
for redundancy, for performance, and so on, that will come at a cost, and some archives might not be able to sustain that cost even if it does not seem to be very large.
Finally, I think we have the opportunity to do a few things. We have to be a little humble and accept that we have to ask for help from computer science, statistics, applied mathematics, and from others who can use our high-dimensional data for their purposes, and to apply their useful algorithms. We should leverage partnerships with those who are already doing this work on the Web scale. There are companies—Microsoft, Google, Amazon, and many others—that are handling and trying to solve these problems, and we should definitely partner with them. They may not have the solution for us, but they will have a big part of some solution.
This page intentionally left blank.
Stephen Friend
-Sage Bionetworks-
There are three points that I would like to focus on today. One concerns the current paradigm shift in medicine, and I am going to make a case for why we need better maps of diseases. The second—and probably most important—point is that it is not about what we do, but how we do it: It is all about behavior. Third, I want to discuss some aspects of building a precompetitive commons.
When we are working with commercially valuable information—unlike the case with astronomy, there is serious money involved with information in the medical area—a number of issues arise. The cost of drug discovery, which is at the intersection between the knowledge and being able to impact people, is around $1 to 2 billion per molecule. There is about $60 billion being spent every year to make drugs, but there are fewer than 30 new drugs that are being approved. People are beginning to realize this lack of efficiency, however, and the good news is that there is information coming along that makes discovery easier. To put it in perspective, even after drugs are approved their rate of effectiveness is low. This is true for cancer drugs and also for the statins used to treat high cholesterol levels. Only 40 percent of patients who take statins have their cholesterol lowered, while more than half do not, and of those who do, less than half see any benefit in decreased occurrence of heart attacks, yet we still give these drugs and say everybody has got to be on a statin.
The problem is that when trying to figure out what is going on inside cells and to understand diseases in terms of pathways, we are using maps that are akin to pre-Kepler models of biology. People get very excited about understanding pathways and believe that if they could just understand the pathway of a disease, they could make sense of it. Almost every single protein has a drug-discovery effort associated with it, and all of these researchers are dreaming that they will be able to make a drug against this or that disease without understanding the context and the circuitry that underlies it.
We have gone through these transitions before. We got beyond the idea that God throws lightning bolts, and we got beyond the concept of Earth being at the center of the universe. In the next 10 to 15 years, we are going to have a change that is similarly dramatic and will be remembered for centuries. It is going to be driven by a recognition that the classical way of thinking about pathways is a narrative approach that we favor, because our minds are wired for the narrative approach.
The reason this change will happen is that technologies are developing—analogous to telescopes—that will allow us to look inside of cells. In the last 10 or 15 years, we have gone from the first sequencing of a whole genome to the point where it will soon be cheaper to get your genome sequenced than it is to get a CT scan or a blood test. Think of that in terms of the amount of information associated with it. It will take a few hours to get your whole genome sequenced, and it will be under $1,000. The cost of CT scans is going up, while the cost of sequencing genomes is going down.
I know of a project in China that is planning to do the whole genome sequence of everyone in a village with a population of 1 million people. They believe they will get whole genome sequences on the village population in the next 8 years.
The problem we have is that most people are still looking at altered component lists, and astronomers have already gone through this. Altered component lists do not equal knowledge of what is happening. While people are excited about the sequencing of genomes and understanding the specifics, the reality is that we are stuck in a world where biological scientists are still generating one-dimensional slices of information and not understanding that those altered one-dimensional slices of information will not add up to understanding altered function, which is ultimately what you have to do. The reason that this is important is that the circuitry in the cell has, over a billion years, been hardwired to buffer any type of changes, and the complexity that the system has in it to tolerate a modification is hardwired together within one cell, within multiple cells within organs, and so on.
A project started in 2003 at Rosetta in Seattle asked how much it would cost to build a predictive model of disease that used integrated layers of information. By integrated, I do not mean raw sequence leads leading through to a genome. I am talking about integrating different layers of information. An experiment done by Eric Schadt represented a fundamental seismic shift in how we think of this science when it is asked: “Why not use every DNA variation in each of us as a perturbation experiment?”
Imagine that we are each living experiments and that if we know all of those variations, we could take a top-down approach similar to the blue key that is on your computer that feeds any error through to Microsoft or other places. Imagine a way in which we could build knowledge not from looking at publications. This would be very important. I will argue that publications are a derivative, and while they are helpful, that is not where we want to be focused. We should be looking at the raw data and building those up.
Imagine that we could look at putative causal genes, that we could look at disease traits, and that we could be zooming across thousands of individuals and asking what happens at intermediate layers and what happens with causal traits. The central dogma in biology, which is that DNA produces RNA, which in turn produces proteins, allows us to build causal maps that enable us to look collectively at changes in DNA, changes in RNA, and effects on behavior and then begin to ask, “What was responsible for doing what to what?”
There are many mathematical models, from causal inferences to coexpression networks, but it does not matter which statistical tools we use to do top-down systems biology. The result is that we do such tasks as taking brain samples and expression, building correlation matrixes and connection matrixes, then clustering them and building network models that use a key driver of a particular effect, not because it was in the literature, but because it was experimented with in different databases.
A key paper that used this approach was published in Nature Genetics in 2005. It was looking for the key drivers of diabetes. The idea behind the paper was to rank the order of the 10 genes most likely to be responsible for going from a normal state to diabetes and from diabetes to normal. Over the next 4 years, that work was validated by the community, which found that 90 percent of the genes on that rank-ordered list were correct. That is unheard of. Those trivial early maps did not have nearly as much power compared to what is coming, but they were good enough to make that type of analysis.
With such an approach, we could look at drugs that are on the market and drugs that are in development and try to identify those that are going to be responsible for certain subtypes of effects on behavior or on cardiovascular function, or we can look for toxicity. This is the type of shift that is emerging in the medical world for understanding compounds. There are already about 80 or 90 papers that have been published in different disease areas. The laboratory of Atul Butte at Stanford is one of the 10 labs that have made an art out of exploring human diseases by going into public datasets and building these types of maps.
The first take-home message is that we have been living in a world where we think of narrative descriptions of diseases—this disease is caused by this one agent. At Sage Bionetworks, we now use maps that are not hardwired narratives, but that look at statistical probabilities and the real complexity. For example, imagine that some components in the cells talk to each other and they need to listen to each other to make a decision on what physiologic module is coming out of that component of the cell that then talks to other parts of the cell. What we are finding is that key proteins aggregate into key regulatory drivers that are able to say, “This is what I need to do.” This is the command language. We are not looking at the genetic language, but rather at a molecular language that determines what is going on within a cell, that says, “I have enough of this,” and interacts with cells in different parts of the body.
The complexity of this approach is outstripping our ability to visualize it and to work with it, but the data are there. Those key ingredients are coming. Maps of diseases will be very accurate for who should get what drugs, and what drugs are able to be developed. This is going to totally revolutionize the way we think of disease. Disease today is still stuck in a symptom-based approach. If you are depressed, you have depression, and you try to find a drug as if all depression were the same. The same is true for diabetes or cancer. The level of the molecular analysis that we will soon be able to do is going to be important, but substantial progress will require significant resources and decades to complete.
Those maps are almost like thin peel-off slices of what is happening, and we realized that the group doing this work could not be one institution or one company. For that reason, a year and a half ago I left the job I had running the oncology division at Merck and started a not-for-profit foundation to do a proof of concept. This foundation, which has about 30 people and a budget of about $30 million, is asking how to do the same work for genomics that has been done for astronomy. We are now trying in genomics to do the virtual equivalents to what has been done in astronomy. The people who are driving it are Lee Hartwell; Hans Wizgell from the Karolinska Institute; Wang Jun, the visionary who is sequencing the million genomes in China; and Jeff Hammerbacher, who was the chief architect of Facebook. We brought Hammerbacher in because we think that this is not strictly about science. We think we need to ask why a young investigator would want to share his or her data. Why would that investigator allow someone else to work on them? We think the social aspects of this are very important.
I will offer four quick examples. In breast cancer research, we are working on ways to identify key drivers by using publicly available datasets and running coexpression and Bayesian networks. We are going to be publishing that work soon. The second concerns cancer drugs. It turns out that virtually all of the cancer drugs that are the standard of care, the first line of drugs that a patient gets, are usually not very effective. We are looking at Cisplatin doublet therapies for ovarian cancer, Avastin for breast cancer, and Sutent for renal cell carcinoma. We realized that we should identify who is not responding to those drugs so that these patients can be given something else. It has been very difficult to get the National
Institutes of Health and the Food and Drug Administration interested, so I had to go to China for funding.
The third example involves getting data from pharmaceutical companies. The richest trove of data is inside these companies, because they often collect genomic and clinical information when assembling information for a drug filing; but they lock all these data up, because they do not want to share what it was that gave them their advantage. Accordingly we took a different approach. We asked to use the data from the comparative arm rather than the data for the investigational drug. This helped, and all of these companies have said they will give us these data.
That brings up the point that Dr. Conti was making earlier—that when we have all these data, we must worry about the annotation and curation. That is what we are doing. We are saying, “You give us the data, and we will annotate, curate, host, and make them available to anyone.” There is a very big difference between accessible and usable data. We tell the pharmaceutical companies that if they make the data accessible, we will make them usable. I am sure we are going to do it in a bad way that has to be redone, but at least we will get it started.
The fourth example is a project in which we took five labs that are supercompetitive with each other and linked them together so that they could share their data, models, and tools. This type of linking opens up new opportunities. We found that we have broken the siloed principal investigator mentality of science. This is part of that fourth dimension. Science is presently practiced like a feudal system: “I have the money, those are my postdocs, and this is my data.” But imagine a world in which scientists could go anywhere they wanted to get anyone’s data before it is published in the same way that astronomers work. We have found that it is the scientists under 35 years old who know what to do. We have been making openly available global datasets, models, and tools.
I want to describe the behavior we have run into. First, clinical genomic data are accessible, but minimally usable, because there is no incentive to annotate and curate the data. Second, people who get the data think they own the data. If someone pays them to generate the data, somehow they think they own them. We are living in a hunter-gatherer community, which is preventing data sharing, and, for the most part, the principal investigators have no interest in changing that situation. This needs to fundamentally change.
Just as Eisenhower in the 1960s talked about the military-industrial complex, I think we now have a medical-industrial complex in which the goals of individual researchers to get tenure, the goals of institutions to improve their reputation, and the goals of the pharmaceutical industry to make money create a situation in which the patients are not benefiting. There is a time for intellectual property, there is a time for innovation, but the current situation is crippling our ability to share the data. The system is not set up to make it easy to reproduce results, as in astronomy. You do not hold people accountable when they publish a paper to explain what they did so that it can be reproduced. We have to figure all of that out.
Finally, I want to ask why we do not use what the software industry has already figured out for tracking workflows and versioning. We have to change the biologic world so that the evolution of a software project is clear, with a code repository and the branches and releases. What we are doing now at Sage Bionetworks is determining the details on the repositories, collaboration, workflows, and cloud computing. We find that the world is saturated with early
efforts, so it would be absurd to think that we ourselves need to do this. We are interested in such efforts as Taverna, Amalga, and work at Google. We are identifying who has what so that we can stitch this together. The hardest thing is that we do not have the support of patients in this area yet. We have to bring the patients and citizens in. Otherwise the data-sharing, privacy, and access issues are going to tear this approach apart.
This page intentionally left blank.
Geoinformatics: Linked Environments for Atmospheric Discovery
Sara Graves
-University of Alabama in Huntsville-
I will be giving this presentation on behalf of Dr. Kelvin Droegemeier, who had an emergency to which he has to attend. Dr. Droegemeier’s area is geosciences. He is from the University of Oklahoma and serves on the National Science Board. He was planning to talk about a project called Linked Environments for Atmospheric Discovery (LEAD). I worked on LEAD as one of the coinvestigators, as did Mohan Ramamurthy. LEAD was a multi-university and multidiscipline research program that was created by the National Science Foundation (NSF).
I heard on the weather report this morning that 100 million people will be affected by a severe weather situation in the Northeast and other parts of the country, so I thought that it was quite fitting to be talking about LEAD. The annual economic losses due to weather are greater than $13 billion per year. Every time we try to get people ready for some weather event, there is a cost to that preparation, and if the weather event does not occur as predicted, there are additional costs.
We have a few weather technologies today that we are familiar with, and we use the output from many of them. According to some meteorologists, however, we still have many technological improvements to make. Radars do not adaptively scan, so one of the keys that we were looking into with LEAD was how to get more adaptation and more dynamic processing of information. We wanted to take into account the complexity of all of the factors and not just approach it linearly.
Operational models run largely on fixed schedules and fixed domains, and the cyberinfrastructure that we often use is quite static. We teach our students about current weather data, but we do not allow them to interact with it. The previous two talks discussed how to get more people involved, not just in an academic setting but also in citizen science. That is certainly something we would like to do in this domain as well. Weather is local, high impact, heterogeneous, and rapidly evolving, yet our technologies and our thinking have not evolved that quickly. One of the fundamental research questions we looked into with this project was whether we can better understand the atmosphere, educate others more effectively about it, and forecast more accurately if we adapt our technologies. Even animals adapt and respond, so why should this not be true for the resources that we have?
This was a multi-university project, and there was an associated project called Collaborative Adaptive Sensing of the Atmosphere (CASA), which I will describe shortly, that interacted with LEAD. The goal of LEAD was to revolutionize the abilities of scientists, students, and operational practitioners. The operational practitioners were a key part of the vision, because we were trying to work not just with the typical student scientist but also with the National Oceanic and Atmospheric Administration and some other practitioners to observe, analyze, predict, understand, and respond to intense local weather by interacting with it dynamically and adaptively in real time. Oklahoma and Alabama have many tornadoes. We did not have this project just for that reason, but severe weather does have a local impact.
What does adaptation mean? Let us take the example of a tornadic storm that occurred in Fort Worth in March 2000. A local TV station radar system and a National Weather Service system with 12-hour computer forecast that was valid at 6 p.m. central time did not explicitly show any evidence of precipitation in north Texas. The reality was quite different, because there was in fact a tornado.
The approach that the project took was to have streaming observations. We wanted to have constant observations and not just frequent static pictures. We did a lot of mining of both the streaming data and some of the archived data to feed into the Weather Research and Forecasting (WRF) Model. We needed on-demand computing, so we were using the NSF TeraGrid computational capabilities. The cloud could be used in that respect too. What comes out of the computation models is fed back into the mining engine, which creates a feedback loop.
What does it take to make all of this possible? Of course, you need adaptive weather tools such as adaptive sensors, adaptive cyberinfrastructure, and a user-centered framework in which these tools can mutually interact. We developed and continued working on these tools with LEAD. What LEAD was doing is creating a Web service. We had our linked environments, our processing capabilities scattered around distributed areas, and Web services to produce some of this information much faster. You also need to link together the services and workflows.
One of the lessons that was learned from this project was the recognition that we can create very elaborate workflows and save them in a special database so that it is possible to go back and recreate some of those problems, both for academic purposes and for practitioner and operational capabilities. We did significant work in creating the workflows and some of the tools. Since all of this work is openly available to others, many of these tools and processes are being used today by various entities.
To develop more accurate forecasts, we needed to come up with better tools. We did a lot of ensemble modeling, and we have continued to develop this capability with WRF.
Figure 2-3 shows the current operational radar system in the United States.
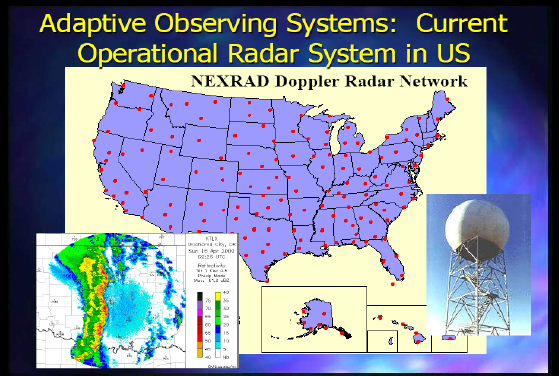
FIGURE 2-3 Current operational radar system in the United States.
SOURCE: Kelvin Droegemeier
There are limitations to NEXRAD (Next-Generation Radar), as it is not set up for some of the issues I have discussed. It is fairly independent of the prevailing weather conditions because of the way it operates, and it is very independent from the models and algorithms that use the data, since the Earth’s curvature prevents 72 percent of the atmosphere below 1 kilometer from being observed. There is also a next-generation capability being developed now.
The other project that we worked on was mainly headquartered at Colorado State University, and Dr. Droegemeier was also involved with it. It was focused on collaborative adaptive sensing of the atmosphere. We would mine and find an event that is occurring and that we could see with the radar data. Then, we would try to steer some of these radars to detect the event through dynamic adaptation. Figure 2-4 depicts the Oklahoma test bed where they had the various capabilities.
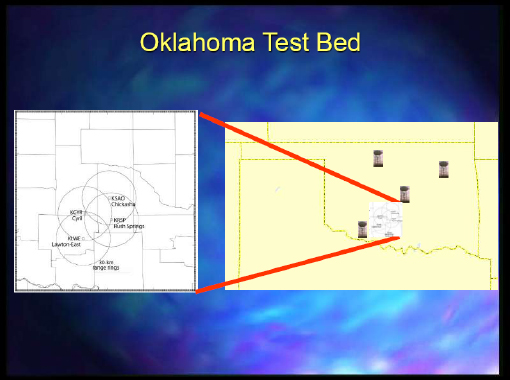
FIGURE 2-4 Oklahoma test bed.
SOURCE: Kelvin Droegemeier
Figure 2-5 is an example of adaptive sampling.
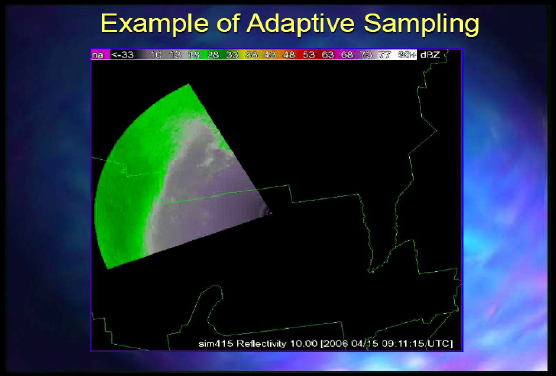
FIGURE 2-5 Example of adaptive sampling.
SOURCE: Kelvin Droegemeier
And finally, Figure 2-6 shows the CASA experiment looking at what they were seeing from the radars and the way they could steer them versus what you could get with NEXRAD.
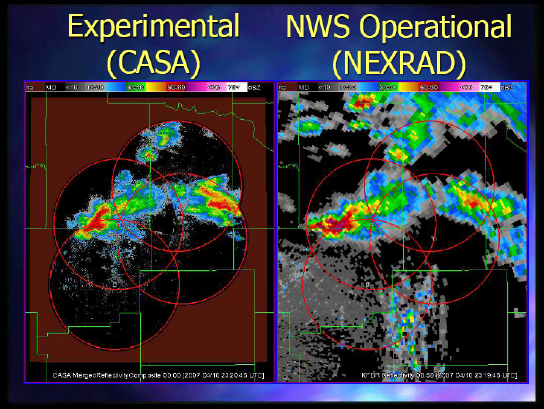
FIGURE 2-6 Comparing CASA with NEXRAD.
SOURCE: Kelvin Droegemeier
The potential, of course, lies with the fact that we are trying to constantly transform and improve our teaching, education, and workforce development.
This page intentionally left blank.
Implications of the Three Scientific Knowledge Discovery Case Studies – The User Perspective
International Online Astronomy Research, c. 2011
Alyssa Goodman
-Harvard University-
Dr. Conti provided a great overview of the challenges that face our field of astronomy. Instead of focusing on only one example, I will draw a map of the way that things work in our field, and then give a few brief examples.
Figure 2-7 shows water droplets on a spiderweb.

FIGURE 2-7 Image of water droplets on a spider web.
SOURCE: Alyssa Goodman
This is the way that I like to think of “the cloud” with the International Virtual Observatory Alliance. Basically each one of those little water droplets is some data repository, service, or person that provides something in this ecosystem of online astronomy research. The way that those kinds of services are connected on the World Wide Web is how I think that online astronomy research should be done—in a slightly less orchestrated fashion that people might have thought of a decade or more ago. In my work, we call this “Seamless Astronomy.”
There is a researcher in the middle of the image in Figure 2-8. Imagine that this researcher is drawing on two kinds of primary sources of information: data on the left, and literature on the right.
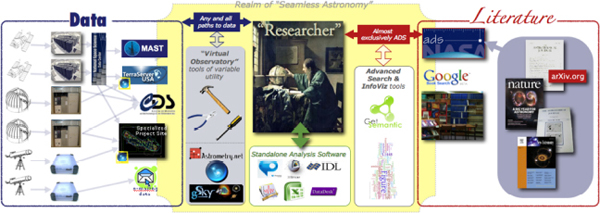
FIGURE 2-8 Realm of Seamless Astronomy.
SOURCE: Alyssa Goodman
In astronomy, our literature is organized very well by a service called the Astrophysics Data System (ADS). Much of what I am going to discuss is how the ADS team is working on connecting the data to the literature through the ADS Labs at the Center for Astrophysics in Cambridge.
I am going to take this Web-like approach and try to explain a few of the interconnections between the two sides of the picture: literature on the right and data on the left. I have reduced this model to just a few services on which I will focus, but as the disclaimer at the bottom says, this slide shows key excerpts from within the astronomy community and excludes more general software that is used, such as papers, graphing and statistic packages, data-handling software, search engines, and so forth. As I mentioned earlier, the literature is very organized, so I have put it all in one accessible repository, and it is accessed through the ADS system. As for the data, there is a set of archives that I will cover shortly.
Figure 2-9 is a view of the international online astronomy ecosystem that I will use to frame the rest of this talk.
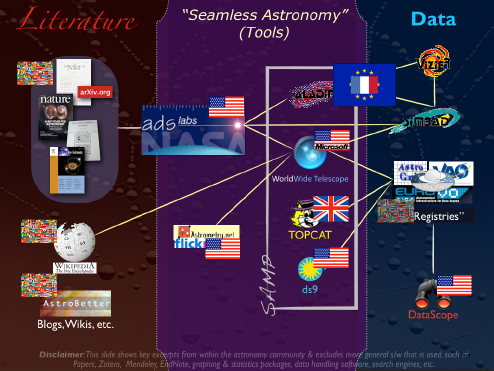
FIGURE 2-9 International online astronomy ecosystem.
SOURCE: Alyssa Goodman
In the middle, I have depicted some tools in a regime called Seamless Astronomy, which offers links between data and literature that are still partly missing, but are now forming and expected to lead to the increase in scientific productivity. The “ADS Labs” is a project in which the ADS team is taking what is already a successful system for accessing the research literature in astronomy and trying to draw that system closer to the data by using the articles themselves as a retrieval mechanism for data. Since ADS already stands for “Astrophysics Data System,” why not develop a way to retrieve articles using the literature as a filter for the data?
There are also linkages between many services that do different work. What you should notice, however, is that there are many yellow lines in the figure, which connect all of these services in a variety of ways. It is important to note that this is not a linear process. There are many different ways that the services can access each other without the user even knowing about it.
Four of these applications—the four highlighted in the “SAMP” box in the figure (SAMP is a message-passing protocol)—are actually connected to each other in a way that allows them to interoperate as a set of services and as a research environment.
I would like to point out that the flags in the figure represent the various countries involved. There is also an international flag that indicates this activity is not associated with any one particular country. Some of the services I am going to talk about are from the United States, but others are not, such as TOPCAT from the U.K. e-Science Program.
Figure 2-10 shows a screenshot of what happens when we run many of these services together as well as the SAMP hub that allows the services to talk to each other.
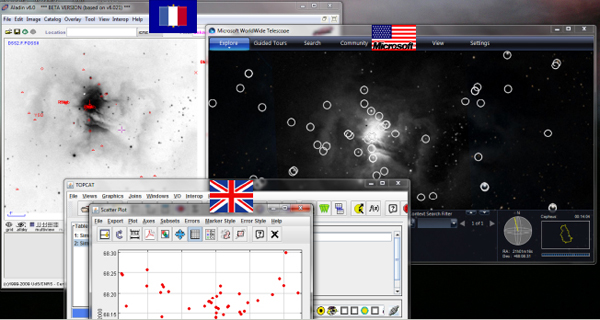
FIGURE 2-10 SAMP hub.
SOURCE: Alyssa Goodman
Suppose that I was looking at some particular region in the sky in one application. I can open another application that looks at the exact same region. Worldwide Telescope can superimpose catalogs in ways that are different from how the first application superimposes catalogs. And TOPCAT, which allows us to statistically manipulate those catalogs, will show the results through live links. It is important to note that these are from three different sets of countries, and the applications are working together through this open-source message-passing protocol. At the moment, my view is that probably only about 5 percent of astronomers know how to do this correctly.
Now let me show you the ADS Labs itself. I should point out that ADS is funded through the U.S. Virtual Observatory funding, provided by the National Aeronautics and Space Administration (NASA). It also has a mirror site in France run by Centre de Données Astronomiques de Strasbourg (CDS). ADS Labs was released in January 2011, at the American Astronomical Society meeting in Seattle. The idea was to take ADS and, instead of just doing a keyword search, do more intelligent searching. There are some interesting semantic aspects of this kind of search. Let me give an example of how the ADS can be used.
I was working on an observing schedule recently to observe an obscure quantum mechanical effect called the Zeeman effect in the molecule CH from a particular telescope. I wanted to make sure that no one has ever done this before, so I checked for related papers. If I use the most recent work function of ADS and type “Zeeman effect,” I get something that looks like the image in Figure 2-11. There is a filter list down the side, and I can choose particular authors, key words, missions, objects, and so on. The list of papers that I get will change depending on what is chosen.
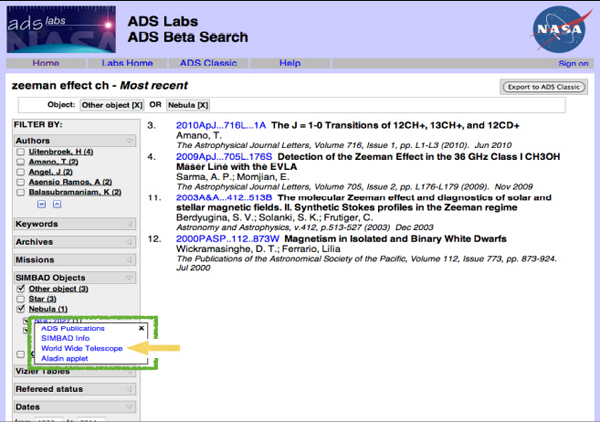
FIGURE 2-11 ADS Labs Beta Search.
SOURCE: NASA
The green box in the figure highlights some linkages between the data and the literature. If I click on one of the particular objects that seems to be mentioned very frequently in these papers, I get four different choices: (1) I can look up all the publications about that object; (2) I can look up data on that object using the SIMBAD data service from CDS; (3) I can see that object in context using Worldwide Telescope; or (4) I can do a similar thing, although it is a narrower context, in anther service called Aladin.
If I clicked on Worldwide Telescope, I would get a view like the one in Figure 2-12. The object turned out to be a planetary nebula that I did not know. Since this is not what I was interested in, I concluded that no one has observed what I was looking at and that this object was irrelevant. But perhaps I became curious about this object, and so I could click on the button at the bottom that says “Research.” By clicking there, I can get another set of choices, linking to yet other services.
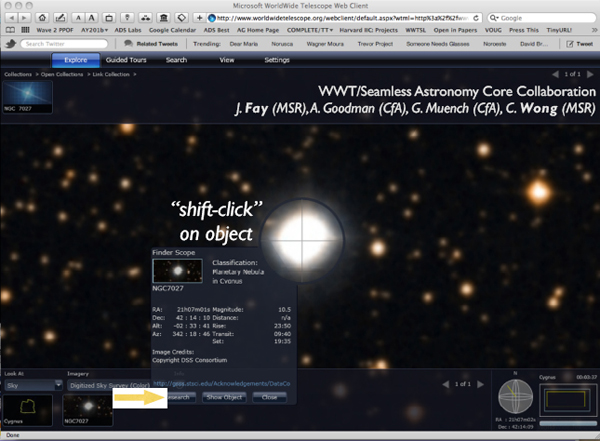
FIGURE 2-12 Worldwide Telescope.
SOURCE: Alyssa Goodman
Basically what we are being offered when clicking on the different options under “Research” is either more data or more literature. The links are such things as “look up all the publications about that particular object,” “look up that object on Wikipedia,” or “look up that object in astronomy databases.” This system offers a lot of flexibility. However, even though the system is supposed to list the most relevant papers, it is employing a very low level of semantics to do the search. For example, because I know this field, I can tell if the list of papers is incomplete. It is partially right, but it is not giving the full picture to a scientist who has never worked in this field, and that can be very misleading.
At this point, I should recognize the people who are involved in this project. Worldwide Telescope was envisioned by Curtis Wong and implemented by Jonathan Fay, who is a software engineer at Microsoft Research. At the Harvard-Smithsonian Center for Astrophysics, Gus Muench and I have been working closely with them since the beginning, but Curtis and Jonathan get all the credit.
Let me give another example. Assume that I find an astronomical image somewhere on the Web or that I go outside on a clear night with a camera and I take a picture of the sky. With this service called astrometry.net, I can submit a picture, and the service will take the image and miraculously calculate exactly where it goes in the sky and, if the picture is good enough, even figure out when it was taken.
There was a service that some researchers were hosting to do this work, but they did not have enough money or servers. So they made a group called “astrometry” on Flickr. As illustrated in Figure 2-13, when you upload a picture to Flickr, not only does it host the image but it also offers some services that, for example, tell you what is in that picture. As soon as a picture finishes loading, if you add it to the astrometry group, you will soon see all sorts of little boxes pop up on the picture, and it will tell you famous objects in the picture that it recognizes. The reason Flickr can do this is because the astrometry.net program went in and blindly solved for the coordinates of the image. On Flickr, after an image is solved by astrometry.net, you will see a button at the bottom of the page that says “View in Worldwide Telescope.” If I click this button, then I will see that object overlaid on the sky where it goes, and I can compare it to 40 different layers of multiwavelength data. I can also use all the catalog-searching tools that I showed before and interact with all the statistical software, plotting software, et cetera, live.
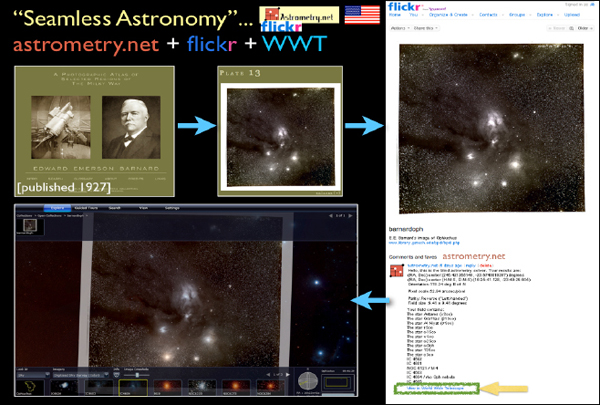
FIGURE 2-13 Seamless Astronomy.
SOURCE: Alyssa Goodman
There are two more projects in which Alberto Pepe is involved. In one of them, we are taking all of the historical images that are online, including all of the literature, and making a historical image map of the sky. With the second project, we can tell where particular papers on the sky are and why those papers were written, so you can get heat maps of articles on the sky.
I will conclude by highlighting a couple of examples where one field of study has borrowed from another. We published a paper (Goodman et al. 2009, Nature) 2 years ago9 that has embedded in it an interactive three-dimensional (3D) diagram; the idea is that you go there, click it, and you can put the data in the literature (Figure 2-14).
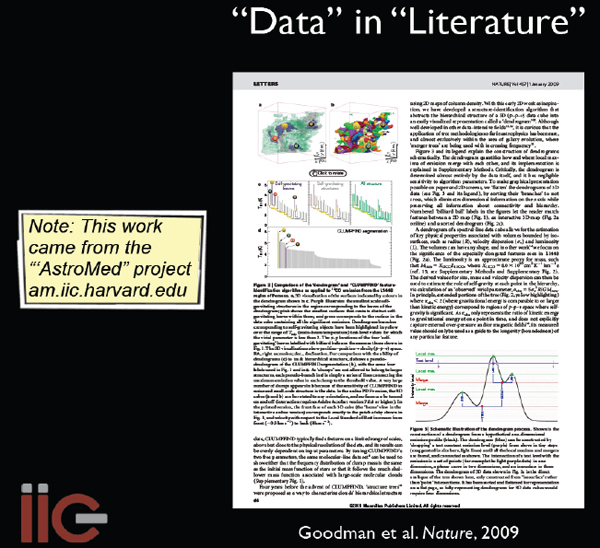
FIGURE 2-14 Data in literature.
SOURCE: Alyssa Goodman
The techniques used to create and study the 3D data were borrowed from medical imaging and used in astronomy. In the new Astronomy Dataverse (theastrodata.org), we have borrowed tools from the social science world as well. The Astronomy Dataverse tools allow individuals or groups to publish their data online in ways that can be retrieved in the future and that are permanent and linked to literature. At present, we are experimenting with the Astronomy Dataverse at the Center for Astrophysics, to let researchers who have small datasets and want to share them with everybody have a way to publish them that is linked to the literature and is persistent.
__________________________
9Nature, Volume 457, January 1, 2009.
Joel Dudley
-Stanford University-
In this talk, I want to offer some tangible examples showing why it is important to integrate data in a certain way. It is not like a seed that you bury in the ground, and then in a few years it is going to bear fruit. Rather, it is a process that produces immediate discoveries. I will present what we have done in just a few years by dealing with data in this way, essentially on a shoestring budget, and hopefully this will motivate larger investments in this area.
We published a paper10 a few years ago describing how we took a gold standard set of Type 2 diabetes and obesity genes and wanted to see how well we could recapture them. This is a classification problem. We looked at different modalities individually, for example, genetic experiments that are measuring Type 2 diabetes, or proteomics, another modality for measuring diabetes
In Figure 2-15, the diagonal is essentially a random chance classifier, and anything worthwhile needs to be above it.
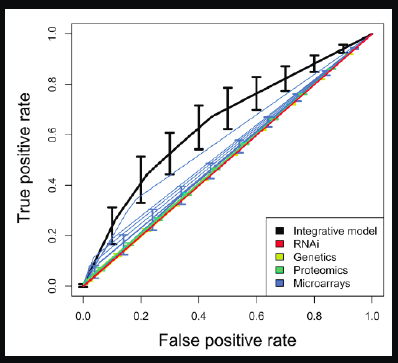
FIGURE 2-15 Rates of Detection for Type 2 Diabetes.
SOURCE: Joel Dudley
We put all the modalities together, which is represented by the black line, and tried to recover a known set of Type 2 diabetes genes. This is a very simple classifier, but what it
__________________________
10 S. English and A. J. Butte, “Evaluation and Integration of 49 Genome-wide Experiments and the Prediction of Previously Unknown Obesity-related Genes,” Bioinformatics (2007) vol. 23 (21), p. 291.
shows is that by putting all the data together, we are able to beat all of the individual modalities. Compared with all the single investigators working very hard on their own single modalities, we were able to surpass each one of them by putting all their data together from the public domain.
One of our main sources of data is the Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information. There is also a European counterpart called Array Express. The GEO is a public repository. If a scientist publishes a paper that incorporates gene expression data, the journals require that this data be submitted into GEO. GEO is growing exponentially and has all kinds of useful data. People deposit their data in there reluctantly, but they are doing it. However, as Dr. Friend pointed out, there is a difference between accessibility and usability. GEO is freely available and it is very accessible (e.g., high school students can access and download all the data they want), but it is not very usable.
There are hundreds of thousands of samples. One sample is essentially data derived from microarrays. This is roughly a 10-year-old technology. The very small chips that measure how much a gene is turned on or off in a biological state by measuring the abundance of its expressed transcripts are roughly a 10-year-old technology. There will be certain levels of mRNA expressed off the DNA, and the chip has probes for each of the genes in the genome. By measuring the responses with a scanning laser, you get an analog readout of how much each gene is turned on or off.
To put these data into GEO in a structured format, they used a standard called MIAME (Minimum Information About a Microarray Experiment), but there were no semantic requirements put on the data. As a result, we have all the data in GEO, but we do not know what is being measured. This means that we could not use GEO to find, for example, all the experiments that measure a disease. It is just not set up that way, so we came up with a solution. Since all these experiments are linked to a publication, we could find the publication in PubMed Central and download the Medical Subject Headings, and then map them onto the Unified Medical Language System, which is a huge medical thesaurus created by the National Library of Medicine. In other words, since a paper linked to an experiment must be related to the experiment in some way, we could use the paper annotations to figure out what the experiment is about. We can indeed do that, and it has worked fairly well.
There are other sorts of annotations related to which state is being measured in an experiment. For example, if we are going to do an experiment in which we measure healthy and sick people, some of the chips will be measuring the healthy people, and some chips will be measuring the sick people. There are some annotations that tell us which chip is which. This is very important information to have if there are remote users of the data. Unfortunately it is free text, so that it is not done to a uniform standard. To refer to a person with diabetes, some cases might say “diabetic,” some might say “Type 2 diabetes,” others might say “insulin resistance,” and so on. I saw an experiment where the annotations read “dead” and “not dead.” To figure out which chips belonged to which subset, we did some text mining.
The next problem we encountered was how to determine which genes were being measured on these chips. There are thousands of different platforms. Some of them are custom made by universities. Also, there are different commercial vendors with different technologies and types of probes. We had to determine which genes were on these chips. This problem had not arisen before, because scientists were not trying to map across these different types of
technologies; they got their chip for their particular study and stayed within their own technology. We also found out that these annotations were changing by 10 percent or so every year. We thus had to build an entire system that would constantly update and remap all these probes to tell us which genes were being measured on the different chips.
We then had to determine the quality of the data. To do that we looked at how well data from different labs matched up. For example, we would look at data from one lab using a particular technology that measured Type 2 diabetes in muscle, and compared that with data from another lab using another technology that measured Type 2 diabetes in muscle in completely different patients. It turned out that they match up fairly well—quite a bit better than random. So even though there are all these possible confounders, the quality of the public data is quite good. This was very encouraging.
Once you have all this data annotated in this way, it can be visualized using a matrix. In such a matrix, you can have 300 or so diseases in the rows and then 20,000 genes in columns, which produces a big grid of all diseases and all genes in a very clean, standardized way, showing how the genes change with the different diseases, as illustrated in Figure 2-16.
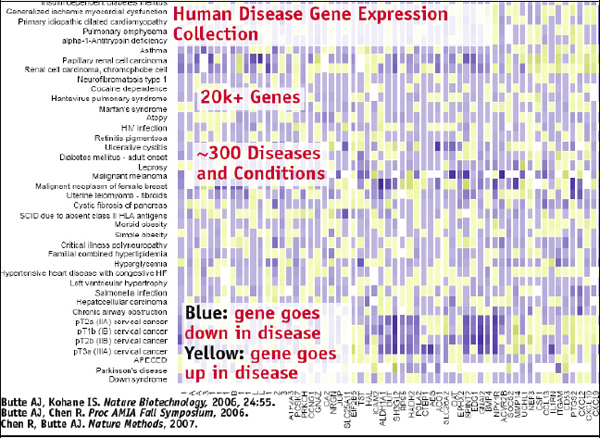
FIGURE 2-16 Human disease gene expression collection.
SOURCE: Joel Dudley
At this point we can start thinking of interesting questions to ask. You could simply cluster that grid and see where the diseases fall with regard to the various molecular bases. This will provide the molecular classification of disease, and then you can see some patterns. Brain
cancers are clustering together, which makes sense, but then you see that pulmonary fibrosis is clustering with cardiomyopathy, which seems strange. It is a lung disease clustering with a heart disease. Typically these are very different diseases, but maybe at their molecular level they have some similarities.
Then you start to wonder what the implications might be for drug treatment. We have all kinds of drugs for cardiomyopathy and hypertrophy and similar cases, but we do not have any drugs for pulmonary fibrosis. Therefore if the molecular basis is so similar, we might be able to borrow drugs from one disease to deal with another.
We do not quite use this representation to figure that out. Instead we have discovered a new way to mine the public data for drug repositioning, as it is called when we borrow a drug from another disease. We have a compendium of clean disease data that is semantically annotated, and we did the same thing for drugs. See Figure 2-17. This gives us both the disease and the drug data. Now we can look at treated versus untreated diseases and get a signature of the disease state versus the healthy state, and the signature in this case is how the genes are changing between the two conditions.
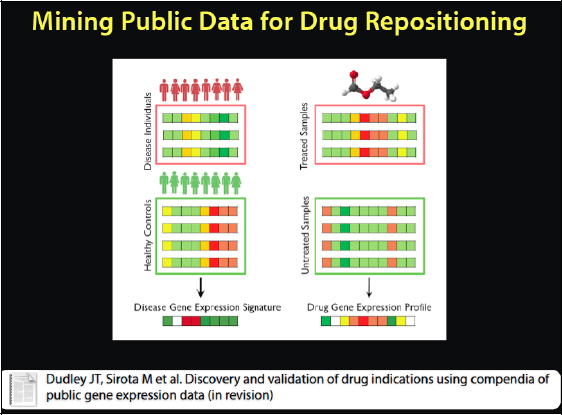
FIGURE 2-17 Mining public data for drug repositioning.
SOURCE: Joel Dudley
Similarly a scientist can look at how the genes are changing in response to different drugs and do pattern matching to match them up statistically. This is exactly what we did, and it produced a representation like the one in Figure 2-18.
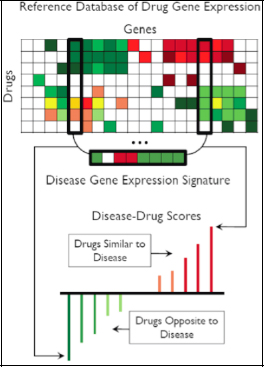
FIGURE 2-18 Reference database of drug gene expression.
SOURCE: Joel Dudley
If a scientist has thousands of drugs and hundreds of diseases, and gives a therapeutic score to each pair, Figure 2-19 shows how the heat map would look.
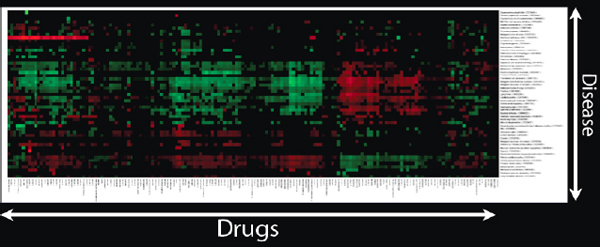
FIGURE 2-19 Heat map of drugs and diseases.
SOURCE: Joel Dudley
After we have done that, we start to see interesting clusters. We might ask why all these drugs and diseases appear in a particular cluster, and check to see if the drugs in the cluster are known to treat the diseases in this same cluster. We might also ask if the drugs that have not been identified as treating those diseases might actually be effective against them.
The first computational prediction that we tested was whether cimetidine, an anti-ulcer drug that anyone can buy inexpensively at any pharmacy, would treat non-small-cell lung cancer. It seemed a very strange prediction, but we thought we would try it (Figure 2-20).
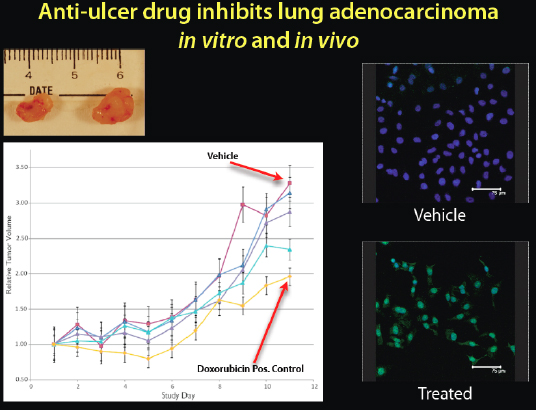
FIGURE 2-20 Anti-ulcer drug inhibits lung adenocarcinoma in vitro and in vivo.
SOURCE: Joel Dudley
When we put human tumors in mice and tested the drug, we got a dose-dependent response. It shrank the tumors, and it was even getting close to a chemotherapeutic drug, doxorubicin. We can see on the top left image the cimetidine-treated tumors on the left and the control on the right. We can also see on the right side the fluorescence microscopy of the actual cancer cells. The green lighting in the bottom right box indicates that when the cells were treated with cimetidine, there was a lot of apoptosis, or cell death, in the cancer cells. We predicted that this over-the-counter anti-ulcer drug would be antineoplastic, or anticancer, and it seemed to be efficacious in this animal and human cell line study.
Another prediction we made was that an antiepileptic drug would treat inflammatory bowel disease, which was another strange connection. We got the rats that have an induced phenotype of inflammatory bowel disease, the TNBS model. A very caustic compound is put in the rat’s colon to induce the inflammatory response. When we induced the disease and treated the rats with the drug, it ameliorated the disease. This is a very harsh phenotype, but the drug appeared to reduce the inflammation. This is an anti-epileptic neurological drug working on the colon. At the bottom of the image in Figure 2-21, you can see the pink colons of the rats that were treated, while in the untreated rats on the top you can see all the necrosis.
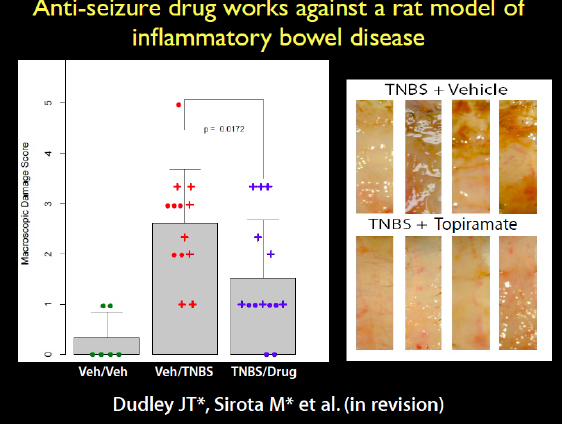
FIGURE 2-21 Antiseizure drug works against a rat model of inflammatory bowel disease.
SOURCE: Joel Dudley
Figure 2-22 provides another recent example. We predicted that a cardiotonic drug, which was researched quite heavily in the 1980s, but never approved for any disease, would also work for inflammatory disorders. We put it in rats that had an inflammatory phenotype. What can be seen in the table are the blood level measurements of a cytokine called TNF-alpha, which is involved in inflammation. On the right you can see that this heart medication substantially reduced TNF-alpha levels and plasma in rats.
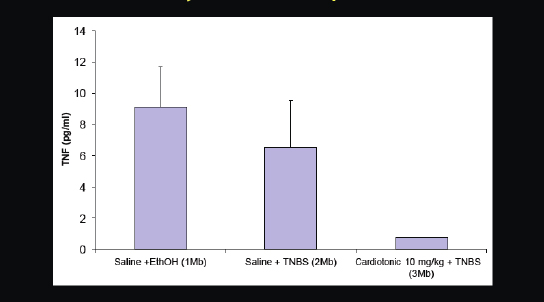
FIGURE 2-22 Cardiotonic drug inhibits or ameliorates inflammatory cytokine TNF-alfa.
SOURCE: Joel Dudley
Also, we took several experiments related to transplant rejection from the public data, intersected them in a certain way, joined them with a number of serum measurements of proteins, and found a biomarker for transplant rejection. This is a particularly important biomarker, because when someone gets a new kidney, for example, medical personnel stick a big needle through the side of the patient’s body, suck out a little tissue around the kidney, look at it under a microscope, and, if this person is lucky, they get the part that is being rejected, and they will try to manage it that way. Just by using the public data and integrating them with public datasets, we computationally predicted a new blood-based biomarker. We are testing it, and it looks like it is also able to be found in the urine. Therefore, instead of putting a big needle into the side of the patient’s body, it should be possible to take a urine sample and tell from that whether the kidney will be rejected. Again, this is a repurposing of public data. We did not generate any new data from the patients at Stanford University to do this, although that is where it was tested in the clinic.
There are many more examples from our lab. We just found a new disease risk variant for Type 2 diabetes, which has been validated in a Japanese population of thousands of cases and thousands of controls. We found a new drug target for Type 2 diabetes and validated it. Indeed, these are all validated experimentally. We found biomarkers for medulloblastoma, pancreatic cancer, lung cancer, and atherosclerosis—all validated—and this is just from our lab, but it should not be just from our lab.
The reason we can do this kind of work is that we have significant computational capabilities. Everyone who works in our lab has professional or graduate-level training in molecular biology and computer science. They can draw cell signaling pathways from memory and also program a compiler. It is unfortunate, however, because much of this technology is an expensive commodity, and the people who actually have the questions cannot do anything with their data.
Another example involves the single-nucleotide polymorphism (SNP) chips, which measure genetic variation on about a million different regions of the genome. We can combine them with the gene expression microarrays, and for relatively little expense, a commissioned researcher can then assemble a group of patients and measure both their DNA variation and their gene expression variation. From that, the researcher can figure out such things as which genetic variants are really behind a disease. The SNP chip contains a million probes, however, and the gene expression microarray measures 50,000 probes, so the researcher is looking at 50 billion array comparisons to determine which are related, which would take several years to do on a desktop computer.
We therefore did a case study that showed it would be possible to do the research in the cloud and pull out 100 nodes with a minimal cost (Figure 2-23). It required a lot of programming that the clinical researcher typically would not be able to do, and there are no tools available that allow the researchers who generate the data and have good questions to answer the questions with their own data.
FIGURE 2-23 In silico research in the era of cloud computing.
SOURCE: Joel Dudley
I think, however, there is a pragmatic first step that can be taken with cloud computing. Many people will disagree with me on this, but there are many people working on perfect workflow systems, and although people in biology do not seem to like to use them, I think those tools will become the standard. One problem is that people like to keep everything in-house—and probably always will—because they are at the forefront of these research areas, and the standard tools do not necessarily allow them to do what they need to do. One valuable aspect about the cloud is that the entire infrastructure is virtualized, so, for instance, I was able to set up a pipeline for doing drug prediction. It is simply a number of Linux servers and our code. What I can do is to freeze that in place, and at the very least I can hand it over to someone in the cloud who can clone it. I am a big believer in simple first steps and rapid iteration versus locking yourself in a room for years to come up with a perfect system that solves all the problems. I think there is some promise there, and we are starting to see it happen. There are some good tools that are being created to facilitate reproducible computing in the cloud.
Let me now focus on some of the lessons learned from the work in our lab. So far, sticks have worked better than carrots. Biologists hate sharing their data. The data are voluminous and although the researchers are supposed to publish them in GEO, there are many microarray experiments that do not get published. When we find that a paper that is funded by the National Institutes of Health and it has microarray data that are not in GEO, we call the Microarray Gene Expression Data Society, and the people there follow up with the researcher and ask that person to post the relevant data there. We constantly have to do that, so we hope that carrots will work better at some point.
In ontology, we have been able to accomplish much by doing very lightweight and simple annotations on gene identifications. We did not need to model biology perfectly in a higher ontology to do what we needed to do.
Computation is a major bottleneck. Right now there is a privileged computational elite. There are people who cannot even answer questions from their own data on their own patients, and, on the other hand, there are the computational elite who have 500 CPU clusters in their labs and the power to do this kind of work. Obviously this is not fair.
Questions first, data second. What I mean by this is that we did not really know the type of system we needed until we knew the questions we wanted to ask. There are many people in informatics who are pattern hunting and trying to find hypotheses, but it was not until we knew the questions we wanted to ask from the data that it became apparent what type of system we needed, and it was very different than what we had. Finally, to sum up, I would like to emphasize that the advancement of new biology and medicine is possible through public data.
Mohan Ramamurthy
-Unidata-
I have been with the Unidata Program for the last 8 years, and before that I was on the faculty at the University of Illinois in the Atmospheric Science Department for 16-plus years. My talk, therefore, will cover the perspectives of the different hats I have worn in the past 25 years or so.
Unidata is a facility funded by the National Science Foundation (NSF) for advancing geoscience education and research. We are part of the University Corporation for Atmospheric Research (UCAR) and a sister program to the National Center for Atmospheric Research (NCAR) in Boulder, Colorado. Part of our mission is to provide real-time meteorological data to users throughout the world. It started with academic universities getting the data, and that data distribution has expanded to include other organizations. We do not give just data to our users; we also give them a suite of tools for analyzing, visualizing, and processing them. One of the software products for which we are best known is NetCDF, because it has become a de facto standard in the geosciences. The data we provide get integrated into research and education in universities, as well as in many high school classrooms. We have more than 30,000 registered users. There are many more who do not register, but get the software. The users come from 1,500 academic institutions and 7,000 organizations in 150 countries around the world, representing all sectors of the geosciences—not just academia, but government, nongovernmental organizations, and the private sector. They get the data and the software from us, because everything we do is available at no cost.
I want to go back almost 25 years to the work that my former colleague, Dr. Robert Wilhelmson, did at the University of Illinois and at the National Center for Supercomputing Applications (NCSA), which ushered in a new era of geoinformatics for my field. That work was the result of a confluence of three factors. First was the availability of high-performance computing. Because the NCSA had been established a few years earlier, he had access to cutting-edge, high-performance computing systems. Second, he had software for visualizing and creating fantastic graphics, which won first prize at the London Computer Graphics Film Festival in 1989 and was also submitted for an Academy Award nomination. Third, there were massive quantities of data coming from state-of-the-art cloud computer models. From those factors, geoinformatics was born, and it revolutionized our understanding of supercell thunderstorms, convective storms that spawn tornadoes, and mesoscale connective systems, such as the one Dr. Graves described earlier.
In February 2011, Science magazine had an entire issue devoted to data, and one of the articles focused on climate data and how they are revolutionizing our understanding of climate change—a point that will be woven into my talk throughout this presentation. We know that data are the lifeblood of our science, but we need to move from creating data to creating knowledge and discovering new frontiers.
In 2008, Wired magazine published a provocative article with the title, “The End of Theory,”11 which argued that the data deluge makes the traditional scientific method obsolete. I am not sure I agree with that assessment, but it was provocative, because it was essentially modeled after what Google has done in the search business, where all search results and page ranks are driven by data rather than an a priori model of how things should be organized and ranked. This was the original Yahoo model, but that became obsolete as a result of Google’s work.
I want to focus now on the multidisciplinary aspects of the geosciences. Whether we are talking about ozone depletion over the South Pole, mud slides in China, forest fires or deforestation of the Amazon, climate change in general, or the impact of El Niño or La Niña—to make advances in any of these areas—scientists have to utilize data from many geoscience disciplines, related social sciences information, and other ecological information. Francis Bretherton, one of the former directors of NCAR, created the famous earth systems science schema, which scientists now refer to as the “Bretherton Diagram.”
Dr. Bretherton got scientists to realize that they should not be looking at just atmospheric systems, hydrologic systems, or ocean systems. All of these different systems are interconnected, whether it is the cryosphere or the hydrosphere, or even ecology and chemistry, and looking at questions from a systems perspective is the key. However, to do systems science and research and get meaningful results, we need to integrate information across all these different systems as well as to synthesize them across the domains. One of the challenges for geoinformatics is providing the right type of data in the right format to the right application in the right place. One of the key aspects of that process is interoperability of data across all the different disciplines that are involved.
The founding president of UCAR and the first director of NCAR, Dr. Walter Orr Roberts, had a famous mantra: science in service of society. We should not stop at just doing science, but we should keep in mind that science has a meaningful impact on society, which means that we should be thinking about geoinformatics and data services that are end-to-end, that are chained through workflows. For example, scientists should not stop at the prediction part, which was the focus of the work that Dr. Graves presented, but they should go beyond that and couple the predictions with the flooding models, ecosystem models, and so on. If scientists are looking at the societal impact of flooding from tropical cyclones and hurricanes, they should integrate data from atmospheric sciences, oceanography, hydrology, geology and geography, and social sciences, and then use the results with decision support systems. In short, we need geoinformatics studies that encompass these end-to-end aspects.
The issues associated with the deluge of data came up earlier. I like the “sea of data” metaphor for reasons explained in a talk I heard by Tim Killeen, the current Assistant Director for Geosciences at the NSF. When we look at it as a sea of data as opposed to a deluge, we have two options: to drown, or to set sail to new land and make new discoveries. That is very appropriate, because data bring us to new discoveries in the world.
Speaking of a sea of data, here is some information about the growth of data that is expected in the future. Today we have less than 15 petabytes total of atmospheric science data, but that is predicted to grow to 350 petabytes by 2030, with the data coming largely from
__________________________
11 Available at http://www.wired.com/science/discoveries/magazine/16-07/pb_theory.
models, particularly those with high resolution, in ensemble mode, and integrated in time scales from days to 100-plus years. There will also be data coming in from the new generation of remote-sensing systems, including radars and satellites. Dr. Graves mentioned the traditional radar systems, but in Norman, Oklahoma, they are developing a new kind of radar that was previously deployed by the military 20–30 years ago. It is called phased array radar, and it can scan an entire thunderstorm in a matter of seconds, as opposed to the 8 to 10 minutes that it takes today to scan a thunderstorm with traditional radar. Furthermore, the new generation of geostationary weather satellites will have 1,600 channels in the vertical for sounding capability. Current-generation satellite gives 20 channels; so we are talking about an 80-fold increase in the number of channels that give us detailed information about the atmosphere.
The polar orbiting meteorological satellites will also provide large volumes of data once the Joint Polar Satellite System Program is finished. It has been delayed many years now because of various difficulties, but once it is deployed, 3 terabytes of data are expected to be produced every day.
With all these data being produced, how do we extract knowledge and make new discoveries? The field of knowledge extraction and data mining addresses the question—going from data to information to knowledge to wisdom—but the field is still very much in its infancy. Some great work is being done at the University of Alabama in Huntsville under Dr. Graves’s leadership, but our field is still mired in these simple-looking plots that produce spaghetti diagrams of ensemble model output or postage stamp–type maps. These may be of some value when there are only 10 ensemble members, but when there are 1,000 ensemble members, there is no meaningful way to understand which data are together, which are clustered, or which have similar patterns and relationships, so an automated capability to extract that knowledge and information is very much needed.
Every 5 years or so, the Intergovernmental Panel on Climate Change (IPCC) assessment reports are written. The group’s Fourth Assessment Report resulted in the Nobel Peace Prize being awarded to the IPCC, along with Al Gore, for work in bringing awareness about climate change issues. One of the simple actions that happened in the geoinformatics world was that the IPCC Fourth Assessment climate modelers were mandated to submit all of their model output using the NetCDF climate forecast convention to the Program for Climate Model Diagnostics and Intercomparison (PCMDI)–Earth System Grid (ESG) Data Portal. The result was approximately 600 publications within 6 months after the IPCC Fourth Assessment Report activity was completed.
All the model runs were submitted, and they were made available by the PCMDI-ESG Data Portal, so everyone could compare their models immediately because they all used exactly the same structure format standard and convention for their data. That mandate had a huge impact on producing new discoveries in a very short period of time.
The next assessment report is now being prepared and the model runs are getting started. The IPCC is looking at the total assimilation of 90,000 years of climate change model results, consisting of many petabytes of output from 60 experiments and 20 modeling centers around the world. They have been mandated to produce the data and output and to have all of the necessary metadata available for the model runs, which was not required for the last assessment. For the next assessment, all of the models need to document, in a consistent way, the metadata associated with how the model was configured, how it was run, and so forth, and
to make the data available through the Earth System Grid Portal, using the NetCDF-CF format. This again raises the question of how we are going to process all of the data
The other type of ensemble work that I did in my previous work was to make ensemble predictions for hurricanes and land-falling tropical cyclones. In this work, a scientist can create ensemble numbers by varying the initial conditions, boundary conditions, physics of the model, sea surface temperatures, and other criteria a scientist has for a prediction system from the external forcing standpoint. This particular system was generating 150 to 200 runs, considering the different permutations and combinations, and we needed an end-to-end efficient workflow system, as well as an informatics system for providing seamless access to all of the data and information that was created. Therefore, we need to be thinking about geoinformatics systems that are capable of facilitating these things and delivering information.
The hurricane research community has proposed a framework for improving hurricane predictions, and, as part of its metrics, the group is attempting to reduce both average track error and intensity error by 50 percent for a 1-to-5-day forecast. Given that the intensity prediction for tropical cyclones and hurricanes has not improved by even 10 percent in the last 20 years, this would be a gigantic leap, and they are expecting to do very high-resolution predictions as well as ensemble-mode predictions to make that possible. All of the data from this endeavor will be openly available. That was mandated by the authors of the 2008 “Proposed Framework for Addressing the National Hurricane Research and Forecast Improvement Initiatives,” because the group figured this is the only way it is going to realize these results and the metrics they are aiming for.
I also want to talk about data visualization as a way of enabling new insights and discoveries. I will focus on one of the software programs that Unidata has developed, the Integrated Data Viewer (IDV). I am not a geophysicist, but I know that the IDV was used by a group called Unavco, in the GEON (GEOscience Network) project, which was another large NSF Information Technology Research project under the Linked Environments for Atmospheric Discovery project (LEAD), described in the previous talk by Dr. Graves, and which got started about 8 years ago. Unavco adopted the Integrated Data Viewer as the framework for visualizing data from the solid earth community.
Figure 2-24 shows the IDV integrating geophysical data. We are looking at seismic plate activity along the west coast.

FIGURE 2-24 Integrated data viewer (IDV).
SOURCE: Unavco
At the bottom left are stress vectors and fluids, the mantle convection assimilation, and Mount St. Helen’s seismic activity underneath. The people who were doing this work told us that they could never see these aspects before this three-dimensional IDV visualization tool became available. It opened their eyes to how the Earth works underneath the surface through plate movements and mantle convection.
I also want to mention geographic information systems (GIS) integration, because I work in a field that is focused on geo-specialties. We need geo-specialty-enabled geoinformatics cyberinfrastructure so that we can integrate location-based information, because many of these processes and events are very much related to the geography. We should not be thinking of GIS as an afterthought. The historical way of thinking about GIS is that atmospheric scientists would do their work and then eventually somebody else would take over from there and put the information into some kind of a GIS framework. Unidata is working to enable NetCDF data models to interface directly with GIS capabilities through the Open Geospatial Consortium standards for Web Coverage Service, Web Features Service, and Web Map Service. As a result, when we have a NetCDF data cube, that data cube becomes available as a Web GIS service, and we can produce maps like the one to the right in Figure 2-24 from that particular capability.
I cannot conclude this talk without talking about data citation, because it has already come up in almost every presentation so far. To me this is the next frontier, I want to give an
anecdotal story to demonstrate that this is not a technical challenge, but a cultural and social challenge. This particular topic came up at the American Meteorological Society. The publication commissioner, who is in charge of all scientific publications of the American Meteorological Society, took a proposal modeled after a Royal Meteorological Society proposal, which was intended to encourage data transparency. He went into the meeting and said that the condition for publication should be that authors be required to agree to make data and software techniques available promptly to readers upon request and that the assets must be available to editors and pre-reviewers, if required, at the time of review. He was basically told that his proposal is a nonstarter, because we will never get the cooperation of the authors. This shows that we still have a lot of work to do.
I also want to talk about globally networked science. The IPCC activity is the gold standard for that, but I also want to mention one other activity that NCAR and Unidata have been involved with, which is the Observing System Research and Predictability Experiment (THORPEX), an interactive and global ensemble project. It is generating 500 gigabytes a day, and the archive is now up to almost a half a petabyte. It brings in data from 10 different modeling centers twice a day. We need to be able to create geoinformatics for these kinds of databases. These are some of the challenges. I want to come back to the point that was made earlier about interfacing social networking systems. As a geoscientist, I have seen GPS, coupled with mobile sensors, revolutionize the geosciences in a major way. I think there are some incredible opportunities, not just for citizens and science, but also for providing workforce development, geospatial awareness, and education for our students at all levels. This is very important, and how we use social networking tools like Facebook for getting the scientific community to provide commentaries, share information and data, and work in a collaborative way is something that is a real opportunity and definitely a challenge.
In closing, I would like to emphasize that we live in an exciting era in which advances in computing and communication technologies, coupled with a new generation of geoinformatics, are accelerating scientific research, creating new knowledge, and leading to new discoveries at an unprecedented rate.
DISCUSSANT: Mr. Dudley, in your chart of supporters, I see some National Institutes of Health (NIH) institutes, but none from the National Science Foundation (NSF). However, you were working at the intersection of what NIH and NSF medicine would fund. I find that curious. When I was at NSF 10 years ago, I discussed with NIH colleagues the cultural differences, such as how NSF will fund soft money research and how NSF likes proposals to be more exploratory, while NIH likes them to be more incremental. We did not come to an agreement, though. There was a division director at NSF who liked to encourage joint work between computer science and medicine. Now that you are doing this kind of work, what organizational steps would you take to help in that area?
MR. DUDLEY: Our funding strategy has always been to involve the computer scientists in whatever we are doing. One way we are trying to solve it at Stanford University is by trying to mix the computer scientists and the biologists more, letting the computer science experts figure out what the important problems are in medicine and how they can engage in those problems, especially for the algorithmic issues. We have been struggling with this problem for a long time, and we have just been pragmatic about it and have gone where we can get the money most easily.
DISCUSSANT: If I am collaborating with the doctor, he writes the NIH proposals and hires my students. Who writes the NSF proposals?
MR. DUDLEY: At Stanford, we now have a lot of rebranding. For example, computer science people are rebranding themselves as bioinformatics experts, because they need funding. Therefore, instead of trying to deal with the problem, they rebranded themselves and went to NIH.
DISCUSSANT: As a lawyer, I sometimes think of myself as a social engineer. Most speakers have described a series of social engineering problems, such as institutional culture affecting funding and institutional culture within the disciplines, so it is a big problem. Imagine this scenario: I am at a midtier institution, and what the presentations showed me is that I am the “sharecropper.” You have got both the brain power and the computing power to utilize all the data, process them, and do different kinds of work with them that I will never be able to do. Therefore, my only competitive advantage against you for funding is to hide the data. How do you respond to that? If you are going to break the logjam, what are the incentives for researchers to share their data with you?
MR. DUDLEY: You are exactly right about that, and that was the point I was trying to make when I said there are computational elites. It should not be that way, however. I think that if we can get some good tools and the cloud, we can level the playing field for costs and accessibility for many researchers at midtier institutions. In fact, we wrote a grant for the NIH to build such a system, which was rejected.
DR. FRIEND: I think the point you made is important. I want to separate researchers into data gatherers and data analyzers. Right now the way it works is that the person who collects the data expects to have the right to take it all the way through to the insight, and then the next person should go back and do it again. That system is wrong. It is just inefficient. I think that having core data zones where people who did not generate the data can mine the data and work with them is important enough that funding institutions and journal publishers need
to determine a way to make it happen. A second issue is the matter of humility. It is very hard to get the “Titans on Olympus” to share their data even among themselves.
DR. HEY: We work with some members of the HIV committee. The moment they send us their data, we send them the results with some tools that we developed. There is no real reason we could not make those tools available in the cloud, and we are planning to do that. However, I do think the cloud can empower scientists in certain cases. We are also working with the Ameriflux community. They have about 150 sensor towers and fields all over America. Typically they are owned by a principal investigator, and they take data and publish it with their graduate and postdoctoral students, so it is generally one professor and two students. By putting the data into a central data server, like the SkyServer, the community is gradually realizing that there is much more value to be gained by publishing not just one tower’s data but several. That is a sociocultural change, because instead of one professor and two students we now can have 10 professors and 20 students, which makes it a 30-author paper. For that particular community, this is a big change in culture, and it takes time.
DR. GOODMAN: In astronomy, the situation is very different, and those of us who like the astroinformatics approach are lucky if we have tenure, because people still do not appreciate that kind of approach. Getting the last three photons out of a Keck telescope is what gets someone the “Olympian” status these days. That is changing slowly, but it is a big battle.
MR. UHLIR: I would like to follow up with a comment and a question. The comment has to do with the issue of incentives. Many people talk about the importance of citation of data and the role of that in giving people incentives to share data, through recognition and rewards, and changing the sociology of how people receive their data and share them. The Board on Research Data and Information is about to start a 2-year project in collaboration with some other groups, in particular, the international Committee on Data for Science and Technology (CODATA), on data attributions and citations. We will start with a symposium in Berkeley, California, in August 2011.
The other issue, which most of you have largely avoided, is the human infrastructure element. Obviously this symposium is about automated knowledge discoveries, so you have appropriately focused on the technical infrastructure and automated knowledge tools, but there seem to be two schools of thought about how this is going to be managed and promoted in the future. One is that the technology will disintermediate many professionals in libraries of science or in information management, because everything is going to be automated and work perfectly, and we will live in a wonderful automated information age. The other school of thought is that the deluge of data will require not only more people to manage it, but also a retraining of the information managers and reorientation of the traditional library and information schools. So my question is, which school of thought is correct? Or perhaps they both are. Also, what do the speakers feel needs to be done to develop this kind of new expertise, because a lot of it seems to be done by people mostly in the middle of their careers? Is there a new approach to education—higher education and training at the outset of careers—that is being implemented, and if not, what would you suggest?
DR. GRAVES: There are new programs starting in data science and informatics. We are initiating one at the University of Alabama in Huntsville. I know Rensselaer Polytechnic Institute (RPI) is also starting some programs, as well as several other universities, and they are attempting to be interdisciplinary.
DR. RAMAMURTHY: The creation of Unidata was intended to address the concern that you just expressed. Atmospheric sciences has been at the forefront of data sharing and of bringing data into the classroom and giving students—at all levels, whether it is an introductory atmospheric science class or master’s and Ph.D. students—access to the same kinds of data and tools that are used by professionals in their operational forecasting or in research. Having that facility that has been sustained for 26 years has made the issue of access to atmospheric science information and data systems almost a nonissue from a workforce development, because literally hundreds and thousands of students go through their labs and classrooms and use state-of-the-art software and data technologies in their daily work. The key is having that kind of a sustained infrastructure that will provide the services that are needed for training the next generation of students and for creating a workforce that will be very skilled in dealing with geoinformatics of the future.
DR. CONTI: There is a need for a paradigm shift in astronomy. The vision that the younger generations bring to this area is very refreshing. They suggest that we can solve this problem from a completely different point of view. At the same time, many of them bring very little knowledge of astronomy combined with an extremely high knowledge of computer science, and those have the ability to solve problems much more quickly and therefore to ask many more questions. Therefore, in our programs, we try to find a good balance between people being trained astronomers, but also having a focus on computer science as well, because the future is going to depend on how they are going to manage data and how they can ask new questions that were not asked before.
The other issue is that once we have all these data, our ability to mine them is extremely important in the sense that we need to be able to understand how the data can be manipulated. Therefore, it is important to change the curriculum to reflect the fact that computer science has to be part of our daily life. We find that younger students are used to managing and producing astronomy and looking at astronomy through the Worldwide Telescope. They do not know the inner working or what it takes to produce plenty of data, but they know what they want to do. They know how to discover new pathways through the data. There is a lot of resistance among senior researchers, but perhaps we just need to learn to say, “I do not really understand how you get to this, but show me how, because we can open a new frontier.”
DR. FRIEND: I want to make three points. The first concerns what we have found to be necessary to bridge the gap between the data mining and the understanding of biology. It is very rare that a single individual does both, and so we split it into two parts. We have people that we call network biologists, who may be mathematical physicists, coming toward the models from one direction, and then we have systems biologists, who understand enough of the math and understand the biology. The people who can do that sort of polymath linkage are rare, and so the education and career paths are split in a way that is similar to what happened previously when someone chose to become either a molecular biologist or an enzymologist. It is important to not expect someone to bridge the entire gap.
The second point is that Dr. Eric Schadt, who drove much of this work, and several others have just started a new journal. The journal will have a section that is just for publishing models that are not validated. A validation often comes later, and the point is to make the model available so that people can work on it.
The third issue relates to the discussion about structuring rewards. In medicine, we are convinced that the patients and citizens themselves must break the logjam between the scientists who do not want to provide the data and those who are trying to get access to large amounts of data. If there are electronically generated data from a patient, the patient can get them back. This is going to be a very fundamental change—shifts in how data are going to flow. The development of the law and the role of the citizen are going to be very important in getting the data flowing, because academic institutions are not necessarily going to do it on their own.
DR. HEY: I have three brief comments. First, I do think that research libraries are in danger of being disintermediated. Librarians are caught in a difficult trap. Subscription costs are clearly a crisis, because the budgets are not increasing at the same rate. There is an interesting move toward iSchools, but the question remains: What should they train the next generation of librarians to do?
Second, training will be important in changing the culture of science. We should work with graduate students in each science, because it is clear we are not going to convince the faculty. I think we should train people to have at least depth in a couple of disciplines. Universities change very slowly, however, so my worry is that we will produce bright graduate students with nowhere to go. I think that is a real issue, although some universities are doing things better.
Third, there are three communities: the scientists doing the real research, the computer scientists who know the technologies, but we also need information technology experts, who can turn a research prototype into a tool that people can actually use.
DISUSSANT: I see the success in bioinformatics and geoinformatics as disciplines, and I see communities of scientists working in multidisciplinary ways with other communities, such as statisticians and computer scientists, but we do not see that much in astronomy. One difference is that there was a tipping point reached—certainly this was true in the medical community—that so many papers are being published that it is impossible for anybody to keep up with what is happening. You therefore need some kind of a summarization or aggregation and an informatics approach to give you the high-level picture. I like the historical example at the University of California, Berkeley, where you can move from a higher-level view down to the exact specific event. Imagine doing the same thing with our databases.
Maybe in astronomy we have not quite reached that tipping point. We have learned with the virtual observatory that if it is built, users will not necessarily come, and so many astronomers are not using this infrastructure. Some are using it, but not as many as we would like.
Then, as I was listening to this education discussion, I thought that perhaps that tipping point could be completely different from the one I was thinking. It would be more the education system that is the tipping point, with the younger people who are familiar with social networking and computer-based tools, whether we call it Web 2.0 or Science 2.0. I would like to call it citizen science, that is, getting people in touch with the science and them being inspired to do science, knowing that they approach it with either the skill sets in using these tools or at least an interest in learning to work with them, because that is the way their lives are, and now they discover they can do science with it.
My university, George Mason University, is one of those places, like RPI and a few others, that are offering this sort of informatics space education. We have found that it is hard to draw the students into the program, especially the undergraduates. Our graduate students already understand it, so the graduate program has a good number of students, but our undergraduates still do not. We talk to students and ask what are they interested in, and they say, “We like biology, but I want to do something with computers.” When we finally get these students into our classes, their eyes open up and they say, “I did not know I could do science with computers.” So, in a sense, the younger generation is reaching that tipping point. They are discovering that they can do what they enjoy doing, such as social networking and online experiences, and do science at the same time.
What I would like to see as a goal is not only doing this kind of work at the graduate level, but moving it to the undergraduate level, as we are doing now, and then moving it even further down to the high school students. I would like to teach data mining to school children. It would not be so much the algorithms of data mining, but it would be evidence-based reasoning and evidence-based discovery—basically detective stories—and in this way we could inspire a generation of new scientists just by using a data-oriented approach to the way we teach these topics.
DR. GOODMAN: Perhaps the core or at least part of the problem is the scientific paper as a unit of publication. I will give an example. Seven first-year graduate students in our department started on their own project called Astro Bites. They go to AstroPH, which is the archive server for astronomy, select the articles that they like, and review them. This is now being read all over the world. It is searchable by any search engine, and they can discover useful materials there. This is kind of the junior version of a blog called AstroBetter, which is produced worldwide by astronomers who are of the mindset that the last discussant was talking about—a kind of astroinformatics community that is evolving in a mostly 30-something world. Those are the same people who would like to take their model or dataset and make it available for people to experiment with. Right now there is an interesting concept of the data paper in astronomy. I did not even know what this was, but fortunately when we finished a very big survey about 5 or 6 years ago, I had a postdoctoral student who said, “We have to publish a data paper now,” and I said, “What is a data paper?” It is basically a vacuous paper that says, “Here are my data. Cite me.” She has gotten hundreds or thousands of citations because of that data paper. This should not be this way.
Going back to Mr. Uhlir’s point about people and training, there is a whole new system that is emerging. I do not know exactly what it looks like, but it does not involve 12-page papers as the currency of everything, and it does not involve even looking at those papers to find the data. Such papers are just one of many resources. How we develop the resources by all these other new people, I do not know.
DISCUSSANT: Part of what we are seeing is the realization that a citation count has become an outmoded way of thinking, and yet it is still the H index that gets people into the academy. Take, for example, Danah Boyd. She has 50,000 Twitter followers. She is not too worried about her citation counts—she does fine in her papers. But, again, what does it mean and why does that work?
DISCUSSANT: The right solution is for those of us who are thinking about this area to engage in that conversation, to look for ways to balance the traditional measures with some
new kinds of metrics. We should look for impact metrics, as well as for sustaining papers. People should still write papers, but we need to get a better mix that looks across a much broader range.
DR. GOODMAN: Where does the data scientist, who maybe does not typically get included in a publication, come in?
DR. CONTI: Most of the time we are penalized for being a data scientist. Perhaps one scientist does not have as many citations as some others, and does not care to publish that many data papers; as a result, the tenure-track path is harder.
DISCUSSANT: One key is that those of us who are involved in other people’s tenure decisions have to rethink what we examine, and not get the junior faculty to rethink what they are publishing.
DR. FRIEND: Some people who have been looking at this have noted that the music industry is fairly advanced in figuring out microattribution and that there are ways to borrow from that experience, by looking at impacts without using citations. We should think about using tools that others have developed to determine impact factor.
DISCUSSANT: I work for the National Ecological Observatory Network, or NEON. It is funded by the NSF to build a network of 60 sites across the United States to collect data on the effects of climate change and land use on the ecosystem. We will be measuring something like 500-plus variables, and we will be delivering these measurements freely to the community. The measurements range from precipitation and soil pH all the way to things like productivity. We have talked about data-intensive science, of bringing a new era of how scientists can work, but what about data-intensive science in forming our policy and in forming how we design our cities, for example? From NEON’s perspective, we will be one of the many data providers, along with the National Aeronautics and Space Administration (NASA), the National Oceanic and Atmospheric Administration (NOAA), and other institutions that will be providing similar data, but somebody else has to do the aggregation of those data into knowledge. Those people will need tools, so who is going to build the tools? And will these tools be used by federal agencies, resource management makers, and policy makers to craft how they will plan the nation’s national resources?
DR. HEY: We have a couple of projects that show there is some hope. One is a project called Healthy Waterways in Brisbane. They had a big drought, now they have too much water, but it is relevant in either case. They have a system for doing ecology reports on their water supplies, and it took several people several days to get a report written and distributed, which is released every week. By putting in some serious information technology, but nothing research grade, one person can now do it in 1 hour or so, and that person can get much more flow of information, so ecologists have a much better evidence base with which to do emergency measures, for example. The other project is an urbanization project in China where they are building 10-million-people cities. They will need to worry about all the environmental impact issues, as well as designing the city. I think there is hope, but I agree it is a challenge. I think that more case studies are needed.
MR. DUDLEY: Arizona State University has a building called Decision Theater. I do not know how successful it has been, but this building has Liquid Crystal Displays around the wall, 360 degrees, and it was built specifically for policy makers to develop evidence-based
views of policy making. I heard rumors that the state legislature wants to defund the entire Arizona State University, so maybe it was not too successful, but it is something to look into.
DR. RAMAMURTHY: As atmospheric scientists, geoscientists, and geoinformatics people, almost all of the data we produce has societal relevance. It is important not to stop with scientific informatics, but to try to determine a way to interface it with decision-support systems and other kinds of policy tools. In the climate change arena, everyone is focusing on mitigation and adaptation, because it is not just whether the globe is going to be two or four degrees warmer in the next 50 or 100 years, but what it means for the watershed, for agriculture, for urbanization, for coastal communities, and so forth. It is absolutely critical that the scientific information systems and geoinformatics systems can interface with the other tools and other systems. We do not build those systems in my program, but we do think about interfacing with geographic information systems, such as what needs to be extracted from climate-change models to help people talk about what the agricultural picture is going to look like 50 years from now, whether for growing wheat or barley or something else.
DR. GOODMAN: I have a question for the people from the funding agencies. Dr. Hey said “some serious information technology, but nothing research grade.” This is a huge problem. Here is a true story. At a meeting I had yesterday we had an undergraduate researcher who works with a postdoctoral student on a project, who said, “I do not know where we are going to put these data, because we need a couple of terabytes, and the computation facility has a lot of security systems, so they will not let us run the right software,” and so on. I knew that my assistant had some extra money from the division and that she bought a stack of 40 boxes of terabyte drives, so I gave a terabyte drive to each of the researchers, and I solved the problem. The point is that there was an obstacle and that there was no person whose job it was to facilitate that and to make the data usable and streamlined for the future. So my question is, How are we going to pay those people in the future? Where are we going to get those people?
DR. BERMAN: If we think about the impact of digital information—what we need to store, how long we need to store it, how we need to curate it, and how we need to respond to data management policies—the university libraries are reconceptualizing themselves and can step into that role, but they need some help. So the question is, How can we jumpstart the system, so that when universities are thinking about infrastructure costs, they consider the data bill along with the water bill, the liability insurance, and so on?
This page intentionally left blank.


















































