“In most cases, the most complex part of developing a therapeutic is to design its appropriate delivery system.”
J. Rubén Morones-Ramírez
“You can kill any bacterial cell on the planet with any antibiotic on the planet, but the real question is whether you can kill any bacteria on the planet with any antibiotic on the planet without killing the patient.”
Mark Smeltzer
In the final session of the workshop, two speakers discussed some additional challenges facing the antibiotics discovery enterprise and possible approaches for addressing those challenges. J. Rubén Morones-Ramírez, Professor of Chemistry at the Universidad Autónoma de Nuevo León, spoke about some of the different tacks that he and his colleagues have taken to design and deliver novel antimicrobial therapies. Mark Smeltzer, Professor of Microbiology and Immunology at the University of Arkansas for Medical Sciences, addressed the issue of biofilms and described methods for getting antibiotics past these physical barriers.
NOVEL APPROACHES TO ANTIMICROBIAL THERAPEUTICS DESIGN
The first approach that J. Rubén Morones-Ramírez discussed in his presentation involved the use of synergistic antibiotic cocktails that include nanoparticulate silver as an active component. Silver has long been known to have broad-spectrum antimicrobial properties, but there was little mechanistic understanding of this property. Using transmission electron microscopy, Morones-Ramírez was able to show that silver nanoparticles bind to the bacterial cell membrane, inducing significant morphological changes. Binding was largely to the sulfur and phosphate groups on the cell membrane glycans and it interfered with the bacterial respiratory system. Further study with nanoparticles of different size showed that only nanoparticles in the 4- to 5-nanometer diameter range interacted strongly with the bacterial membrane, which he pointed out correlates with the fact that the larger surface-to-volume ratio of smaller nanoparticles makes them more active catalytically.
Using energy-dispersive x-ray spectroscopy, Morones-Ramírez and his collaborators were able to identify silver nanoparticles inside the bacteria, demonstrating that they are able to not only interact with the cell membrane, but penetrate it as well. They also found that nanoparticles were able to interact with bacteriophage that happened to be infecting some of the bacteria. This prompted a study looking at the effects of silver nanoparticles on HIV infection that showed that silver nanoparticles were able to bind to the gp120 protein on the HIV capsid (Figure 4-1) and thereby interfere with the entry mechanism that the virus uses to enter cells.
Wishing to pursue the mechanistic aspects of the antimicrobial properties of silver nanoparticles, Morones-Ramírez employed some of the tools of systems biology. At a blood concentration of 30 micromolar, which preliminary studies showed was nontoxic to mice, silver nanoparticles were able to reduce gram-negative bacterial load by about 3,000-fold. Metabolic network analysis using microarray expression data showed that nanoparticulate silver was upregulating iron metabolism, triggering a membrane stress response and disrupting metabolic regulation. Followup gene knockout studies supports the hypothesis that silver nanoparticles disrupt the integrity of the outer and inner membrane of gram-negative bacteria, triggering misregulation in the Krebs (or TCA) cycle and electron transport chain and the breakdown of iron-sulfur clusters, producing an increase in reactive oxygen species that ultimately causes cell death .
From this mechanistic understanding came the prediction that since many antibiotics have at least some effect on the intracellular concentration of reactive oxygen species, it may be possible to potentiate that effect with agents that could shift the intracellular oxidation state in bacteria. In fact, adding silver increased the antimicrobial effect of a wide range of antibiotics and even rendered active antibiotics that were normally ineffective against gram-negative bacteria. In addition, Morones-Ramírez and his collaborators were able to take advantage of the fact that nanoparticulate silver impacts the permeability of the gram-negative bacterial
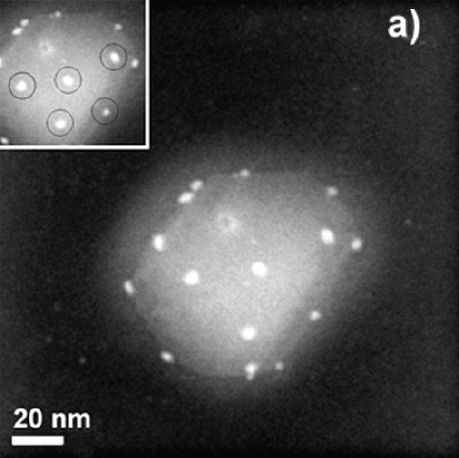
FIGURE 4-1 The interaction of silver nanoparticles (bright white spots) with HIV-1.
SOURCE: Elechiguerra (2005).
Morones-Ramírez then discussed the microbial competition project his group is conducting in an attempt to harness microbial ecosystems as sources of novel antimicrobial agents. One set of experiments in which the cocultured E. coli and Candida albicans showed that E. coli produces a novel antifungal agent that enables it to have a growth advantage over Candida. Though efforts to isolate and identify this antifungal compound have eluded him so far, he believes that the concept of co-culturing microorganisms to look for new antimicrobial compounds is sound and he plans to pursue this idea to search for antibiotics that would be effective against resistant strains.
Noting that the most difficult part of a drug discovery effort often involves solving a drug delivery problem, Morones-Ramírez concluded his talk with a description of the antibiotic delivery system his group is developing using biocompatible microbial exopolymers. When stressed by exposure to toxic metals, some microorganisms respond by producing a protective polymer coating. Working with a group at the University of New Orleans, Morones-Ramírez and his team have used these exopolymers as stabilizers to control silver nanoparticle production and create a polymer-encapsulated nanoparticle assembly that exhibits potent antimicrobial properties. He also briefly described work just beginning in his laboratory on a light-activated?1 polymeric nanoparticle delivery system for some infections.
OVERCOMING ANTIBIOTIC CHALLENGES IN BIOFILM-ASSOCIATED INFECTIONS
In the workshop’s final presentation, Mark Smeltzer discussed his group’s efforts to develop antibiotics to treat chronic osteomyelitis, a limb-threatening infection associated with biofilm formation that can develop after bone injuries and joint replacement surgery. He explained that he first became interested in this problem when a patient with chronic osteomyelitis who was about to undergo his sixth revision surgery demanded amputation instead. What struck Smeltzer about this patient’s infection was that it was caused by a Staphylococcus aureus isolate that was only resistant to penicillin and was not MRSA, yet it was still persisting in the face of repeated local treatments with antibiotic-eluting polymer beads packed into the wound after each surgery.
Osteomyelitis is a good example of the broader issue of biofilm formation as a mechanism of antibiotic resistance. There are many additional examples of biofilm-related infections; however, this discussion focuses on the Staphylococcus aureus biofilm as an exemplar.
As a bacteriologist who specialized in Staphylococcus aureus, Smeltzer was well aware of the ability of this bacterium to form a biofilm, which he characterized as multiple layers of bacteria growing within a glycocalyx, a polysaccharide mixture secreted by the growing bacteria. The effect of the biofilm, he explained, is that it reduces antibiotic delivery to the underlying organisms and creates an intrinsically resistance phenotype without other genetic or biochemical changes, in large part because organisms growing within a biofilm are metabolically inert. To illustrate this point, he discussed the results of an experiment in which a catheter colonized with biofilm-embedded Staphylococcus aureus was exposed to vancomycin, linezolid, and daptomycin, at concentrations 5, 10, and 20 times the breakpoint minimum inhibitory concentration of each antibiotic. “After three days of direct exposure, changing the antibiotic every day, we still had not cleared the catheter. That’s the problem with a biofilm,” he explained (Figure 4-2).
One approach to addressing this problem is to improve local antibiotic delivery. Current practice is to mix daptomycin with poly(methyl methacrylate) (PMMA), also known in
_________________
1 Light-activated polymer delivery systems have been use for topical and subsurface infections, as well as in surgery.
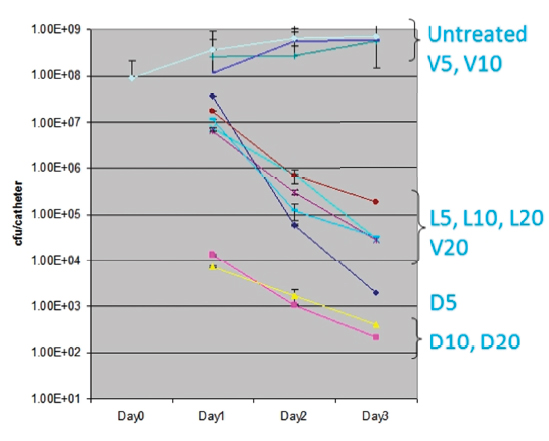
FIGURE 4-2 The intrinsic resistance of Staphylococcus aureus biofilms. D, L, and V refer to daptomycin, linezolid, and vancomycin, respectively, at 5, 10, and 20 times the minimum inhibitory concentration.
SOURCE: Weiss (2009a).
the biomedical community as bone cement. Bone cement, as its name implies, is not porous and there is little diffusion of antibiotics out of this material. Nonetheless, orthopedic surgeons routinely load some amount of daptomycin in pellets made from this material in the operating room and implant the pellets into the wound. Smeltzer noted that if the amount of daptomycin that his studies show are needed to be effective, it would require almost $20,000 worth of antibiotic alone. As a solution, his team added the polysaccharide xylitol and daptomycin to PMMA, producing a porous structure. This porous structure released levels of daptomycin that remained 10 times higher than the breakpoint minimum inhibitory concentration for over a week. Tests in animals with osteomyelitis showed that the xylitol plus daptomycin combination was highly effective at eliminating the Staphylococcus aureus infection.
Another approach that Smeltzer and collaborator Paul Dunman at the University of Rochester have taken is to develop new antibiotics with a focus on efficacy in biofilm-associated infections. The target of this effort is ribonuclease RnpA, which has low homology to mammalian proteins, is conserved across bacterial pathogens, and is involved in two essential cellular processes. Conventional screening identified a number of hits, and these were then tested against Staphylococcus aureus biofilms. Two compounds from this second screen showed significant activity, though these compounds were associated with unacceptable cytotoxicity in a human hepatic cell line assay. Nonetheless, these compounds may serve as the starting point for further medicinal chemistry derivatization.
In the final portion of his presentation, Smeltzer discussed studies aimed at finding targets that would impact biofilm formation, which would eliminate some of the issues with treating biofilm-based infections in the first place. He noted that there is a substantial body of literature on this subject, but much of it is contradictory. His group chose to focus on the transcriptional regulator SarA and was able to show that SarA-negative Staphylococcus aureus mutants do not form biofilms. Further study showed that protease production rose substantially in this SarA-negative mutant and that some combination of four extracellular proteases were inhibiting biofilm formation by the mutant.
These findings raise the possibility of treating osteomyelitis with a combination of an antibiotic and a suitable protease, but delivery becomes the problem. “If you can’t get vancomycin out of bone cement, you’re not going to get a protease out either,” explained Smeltzer. He noted that his group plans on exploring various biocompatible and biodegradable polymers, such as chitosan, as delivery vehicles that could be formed into implantable pellets and used to release proteases locally at the site of infection. Alternatively, it may be possible to identify a small molecule that would repress SarA activity and increase expression of protease genes by the bacteria themselves, which in essence is similar to the approach taken to develop inhibitors of β-lactamases to restore antibiotic susceptibility. So far, Smeltzer’s group has identified several hits and has shown that one of them has a marked effect on SarA expression and biofilm formation in two different strains of Staphylococcus aureus isolated from patients with osteomyelitis.
In response to a question about the toxicity of silver nanoparticles, Morones-Ramírez said that silver is toxic to mammals, but at much higher concentrations than for bacteria. He noted that mammalian cells appear to have a mechanism absent from bacteria for eliminating silver at the concentrations that are toxic to bacterial cells.
Smeltzer said in response to a question about biofilms in conditions other than osteomyelitis that there are bead-based delivery options that would be applicable to soft tissue infections, though probably not for the lungs in the case of tuberculosis. He also acknowledged that the SarA inhibitors are likely to be specific for Staphylococcus aureus infections, but that most biofilm-forming organisms would have some other molecular Achilles heel that could be attacked. He added that his group is exploring a nanoparticle delivery system that could be irradiated with a laser to also deliver heat to the site of infection, which could amplify the effects of antibiotic therapy.
Beenken, K. E., L. Bradney, W. Bellamy, R. A. Skinner, S. G. McLaren, M. J. Gruenwald, H. J. Spencer, J. K. Smith, W. O. Haggard, and M. S. Smeltzer. 2012. Use of xylitol to enhance the therapeutic efficacy of polymethylmethacrylate-based antibiotic therapy in treatment of chronic osteomyelitis. Antimicrobial Agents and Chemotherapy 56(11):5838-5844.
Elechiguerra, J. L., J. L. Burt, J. R. Morones, A. Camacho-Bragado, X. Gao, H. H. Lara, and M. J. Yachman. 2005. Interaction of silver nanoparticles with HIV-1. Journal of Nanobiotechnology 3:6.
Morones-Ramirez, J. R., J. A. Winkler, C. S. Spina, and J. J. Collins. 2013. Silver enhances antibiotic activity against Gram-negative bacteria. Science Translational Medicine 5(190):190ra81.
Morones, J.R., J. L. Elechiguerra, A. Camacho, K. Holt, J. B. Kouri, J. T. Ramírez, and M. J. Yacaman. 2005. The bactericidal effect of silver nanoparticles. Nanotechnology 16(10):2346-2353.
Olson, P. D., L. J. Kuechenmeister, K. L. Anderson, S. Daily, K. E. Beenken, C. M. Roux, M. L. Reniere, T. L. Lewis, W. J. Weiss, M. Pulse, P. Nguyen, J. W. Simecka, J. M. Morrison, K. Sayood, O. A. Asojo, M. S. Smeltzer, E. P. Skaar, and P. M. Dunman. 2011. Small molecule inhibitors of Staphylococcus aureus RnpA alter cellular mRNA turnover, exhibit antimicrobial activity, and attenuate pathogenesis. PLoS Pathogens 7(2):e1001287.
Tsang, L. H., J. E. Cassat, L. N. Shaw, K. E. Beenken, and M. S. Smeltzer. 2008. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS One 3(10):e3361.
Weiss, E. C., H. J. Spencer, S. J. Daily, B. D. Weiss, and M. S. Smeltzer. 2009. Impact of sarA on antibiotic susceptibility of Staphylococcus aureus in a catheter-associated in vitro model of biofilm formation. Antimicrobial Agents and Chemotherapy 53(6):2475-2482.
Weiss, E. C., A. Zielinska, K. E. Beenken, H. J. Spencer, S. J. Daily, and M. S. Smeltzer. 2009a. Impact of sarA on daptomycin susceptibility of Staphylococcus aureus biofilms in vivo. Antimicrobial Agents and Chemotherapy 53(10):4096-4102.
Zharov, V. P., K. E. Mercer, E. N. Galitovskaya, and M. S. Smeltzer. 2006. Photothermal nanotherapeutics and nanodiagnostics for selective killing of bacteria targeted with gold nanoparticles. Biophysical Journal 90(2):619-627.




