EMERGING VIRAL DISEASES—THE ONE HEALTH CONNECTION
Viruses have caused some of the most dramatic and deadly disease pandemics in human history. Before it was declared to be eradicated in 1980, smallpox, a highly contagious human disease caused by the Variola virus, killed 300 to 500 million people worldwide in the 20th century alone (Koplow, 2003). The 1918–1919 “Spanish flu” pandemic infected roughly one-third of the world’s human population and caused an estimated 50 to 100 million deaths. In the past half century, deadly disease outbreaks caused by novel viruses of animal origin—Nipah virus in Malaysia, Hendra virus in Australia, hantavirus in the United States, Ebola virus in Africa, along with HIV (human immunodeficiency virus), several influenza subtypes, and the SARS (severe acute respiratory syndrome) and MERS (Middle East respiratory syndrome) coronaviruses—have underscored the urgency of understanding factors influencing viral disease emergence and spread.
The world’s current leading infectious killer, HIV, has caused an estimated 36 million deaths since the first cases were reported in 1981. In 2012, more than 2 million people were newly infected with the virus, and 1.6 million died of HIV/AIDS. In 2009, a novel swine-origin H1N1 strain of influenza A rapidly spread to over 213 countries in the first declared pandemic of the 21st century. And now, on
_______________
1 The planning committee’s role was limited to planning the workshop, and the workshop summary has been prepared by the workshop rapporteurs (with the assistance of Rebekah Hutton, Katherine McClure, and Priyanka Nalamada) as a factual summary of what occurred at the workshop. Statements, recommendations, and opinions expressed are those of individual presenters and participants and are not necessarily endorsed or verified by the Forum, the Institute of Medicine, or the National Research Council, and they should not be construed as reflecting any group consensus.
August 8, 2014, the World Health Organization (WHO) Director-General Margaret Chan declared the Ebola outbreak in West Africa a “public health emergency of international concern,” triggering powers under the 2005 International Health Regulations (IHR). The IHR require countries to develop national preparedness capacities, including the duty to report internationally significant events, conduct surveillance, and exercise public health powers, while balancing human rights and international trade.
Emerging infectious diseases (EIDs) were both anticipated and studied by the late Joshua Lederberg, Nobel laureate and a founder of the Forum on Microbial Threats. He recognized microbes as humanity’s competitors and appreciated their disregard for human sovereignty over Earth’s creatures (Lederberg, 2000). The same wisdom, plus a dose of reality delivered by SARS and avian influenza A (H5N1), informed the 2005 revisions to the IHR. The IHR are legally binding regulations (forming international law) that aim to (1) assist countries to work together to save lives and livelihoods endangered by the spread of diseases and other health risks, and (2) avoid unnecessary interference with international trade and travel.
The purpose and scope of the IHR 2005 are to prevent, protect against, control, and provide a public health response to the international spread of disease in ways that are commensurate with and restricted to public health risks, and which avoid unnecessary interference with international traffic and trade (Art. 2, IHR 2005).
Statement of Task
Over the course of more than two decades, beginning with the landmark report Microbial Threats to Health in the United States (IOM, 1992), the Forum and its predecessors within the Institute of Medicine have examined the growing body of research on EIDs and the growing list of diseases that fit this description (IOM, 2003).2
In this tradition, on March 18 and 19, 2014, the Forum hosted a public workshop in Washington, DC, to examine factors driving the appearance, establishment, and spread of emerging, reemerging, and novel viral diseases; the global health and economic impacts of recently emerging and novel viral diseases in humans; and the scientific and policy approaches to improving domestic and international capacity to detect and respond to global outbreaks of infectious disease.
Organization of the Workshop Summary
This workshop summary was prepared by the rapporteurs for the Forum’s members and includes a collection of individually authored papers and
_______________
2 See http://www.iom.edu/Reports.aspx?Activity={C8EA50BF-D234-4E44-9E42-9636B7FC2D22} for a complete list of Forum workshop summary reports.
commentary. The contents of the unattributed sections of this summary report provide a context for the reader to appreciate the presentations and discussions that occurred over the 2 days of this workshop.
The summary is organized into sections as a topic-by-topic description of the presentations and discussions that took place at the workshop. Its purpose is to present information from relevant experience, to delineate a range of pivotal issues and their respective challenges, and to offer differing perspectives on the topic as discussed and described by the workshop participants. Manuscripts and reprinted articles submitted by workshop participants may be found, in alphabetical order by participant, in Appendix A.
Although this workshop summary provides a description of the individual presentations, it also reflects an important aspect of the Forum’s philosophy. The workshop functions as a dialogue among representatives from different sectors and allows them to present their views about which areas, in their opinion, merit further study. This report only summarizes the statements of participants over the course of the workshop. This summary is not intended to be an exhaustive exploration of the subject matter, nor does it represent the findings, conclusions, or recommendations of a consensus committee process.
IMPACT OF EMERGING VIRAL DISEASES
In addition to causing nearly one in five human deaths worldwide, infectious diseases impose a heavy societal and economic burden on individuals, families, communities, and countries (Lozano et al., 2012; Murray et al., 2012). The appearance of new infectious diseases has been recognized for millennia, well before microbes were recognized as their causes (Morens and Fauci, 2013). EIDs comprise a substantial fraction of important human infections, and they have caused the deadliest pandemics in recorded human history, including the 14th-century Black Death (during which 75 to 200 million people in what is now Europe died of bubonic or pneumonic plague); the 1918–1919 Spanish influenza pandemic (at least 50 to 100 million deaths in a span of 18 months); and the ongoing HIV/AIDS pandemic, in which more than 35 million people have perished (Morens and Fauci, 2013).
Jones and coworkers described the emergence of 335 infectious diseases in the global human population between 1940 and 2004, of which nearly two-thirds originated in wildlife (Jones et al., 2008). These can be further characterized as either newly emerging or reemerging infectious diseases, that is, caused by pathogens infecting a new host species, or caused by pathogens that historically have infected the same host species, but continue to appear in new locations or in drug-resistant forms, or that reappear after apparent control or elimination (Fauci and Morens, 2012). Figure WO-1 illustrates the global distribution of key emerging and reemerging diseases including the anthrax-laced letters of fall 2001.
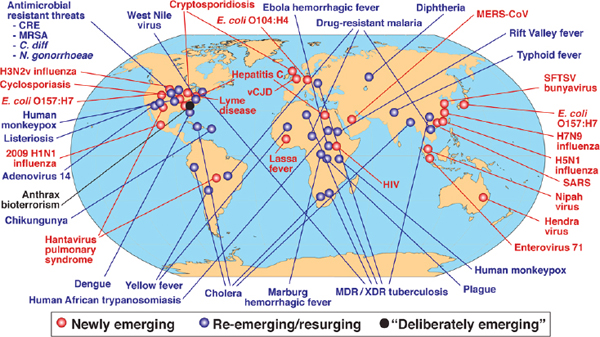
FIGURE WO-1 Global examples of emerging and reemerging infectious diseases.
SOURCE: Morens et al., 2004.
Zoonotic3 viruses pose a particularly serious threat to human health as populations grow and expand geographically, increasing opportunities for contact with wildlife, disturbing habitat, and requiring intensified agriculture to meet increased demand for food; meanwhile, the precipitous rise in global travel and trade have vastly expanded transmission opportunities for emergent pathogens (Bean et al., 2013; IOM, 2014). As presented in Figure WO-2, a century of such global environmental change has produced a legacy of emerging viral diseases. HIV/AIDS is thought to have emerged a century ago through a complex transition from chimpanzees to humans, after which a combination of social and demographic factors eventually propelled it to pandemic status. Meanwhile, global environmental change allowed the formerly range-restricted dengue, chikungunya, and West Nile viruses to reemerge among major populations worldwide (Morens and Fauci, 2013, 2014).
Just as air travel has increased the variety of viruses to which humans are exposed, flying animals are particularly adept at dispersing viruses to new locations and hosts. As shown in Table WO-1, bats and wild birds predominate as primary hosts of important zoonotic viruses (Bean et al., 2013). This phenomenon was raised in several workshop presentations and discussions summarized in this overview, particularly with reference to Middle East respiratory syndrome coronavirus (MERS-CoV), for which bats appear to serve as a reservoir species, and for influenza A (H7N9), now largely limited to poultry, but with the potential to become a serious threat should it make the transition to wild birds.
Growing knowledge of the nature and severity of the threat posed by emerging viral diseases has spurred a range of responses from multiple sectors, described in several workshop presentations. Technical efforts to address emerging viral diseases encompass pandemic prediction; risk assessment; surveillance and detection; descriptive and analytic epidemiology; pathogen characterization; public health interventions; and drug and vaccine development. Legal and political means to reconcile the “borderless world” of microbes with the macroscopic structures of sovereignty continue to be developed and debated. All such work may be productively united under the One Health paradigm: “the collaborative effort of multiple disciplines—working locally, nationally, and globally—to attain optimal health for people, animals, and the environment” (AVMA, 2008).
Global Challenges and Trends in Emerging Viral Diseases
Keiji Fukuda, WHO’s Assistant Director-General for Health Security, opened the workshop with a keynote address on global public health issues related to emerging infectious diseases, and more specifically, to the emerging viral diseases MERS-CoV and H7N9 influenza. At the time of this workshop, MERS-CoV and the H7N9 strain of avian influenza were under active surveillance for their
_______________
3 Zoonotic diseases, or zoonoses, are diseases that can be transmitted from animals to humans.
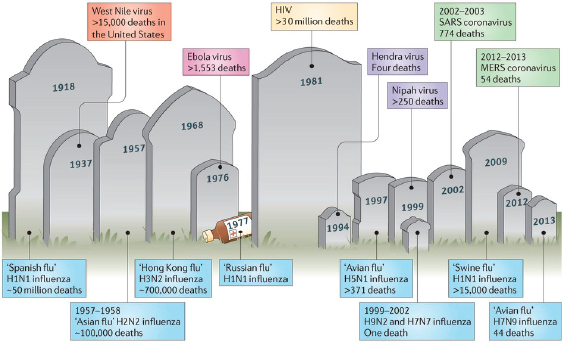
FIGURE WO-2 Emergence of zoonoses. Over the past century, humanity has witnessed the emergence of numerous zoonotic infections that have resulted in varying numbers of human fatalities. Influenza viruses that originate from birds account for an important proportion of these deaths, and recently many new zoonotic viruses that originate in bats, such as Hendra virus, Nipah virus, and the SARS coronavirus, have caused outbreaks with high mortality rates.
NOTE: As of June 2, 2014, the Centers for Disease Control and Prevention (CDC) reports that there were 39,557 cases of West Nile virus in the United States resulting in 1,668 deaths between 1999 and 2013. Source: http://www.cdc.gov/westnile/resources/pdfs/cummulative/99_2013_CasesAndDeathsClinicalPresentationHumanCases.pdf (accessed February 19, 2015).
SOURCE: Bean et al., 2013.
pandemic potential. Instead, the current outbreak of Ebola virus disease in West Africa has infected and killed more people than all previous outbreaks combined (Salaam-Blyther, 2014). According to the latest figures released by WHO on February 25, 2015, the total number of cases had risen to 23,694, with 9,589 deaths, in six West African countries—Guinea, Liberia, Mali, Nigeria, Senegal, and Sierra Leone—as well as one case in Spain and four cases in the United States (CDC, 2015).4 Though the disease was identified in March, “more than 40 percent of the total number of cases have occurred within the past 21 days,” according to WHO. “However, most cases are concentrated in only a few localities” (WHO, 2014e). The West African Ebola epidemic both epitomizes many of the concepts put forth by participants in the workshop, as well as highlights our current inability to successfully predict in almost any way what will next emerge.
Health, Fukuda noted, has reached a level of geopolitical significance that raises new challenges and opportunities for addressing infectious disease. While public health’s traditional and effective focus has been disease prevention and control (through such measures as sanitation, immunization, and clinical care), a range of global trends now demands attention: climate and environmental change; population growth; urbanization; globalization (encompassing trade, travel, and migration); the predominance of poverty among the populations of middle-income countries leading to gaps in health care; and the deterioration of and declining investment in public health infrastructure (IOM, 2003, 2008, 2010, 2014).
As these trends have advanced so too have the global expectations for health care, Fukuda observed. “People have begun to take it for granted that food will be safe, water will be safe, that they will somehow be protected from epidemics and pandemics,” he said. When that does not occur, the reaction—shaped and amplified by social communications—can be severe. Health issues therefore overlap with development, foreign policy, trade, sovereignty, and intellectual property—a phenomenon that is reflected in a policy transition from the Millennium Development Goals5 to a “sustainable development agenda,” a process that Fukuda identified as central to the future of global public health. “This is probably the single largest discussion in global public health taking place right now,” he explained, and its broad scope includes health systems, determinants of health, sustainable development, environment, poverty reduction, and education. Much attention is focused on the implementation of universal health coverage that—while a topic of debate within the United States—is a widely accepted global goal.
_______________
4 On September 30, 2014, the CDC confirmed the first laboratory-confirmed case of Ebola in the United States in Thomas Eric Duncan, a Liberian national who had traveled to Dallas, Texas. Mr. Duncan passed away on October 8, 2014.
5 The eight Millennium Development Goals—which range from halving extreme poverty to halting the spread of HIV/AIDS and providing universal primary education, all by the target date of 2015—form a blueprint agreed to by all the world’s countries and the entire world’s leading development institutions. Source: http://www.un.org/millenniumgoals/bkgd.shtml (accessed February 19, 2015).
TABLE WO-1 Natural Reservoir Hosts and Susceptible Hosts Involved in Transmission of a Selection of Emerging Zoonotic Diseasesa
| Disease (virus) | Known reservoir hosts | Other susceptible hosts | Transmission host to humans | References |
| Avian influenza (H5N1, H7N9, H7N7, H9N2, H3N2, and others) | Waterfowl and wild birds | Bats, cats, dogs, ferrets, pigs, poultry (chickens, ducks, and turkeys), and marine mammals | Chickens | Centers for Disease Control and Prevention,b Zoonoses,c Animal Disease Information Summaries,d Reperant et al., 2012; Swenson et al., 2010; Tong et al., 2012 |
| “Swine flu” strains (H1N1 and H3N2) | Pigs | Ferrets, foxes, cats, dogs, poultry (chickens, ducks, and turkeys), and marine mammals | Pigs | Centers for Disease Control and Prevention,b Zoonoses,c Animal Disease Information Summaries,d Reperant et al., 2012; Swenson et al., 2010; Tong et al., 2012 |
| SARS (SARS coronavirus) | Bats | Civet cats | Civet cats | Centers for Disease Control and Preventionb and Shi and Hu, 2008 |
| Dengue fever (dengue virus) | Primates | Unknown | Mosquitoes | Centers for Disease Control and Preventionb and Carver et al., 2009 |
| Hendra (Hendra virus) | Bats | Horses and ferrets | Horses | Centers for Disease Control and Preventionb and Clayton et al., 2013 |
| Rabies (rabies virus and other lyssaviruses) | Bats | Cats, cattle, coyotes, dogs, foxes, horses, mongooses, primates, raccoons, sheep, skunks, and wolves | Bats and dogs | Centers for Disease Control and Prevention,b Animal Disease Information Summaries,d Hatz et al., 2012; Rupprecht et al., 2011 |
| Ebola viral haemorrhagic fever (Ebola virus) | Bats | Primates | Primates and bats | Centers for Disease Control and Preventionb and Zoonosesc |
| Crimean–Congo haemorrhagic fever (Crimean–Congo haemorrhagic fever virus) | Rodents, hares, hedgehogs, and ostriches | Cattle, goats, horses, pigs, and sheep | Ticks | Centers for Disease Control and Preventionb and Zoonosesc |
| Ross River fever (Ross River virus) | Kangaroos and wallabies | Bats, birds, cats, dogs, horses, and possums | Mosquitoes | Carver et al., 2009; Kay et al., 2007; Tong et al., 2008 |
| Japanese encephalitis (Japanese encephalitis virus) | Pigs and wild birds | Horses | Mosquitoes | Centers for Disease Control and Preventionb |
| West Nile virus encephalitis (West Nile virus) | Domestic and wild birds | Bats, camels, horses, marine mammals, reptiles, and >30 vertebrate species | Mosquitoes and birds | Centers for Disease Control and Preventionb and Animal Disease Information Summariesd |
NOTE: SARS = severe acute respiratory syndrome.
a Supplementary information S1 (table in Bean et al., 2013) lists numerous other zoonotic diseases, including bacterial, prion, parasitic, and other viral diseases.
b See the Centers for Disease Control and Prevention at http://www.cdc.gov.
c See Zoonoses on the World Health Organization website at http://www.who.int/zoonoses/diseases/en (accessed February 19, 2015).
d See Animal Disease Information Summaries on the World Organization for Animal Health website at http://www.oie.int/en/for-the-media/animal-diseases/animal-disease-information-summaries (accessed February 19, 2015).
SOURCE: Bean et al., 2013.
Having established the global context within which public health operates to address emerging infectious diseases, Fukuda proceeded to describe the direction of these efforts both in general and as applied to MERS-CoV and H7N9 influenza.
Lessons from Emerging Infectious Diseases
Global efforts to address emerging infectious diseases, as noted by Fukuda, have been shaped by experience in several ways:
- The consequences of slow response to threats such as HIV/AIDS has led countries to shift infectious disease efforts that once focused primarily on acquiring resources such as laboratories, physicians, and public health scientists to combat ongoing infectious diseases toward the goal of establishing active approaches and capabilities to identify and respond to outbreaks caused by emerging infectious diseases with epidemic or pandemic potential.
- The predominance of zoonoses among emerging infectious diseases illustrates the central role of the animal–human–ecosystem interface and informs the One Health paradigm. “Dealing with these kinds of diseases and responding to them, whether they are zoonoses or whether they are phenomena such as antimicrobial drug resistance can’t be handled by single sectors anymore,” Fukuda observed. “We live in a world where thinking about [infectious disease] . . . as a health issue alone has become outdated.”
- Ongoing tensions involving the sharing of pathogen specimens and the benefits (e.g., vaccines) that result from the characterization of those pathogens must be balanced in global efforts to control emerging disease threats. If new technologies, vaccines, or countermeasures are derived from research on these samples, what is the appropriate quid pro quo? This dilemma sets up “a major balancing act, internationally,” Fukuda stated.
The above considerations are reflected in major international agreements or frameworks governing responses to emerging infectious diseases, Fukuda continued. These include the 2005 revision to the IHR,6 which was spurred by the emergence of both SARS and avian influenza H5N1 and was intended to accelerate
_______________
6 See http://www.who.int/ihr/about/FAQ2009.pdf (accessed February 19, 2015).
the global response to “public health emergencies of international concern”7 including emerging infectious disease threats. A rare, binding, treaty-level agreement among the 196 countries represented by WHO, the IHR were designed to facilitate the detection of emerging diseases of international concern, most but not all of which are infectious. “The IHR place a great deal of emphasis on detection and notification, verification and risk assessment,” Fukuda explained, and they define a mechanism to coordinate the flow of information internationally during health emergencies. The IHR also attempt to avoid or reduce interference with international travel and trade. The regulations also specified the development of core capacities for health security (e.g., disease surveillance and laboratories) within each country by 2014—a deadline that fewer than 20 percent of the countries are on track to meet; this deadline has now been extended 2 years to 2016. “Right now there is a tremendous push to try to do whatever can be done to help countries attain those kinds of capacities,” he stated. Similarly, the global Pandemic Influenza Preparedness Framework8 has been developed, through “very difficult and long negotiations,” to address concerns over equity in pathogen sample sharing and its resulting benefits, according to Fukuda (WHO, 2011a).
Another, less well-known agreement, the Convention on Biological Diversity,9 has recently been adopted by environmental agencies within WHO countries. This convention, which was originally intended to promote sustainable development, will likely have major, unanticipated implications for health, in part because it establishes agreements for moving pathogen samples between countries, Fukuda added. How these agreements will affect the work of laboratories collaborating in response to emerging infectious disease outbreaks of international concern “is not very clear right now,” he observed.
MERS and H7N9 Influenza
Fukuda provided a brief summary of the epidemiological findings on MERS and H7N9 influenza. As of mid-March 2014, the majority of MERS cases have
_______________
7 On August 8, WHO Director-General Margaret Chan declared the West Africa Ebola crisis a “public health emergency of international concern,” triggering powers under the 2005 International Health Regulations (IHR). The IHR requires countries to develop national preparedness capacities, including the duty to report internationally significant events, conduct surveillance, and exercise public health powers, while balancing human rights and international trade. Until last year, the director-general had declared only one such emergency—influenza A H1N1 (in 2009). Earlier this year (2014), she declared poliomyelitis a public health emergency of international concern and now again for Ebola, signaling perhaps a new era of potential WHO leadership in global health security (Gostin et al., 2014).
8 The WHO Pandemic Influenza Preparedness (PIP) Framework, effective May 2011, is intended to improve and strengthen the sharing of influenza viruses with human pandemic potential, and to increase the access of developing countries to vaccines and other pandemic-related supplies. Source: http://www.who.int/influenza/pip/en (accessed February 19, 2015).
9 Signed by 150 government leaders at the 1992 Rio Earth Summit, the Convention on Biological Diversity is dedicated to promoting sustainable development. Source: http://www.cbd.int/convention (accessed February 19, 2015).
been associated with Saudi Arabia, with a few additional cases reported in the region (updated information on MERS cases appears in the section, “Emergence of MERS-CoV”). Primary and secondary cases of the disease appeared distinct, with primary cases tending to be older and male, as compared with their secondary counterparts; fatalities had been higher in the primary cases than the secondary cases, who tend to be younger and healthier. People sickened by MERS tended to have significant underlying chronic diseases such as diabetes, heart disease, or hypertension, he noted. Fukuda added, however, that “the degree to which that reflects the background population versus some unusual predilection is not so clear yet.”
Like SARS—and unlike H7N9 influenza—MERS case clusters have occurred within households and health care facilities; the latter comprised more than half of all secondary cases, Fukuda stated. “Most of the health care worker infections have generally been mild—some have been detected on contact tracing—but there have been deaths among them,” he added. “We’re not positively sure about what the mode of transmission from person to person is in these settings.”
Recent efforts have attempted to identify possible animal reservoirs of MERS-CoV (see the section “Emergence of MERS-CoV”). “Much of the attention has been focusing on camels because of serologic, polymerase chain reaction (PCR), and virus studies identifying this virus in camels,” Fukuda said. “There have been extensive efforts to look at whether there are other potential reservoirs, and so far nothing has really panned out.”
As of mid-March 2014, 390 laboratory-confirmed human cases of H7N9 influenza had been reported, resulting in 121 deaths,10 Fukuda said; most infected people who had been interviewed had been exposed to either live poultry or poultry markets. The cases occurred in two distinct waves that occurred in 2013, followed by a larger one in early 2014. Six small family clusters were associated with emergence, primarily in the second wave, he added.
First-wave cases occurred on China’s eastern seaboard, Fukuda continued. During the second wave, the range expanded slightly to the north and significantly to the south, near the borders of Vietnam and Cambodia—a situation that is being carefully watched. H7N9 influenza cases encompassed a broad age range, but most involved middle-aged to older people, predominantly males, Fukuda reported; approximately 30 percent of cases were fatal.
Characterization of viral samples that revealed antigenic similarity among birds and humans led WHO to identify a recommended vaccine strain, Fukuda said. The viruses are uniformly resistant to one class of antiviral drugs, M2 inhibitors, and nearly all are sensitive to neuraminidase inhibitors, he said.
_______________
10 As of the beginning of February 2015, 597 cases were reported, including 2 in Canada and 1 in Malaysia, with 207 deaths. Source: Hong Kong Centre for Health Protection: Avian Influenza Report. http://www.chp.gov.hk/files/pdf/2015_avian_influenza_report_vol11_wk07.pdf (accessed February 23, 2015).
Several similarities—beyond their near-simultaneous occurrence—unite the emergence of MERS-CoV and H7N9 influenza A, Fukuda observed:
- Both viruses were relatively limited in terms of their geographic spread. MERS-CoV cases in Europe, where the virus has failed to take hold, have clearly been imported or resulted from close contact with an imported case. At the time of this writing, while human infections had been reported from several countries both within and beyond the Middle East, those outside the region had recently traveled there (WHO, 2014d). No human cases of H7N9 influenza A had been reported outside China at the time of this writing (WHO, 2014c).
- Both viruses are zoonoses11 with limited person-to-person transmission, resulting in sporadic cases and clusters, rather than community-wide spread.
- Future transmission patterns for either virus are uncertain. “Like all emerging infectious diseases, before something has actually happened, we are never quite sure what the potential is for these to change and escalate [or burn out] beyond the current patterns,” Fukuda observed.
Several key features also separate MERS-CoV and H7N9 influenza, according to Fukuda:
- The viruses are not related and are geographically restricted to different regional locations.
- MERS-CoV is linked with camels and bats; H7N9 is linked with poultry.
- Clusters of MERS-CoV cases have primarily occurred in communities or health care settings, while H7N9 clusters have occurred primarily among people in contact with poultry, including families.
- Vaccine development against MERS-CoV is in the investigational stage, whereas production of an H7N9 influenza vaccine could be quickly launched if the need arose.
- Therapeutics for MERS-CoV are currently under investigation and include drugs and antisera; for H7N9 influenza, the value of antiviral drugs has been well established.
Fukuda elaborated on WHO’s current priorities for MERS-CoV, beginning with regional surveillance. To better understand the risk factors associated with
_______________
11 Zoonotic diseases are contagious diseases spread between animals and humans. These diseases are caused by bacteria, viruses, parasites, and fungi that are carried by animals and insects. Examples are anthrax, dengue, Ebola hemorrhagic fever, Escherichia coli infection, Lyme disease, malaria, plague, Rocky Mountain spotted fever, salmonellosis, and West Nile virus infection. Source: CDC factsheet: Zoonotic Disease: When Humans and Animals Intersect. http://www.cdc.gov/24-7/cdcfastfacts/zoonotic.html (accessed July 23, 2014).
human disease, an international case-control study is under discussion but has yet to be launched. In addition, there has been a great deal of discussion about the need for validating the serologic tests used to confirm exposure(s) to the MERS-CoV, but progress toward that goal has been slow. Therapeutics and vaccines for MERS-CoV are in the early stages of development. Finally, he said, controlling the threat of MERS-CoV will require coordination and discussion between human and animal health sectors, which so far has proven relatively difficult.
In the case of H7N9 influenza A, WHO’s highest priorities include monitoring the regional spread in humans and animals, transmission patterns, and drug resistance; vaccine development and deployment planning; and developing strategies for preventing and controlling the spread of disease, Fukuda said. Unlike highly pathogenic H5N1 avian influenza, H7N9 influenza A is asymptomatic in poultry, making its spread among animals difficult to monitor, and increasing the need to anticipate its potential shift from its current status as a zoonotic infection to person-to-person transmission.
While vaccine development is well under way for H7N9, Fukuda noted that deployment planning is a concern, given the political, legal, and operational experiences with vaccine distribution during the H1N1 influenza pandemic of 2009.
Global Response to Emerging Infectious Diseases
As public health broadens its goals, perspectives, and connection to other sectors, a concomitant transition will change approaches to addressing emerging infectious diseases, Fukuda predicted. “We no longer have health discussions,” he said. Today, global health deliberations encompass population growth, globalization, communications, economics, or social justice—all of which must be addressed in developing sustainable solutions to infectious threats. By providing “the accepted foundation for health security,” the IHR represent one such solution, Fukuda observed. Likewise, the Global Influenza Surveillance and Response System12 supports both virus detection and vaccine development worldwide. The acceptance of the One Health concept further extends global connections and demonstrates that “the need for intersectoral coordination is moving past the stage of rhetoric to actually being acted upon,” he asserted.
More concretely, recent alliances among the Food and Agriculture Organization of the United Nations (FAO), World Organization for Animal Health (OIE),
_______________
12 Global influenza virological surveillance has been conducted through WHO’s Global Influenza Surveillance and Response System (GISRS) for over half a century. Formerly known as the Global Influenza Surveillance Network, the new name came into effect following the adoption of the PIP Framework (see above) in May 2011. GISRS monitors the evolution of influenza viruses and provides recommendations in areas including laboratory diagnostics, vaccines, antiviral susceptibility, and risk assessment. It also serves as a global alert mechanism for the emergence of influenza viruses with pandemic potential. Source: http://www.who.int/influenza/gisrs_laboratory/en (accessed February 19, 2015).
and WHO signal a “sea change in the degree to which the organizations work together on a functional basis,” Fukuda observed. This collaboration (discussed in greater detail in the section “International and Domestic Responses to Emerging Viral Diseases”) was initiated in response to economic and social concerns associated with emerging pathogens. It was formalized through a memorandum of understanding signed by the three directors-general of these organizations to (1) work together closely, (2) meet annually, and (3) strategically plan for the coming year together. In addition, he noted, FAO and WHO have agreed to collaborate on the next international conference on nutrition. Meanwhile, OIE and WHO are attempting to harmonize the ways by which they measure gaps in capacity.
Despite these indicators of progress toward global cooperation and coordination in addressing emerging infectious diseases, according to Fukuda significant challenges remain. These include the lack of core capacities for surveillance and detection in most countries, as previously noted—leading to incomplete information on the spread of emerging pathogens such as MERS-CoV. This is less the case with H7N9, he added, attesting to the fact “that we are at different stages in different parts of the world in implementing the concepts or the spirit of IHR.”
There is no overriding reason for the gap in implementing the IHR, Fukuda explained during the discussion that followed his presentation. Some countries are simply too poor to build core capacity for infectious disease surveillance and detection, he observed, whereas others are “concerned that if they indicate that they have all core capacities, perhaps their funding or their support may go down.” But most countries, he said “with a reasonable amount of support, are going to get there,” at least over the long term. While the development of core capacities is the responsibility of national governments, he insisted, WHO can support them in implementing quality assurance and assessment.
While intersectoral collaboration has improved over recent years, it is not routine, particularly at the national level and below, Fukuda said. At the same time, the rapid pace of scientific development (e.g., high-throughput sequencing) has begun to strain existing frameworks and approaches, and even the concept of what constitutes a pathogen—a definition central to such frameworks as pandemic preparedness and disease eradication. “Right now we don’t quite know how to handle . . . [such] questions,” he acknowledged, noting that both MERS-CoV and H7N9 influenza are raising them anew. In particular, with regard to the Convention on Biological Diversity, Fukuda noted that its potential to inhibit sample sharing during a pandemic has been addressed in part through the ability to create special agreements, but he nonetheless predicted that its implementation would cause “a great deal of uncertainty and confusion” that would not be resolved without challenge (and resultant failure) during a major disease outbreak. “I don’t think it’s going to be orderly,” he said of that transition.
Many of the same issues WHO confronts in addressing MERS-CoV and H7N9 are also relevant to antimicrobial resistance, which Fukuda characterized as a “super-emerging infectious disease.” Although recognized by scientists
since penicillin’s initial release in the mid-20th century, public awareness of this phenomenon has only recently become widespread. The impact of antimicrobial resistance extends well beyond the health of individuals, to development, foreign policy, and economics—a recognition that was articulated by President Obama’s mention of antibiotic resistance in his 2014 State of the Union address.
“In the next year, there will be the development of a global action plan, which is an attempt to bring all of this together and to move this at a more coherent and coordinated level than is currently available,” Fukuda announced. The plan will define the scope of the problem and functional issues to be tackled, describe a sustainable research base, and propose a new, sustainable and appropriate model for marketing antimicrobial drugs, he said. The latter innovation is important, he noted, given that “much [of the] effort is focused on how to stimulate research into new antibiotic modalities, and not on the overarching question of how risk will be distributed.” The global plan should also, eventually, identify benchmarks for success and clarify actions that can be taken to achieve them, providing a blueprint that can be adopted, in whole or in part, by anyone in the world, he concluded.
When asked in the subsequent discussion to define the “hot button” issues the global action plan intends to address, Fukuda replied that an immediate concern is “future fights that you can almost predict” between poor and wealthy countries, and between the pharmaceutical industry and the public. “Probably the biggest sensitivity is the use of antibiotics for nonhealth (e.g., agricultural) uses,” he observed, noting the paucity of relevant data on the risk of such uses, given their widespread occurrence. Further priorities include regulating antibiotic prescription practices, supporting antimicrobial research and development through marketing-independent mechanisms, and improving surveillance for antimicrobial resistance. “We can’t think about antibiotics as products; we have to think about them as global goods,” he insisted.
As Fukuda noted in response to questions from members of the Forum, the crafting of the global action plan to address antimicrobial resistance involves considerations of multiple sectors, including the private sector and the research community. “There will be a very concerted effort to bring in all of those viewpoints, including from the research community, but also from industry, from civil society, from a lot of the scientific groups that have been working on this for decades,” he stated.
Relman noted a similar initiative, in February 2014, when the White House and the Secretary of Health and Human Services, together with WHO, FAO, and other organizations, announced a new global health security agenda intended to catalyze international action on this issue—for example, by completing the core capacity building specified by the IHR. Relman wondered how this effort might be directed to have maximum beneficial impact. Fukuda replied that that will happen if all of the involved parties move together in a coherent and coordinated way—a concept that is “very clear . . . [but] still challenging to implement.”
Instead of a wasteful sector-by-sector approach, implementation should proceed in ways that link both countries and agencies, he urged. “This, in essence, is what things like the IHR were meant to try to get at,” Fukuda observed. The global health security agenda strives for a new level of international coherence in responding to EIDs, he said, yet it remains to be determined how to reach that goal.
LESSONS LEARNED FROM THE 2009 H1N1 INFLUENZA A PANDEMIC
In his opening remarks to the workshop, Forum chair David Relman, of Stanford University, noted that EIDs—especially those caused by influenza viruses and β-coronaviruses—have greatly inspired expanding curiosity about and understanding of the origins and consequences of infectious diseases. If EIDs are teachers, pandemics provide a particularly intense educational experience. Keynote speaker Harvey Fineberg, president of the Institute of Medicine,13 recounted lessons learned from the 2009 H1N1 influenza A pandemic—the first declared pandemic of the 21st century (Dr. Fineberg’s contribution may be found on pages 152–165 in Appendix A). The international response to this pandemic was strongly influenced by the global experience with SARS.
Between November 2002 and July 2003, the introduction of SARS—primarily through international travel—wreaked havoc in 26 countries, resulting in more than 7,000 cases and more than 700 deaths, Fineberg recalled. While the epidemic was extinguished largely through the isolation and management of hospitalized patients, “It was a warning sign,” he said. “MERS is another threat,” Fineberg continued, “but among the many threats for pandemics, in terms of versatility, the persistence, the rapidity, and the possibility for extremely severe consequences, there’s nothing that quite rivals influenza.” These viruses, he noted, have caused some of the most catastrophic pandemics in history, most particularly the Spanish flu pandemic of 1918–1919. There are many lessons to be learned from the history of influenza and from the experiences of and with pandemics, he observed.
Fineberg took a global perspective in his analysis of the international response to the 2009 H1N1 influenza A pandemic (Fineberg, 2014). He focused on the role and actions of WHO, a topic he studied as the chair and member of the WHO Review Committee on the Functioning of the International Health Regulations (2005) and on Pandemic Influenza (H1N1) 2009 (WHO, 2011b).
_______________
13 Dr. Fineberg’s presidency ended on June 30, 2014. His current affiliation is with the Gordon and Betty Moore Foundation.
The Global Experience
History
As Fineberg described it, the 2009 H1N1 influenza A pandemic unfolded as follows:
In late March 2009, the cause of flu outbreaks in Mexico was discovered to be a previously unrecognized H1N1 virus; it subsequently was associated with prior cases in California. By the end of April 2009, H1N1 had already been recognized in a number of states within the United States, as well as in Canada, New Zealand, Spain, the United Kingdom, Germany, and Israel. Invoking its authority under the 2005 IHR, WHO on April 25, 2009, declared a public health emergency of international concern and convened the emergency committee called for in the regulations. WHO also established a dedicated internal group to coordinate the international response to the widening outbreaks. (Fineberg, 2014)
By June 9, 2009, more than 70 countries had isolated more than 26,000 laboratory-confirmed cases. WHO declared on June 11, 2009, that a full-fledged pandemic was under way. This strain of influenza spread so rapidly, that by late July 2009, virtually every country in the world had identified and isolated laboratory-confirmed cases of H1N1 influenza.14
Burden of Disease
Two recent estimates (Dawood et al., 2012; Simonsen et al., 2013) suggest that between 100,000 and 400,000 deaths were attributable worldwide to the 2009 H1N1 influenza A pandemic, as compared with as many as 500,000 deaths in a typical interpandemic influenza season. Yet, the distribution of mortality was unlike that of seasonal flu, in which most illness and death occurs among the very young and the elderly. For H1N1, mortality among children, young adults, and pregnant women was especially high compared with a typical flu season. In considering the burden of disease in terms of years of life lost, according to Fineberg, the 2009 H1N1 influenza pandemic strain represented a more serious threat to global health than seasonal influenza.
_______________
14 In March and early April 2009, 2009-H1N1 influenza A emerged in Mexico and the United States. During the first few weeks of surveillance, the virus spread worldwide to 30 countries by human-to-human transmission, causing the WHO to raise its pandemic alert to level 5 of 6. On June 11, 2009, WHO raised the worldwide pandemic alert level to level 6 in response to the ongoing global spread of the 2009-H1N1 influenza A virus. This virus has now become the first influenza pandemic of the 21st century. The third public health emergency of international concern (PHEIC) was declared on August 8, 2014, for the Ebola virus outbreak in Africa. In both cases, the scientific, public health, security, and policy communities are moving quickly to learn more about the nature and potential impact of these viral diseases on human and animal health.
Testing the 2005 IHR
To mount an effective response to an emerging and evolving pandemic public health authorities must act rapidly and authoritatively on incomplete knowledge of the disease they are attempting to address. This situation, which Gostin (2004) has aptly named “the public health paradox,” lies at the heart of the ethical and legal response to pandemic disease. “There is no way to avoid the dilemmas posed by acting without full scientific knowledge,” he writes. “The only safeguard is the adoption of ethical values in formulating and implementing public health decisions.” Similar arguments have been made in favor of an ethical framework for decision making concerning biodefense and bioterrorism.
The IHR provides the legal framework for international cooperation on infectious disease surveillance. The IHR were adopted by the World Health Assembly in 1969, having evolved from the International Sanitary Regulations adopted in 1951 (which, themselves, were direct descendants of the international sanitary conventions adopted from the 1890s through the 1940s) (IOM, 2007). The IHR (1969) were intended to monitor and control six diseases—cholera, plague, smallpox, relapsing fever, typhus, and yellow fever—yet revisions in 1973 and 1981 resulted in only three reportable diseases—cholera, plague, and yellow fever—whose occurrence required WHO notification (WHO, 2009). In the mid-1990s it became clear that the IHR (1969) had become outdated given the vast number of global microbial threats that had emerged and reemerged, including those which were not deemed “notifiable” in the original set of guidelines (i.e., Ebola hemorrhagic fever) (IOM, 2007; WHO, 2009). There was also concern that the IHR’s dependence on “official” country notification, along with a lack of a formal internationally coordinated mechanism to contain international disease spread, might prove problematic in effectively containing disease with pandemic potential (WHO, 2009). Several resolutions passed by the World Health Assembly (in 1995, 2001, 2003) encouraged revision of the IHR, with the final resolution WHA58.3 formally adopting the IHR (2005) on May 23, 2005.
When the revisions to the IHR came into force on June 15, 2007, member nations of WHO were required to report all new and reemerging diseases with epidemic or pandemic potential, irrespective of their origin or source (WHO, 2008). These revisions also stipulated that member nations were to assess their disease surveillance capacity and develop national action plans within 2 years, and meet the IHR requirements within 3 years regarding their national surveillance and response systems, as well as requirements at designated airports, ports, and ground crossings (although extensions may be obtained) (WHO, 2008).
The 2009 influenza A pandemic was the first real-world test of the IHR revisions (2005) in a PHEIC, and, Fineberg noted, it exposed vulnerabilities in public health capacity and response. The experience also revealed the limitations of available scientific knowledge in understanding and coping with pandemic disease in general. Decision making under these conditions of uncertainty, which inevitably occur during an evolving pandemic, was predictably difficult, he
observed. There were also many communications challenges between and within the many organizations involved in addressing the crisis.
A number of criticisms were leveled at decisions made by WHO in the course of the 2009 pandemic, Fineberg stated. Some of these criticisms were reasonable, and some were unjust, but it was difficult to discern between them until the consequences of the decisions in question were clear, he said; in some cases, that was not until the pandemic was long extinguished. To productively analyze these concerns, WHO convened the committee that Fineberg chaired; he described its findings to the workshop (Fineberg, 2014; WHO, 2011b).
Report of the Review Committee
In 2010, WHO formed an international committee to review the regulations of the IHR in their performance in the pandemic, and also the performance of WHO itself, Fineberg said. The committee consisted of 24 members, each from a different country. In response to criticism regarding WHO’s secrecy in decision making during the pandemic, the committee held all fact-gathering sessions open to representatives from its member states, as well as to the public and the press, he reported. The committee did deliberate in closed sessions, he added, and there were subgroups of the committee that worked privately.
With its diverse makeup, the committee was often difficult to manage, according to Fineberg. At the same time, these challenges paid off since the recommendations of the committee were viewed as representing a broad spectrum of consensus opinions of WHO member states, and therefore readily endorsed at the World Health Assembly in 2011, stated Fineberg. The committee reached three key conclusions about the IHR, WHO operations, and global pandemic response(s)—which Fineberg discussed along with additional relevant findings, implications, and recommendations.
The IHR
Implementation of the IHR (2005), which were born of the 2003 SARS pandemic, clearly helped the world prepare to cope with the public health emergency presented in 2009 by the H1N1 influenza A pandemic, Fineberg stated. However, he noted, the committee found that core capacities specified by the IHR were not yet fully operational in many member states, and that many countries were not even on a path toward successful implementation of these capacities.
The IHR call for consistent communication and cooperation was in fact the case in many countries throughout the pandemic, Fineberg reflected. WHO provided needed and appreciated technical support to many countries. Provisions of the IHR that address the impact of public health emergencies beyond the health sector (e.g., social and economic impacts) “were very salutary,” he observed.
These recent revisions to the IHR enabled more flexible, dynamic, and adaptive responses.
But for all of the virtues of the IHR (2005), there were a number of shortcomings, Fineberg continued. “In 2011, WHO surveyed all of its 194 member state signatories to assess where were they on the path towards successful implementation of the core capacities called for in the IHR,” he stated; only 58 percent responded. Of them, only 11 percent had completed the core capacities called for by 2012. Moreover, as Fineberg noted, if a signatory to the IHR fails to comply with its provisions—for example, by taking unilateral action that interferes with travel and trade—that state must provide a rationale for its breach of regulations. Yet such a state faces no consequences if fails to do so, he said. “There are not provisions for any enforcement of these expectations and agreements,” he observed.
Among the lessons learned regarding the IHR in the course of the pandemic is the recognition that the regulations, as they stand, are insufficient to ensure that countries will fully implement the required capacities for addressing public health emergencies of international concern. The case must be made that it is in the interest of each country to implement these capacities for the protection of their own citizens, as well as for the global good. Fineberg continued by stating that implementation of the IHR-required capacities must be made easier. “Mobilizing agencies willing to provide technical assistance to those countries that require it would be a help,” he suggested, as would better-organized channels by which specialized resources could be shared internationally. Along these same lines, the committee recommended that WHO’s information sites be geared to meet specific countries’ needs under the IHR.
Finally, Fineberg noted that the requirements of the IHR must somehow be enforced, in order to encourage countries to support and comply with their effective implementation. “It is also going to be useful to try to clarify the effect and measure the consequences of various actions that may be taken in future public health emergencies,” he concluded.
WHO Operations
The committee agreed that WHO achieved a number of successes as it led the international response to the 2009 pandemic, Fineberg stated. Be that as it may, systemic problems hampered the organization’s performance, and the committee identified several operational shortcomings that occurred. During the pandemic, some had accused WHO of purposefully misleading countries and of making distorted decisions, Fineberg recalled. Based on their analysis of WHO’s complete files on its actions, as well as interviews of all the relevant parties, they could find no evidence of malfeasance, only of error.
Overall, the committee determined that WHO provided very timely guidance in the face of the emerging pandemic to countries worldwide, according to Fineberg. “Preparedness plans specifically were in place in 74 percent of countries by
the time the pandemic emerged. Once the public health emergency was declared under the authority of the IHR, WHO immediately convened an emergency committee that was called for under the provisions,” he reported. WHO provided rapid and appropriate field support as well as early recommendations on vaccine target groups and optimal dosages, he continued, and the organization gathered and disseminated information on laboratory-confirmed cases from countries around the world. WHO also strived to work with sister agencies of the United Nations and, in concert with national organizations, was largely successful in that effort, Fineberg observed.
Candidate vaccine strains of H1N1 were identified early—within weeks—and seed strains were quickly made available along with appropriate reagents, according to Fineberg, another success driven by WHO. Likewise, the organization oversaw the efficient deployment of antiviral drugs, a possible means of containing the pandemic, to high-risk populations in 72 countries.
“There’s no substitute for WHO when it comes to capacity and role in managing a global emergency in the health sphere,” Fineberg insisted. He also noted, however, that structural constraints within the organization led to some problems in its response to the 2009 pandemic. For example, reflecting its dual capacity as the moral voice for health equity in the world and as the servant of the member states, WHO encountered conflicts between the interests of individual member states and that of its global responsibilities.
Those responsibilities, Fineberg noted, vastly dwarf WHO’s budget, which is only partially (25 percent) funded by its member states. Most of its funding is directed not by the organization, but by individual member states, or by foundations or other donors, he explained. “Organizationally, WHO is pretty well equipped to do two things,” he said: (1) managing multiyear disease control programs (e.g., for malaria), and (2) mounting emergency responses to investigate emerging infectious disease outbreaks (e.g., of hemorrhagic fever in sub-Saharan Africa). The organization was not able to sustain the focused, global effort required to address a pandemic, Fineberg concluded.
Compounding these structural barriers within WHO was ambiguity about the definition of a pandemic, Fineberg reported, particularly in terms of severity. The pandemic preparedness plan in place when H1N1 arrived was designed to address H5N1, with its high fatality rate. But a pandemic is defined by its geographic spread, not its severity—which is arguably more important to determining an appropriate response, he stated, and as illustrated in Figure WO-3. “Failure to have a consistent, measurable, and explicable measure of severity was a real handicap throughout the [H1N1] pandemic,” he observed.
WHO’s six-level pandemic phase alert system was overly complex, Fineberg continued. Moreover, when WHO declared phase six, they ceased press conferences, which he deemed “a rather odd response to having now reached a full-fledged pandemic.” Other practices raised public suspicions, he noted. For example, unlike standard 2-day WHO consultative committees, the emergency
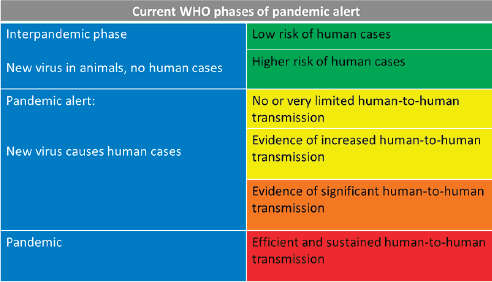
FIGURE WO-3 WHO phases of pandemic alert at the time of the H1N1 pandemic.
NOTE: As of April 18, 2007.
SOURCE: WHO, 2008.
committee invoked according to the IHR remained in place until the pandemic ended. When WHO convenes consultative committees, it does not publicize the identities of its members until their report is made public, in order to protect them from commercial or political pressure and obtain their best judgment—but this confidentiality strategy is only effective over short periods, he said. Also, the emergency committee shared little information with the public as to how it managed conflicts of interest among its members, he observed. For instance, it did not reveal whether any of its members were associated with pharmaceutical companies that were producing vaccines.
WHO generated tremendous amounts of guidance in response to the pandemic, but it lacked a system for collating, coordinating, and prioritizing that information, Fineberg stated, as well as a means to clearly communicate the pandemic’s scope and severity. Delays in translation into all of WHO official languages rendered their guidance even less useful to national decision makers, he added.
Global Pandemic Response
The delayed distribution of vaccine represents a fundamental shortcoming in the otherwise strong global response to the 2009 H1N1 influenza A pandemic, according to Fineberg. In most countries, he observed, that delay would
have rendered the vaccine less useful had the pandemic been more severe. This shortcoming stems from the fact that vaccine production remains predominantly dependent on egg viral culture, he pointed out. Regulatory, legal, and logistical complexities in vaccine distribution further slowed its release. Most importantly, he added, arrangements with vaccine manufacturers were not in place when the pandemic struck, so negotiations began during the initial outbreak—a lesson in the importance of advance preparation for emerging infectious disease events.
Lessons for WHO
Fineberg described these recommendations by the review committee aimed at improving pandemic response by WHO:
- Clarify and strengthen pandemic processes and guidelines. This includes clarifying guidance around severity, simplifying the pandemic phase structure, and more flexibly responding to pandemics as they unfold, learning from and applying lessons learned from the experiences in different countries. There is also a need to develop practices to identify conflicts of interest and to encourage transparency and openness in the appointment of emergency committees.
- Strengthen internal capacity. A contingency plan for mobilizing and sustaining relevant expertise during a public health emergency needs to be developed, along with the increased financial support for WHO that would make such a plan possible.
- Improve communications. Sustain active communications throughout the emergency, and acknowledge mistakes appropriately, which strengthens refutations of unwarranted accusations. Track and archive changes in the web. Use social media to reach a wider public.
- Encourage advance agreements. Since the time of the 2009 influenza pandemic, a protocol has been adopted by many countries to support the solicitation of donated vaccine in advance of a pandemic. These PIP agreements preposition relevant vaccine, make seed strains widely available to manufacturers, and extend technology to developing countries, enabling them to produce their own vaccine. Such distributed capacity, Fineberg argued, is the only long-term solution to the vaccine-sharing dilemma that pits the interests of countries with manufacturing capacities against global needs when supplies are limited—as likely occur in a pandemic.
Lessons for the Global Pandemic Response
The overall conclusion of the review committee was that the world was ill prepared to respond to a severe influenza pandemic, or to any similarly global, sustained, and threatening public health emergency, Fineberg stated. Much
remains to be done, including building public health capacity and pursuing research to improve response to pandemic threats. “If we had a severe pandemic today, with the vaccine capacity that we have today, and the distribution of methodology and production that we have today, with a total global capacity for about one-third of the world’s population, we could have tens of millions of people who would perish,” he warned.
Fineberg described the following recommendations for global efforts to address future pandemics, derived from experience in the 2009 H1N1 influenza A pandemic:
- Mobilize during emergencies, and deploy to countries that need assistance.
- Create a contingency fund for public health emergencies.
- Prepare distribution agreements between industry, WHO, and countries.
- Make seed virus strains widely available to vaccine manufacturers.
- Require vaccine manufacturers to contribute vaccine to a global pool.
- Encourage countries to provide immunizations to high-risk populations.
Fineberg observed that research on the following key topics will enable even more significant improvements in pandemic preparedness:
- Detection, characterization, and monitoring of new viruses;
- Viral and host determinants of transmissibility and virulence;
- Point-of-care diagnostic tests;
- Accuracy and timeliness of modeling projections;
- More effective, safer, and long-lasting vaccines;
- Antiviral drugs;
- Protective equipment, personal hygiene, and social interventions; and
- Effectiveness and costs of border control measures.
The World Health Assembly endorsed the review committee’s report and accepted their recommendations in May 2011. Within the next year, several WHO units incorporated recommendations from the report into their annual working plans, Fineberg stated. Even so, structural impediments and scientific shortcomings remain that can impair the world’s ability to prepare for and respond effectively to the next pandemic, he concluded. For example, in discussion following his presentation, he observed that while WHO may have developed contingency plans to improve their capacity to mobilize response to a pandemic, he did not believe that they had been implemented to the extent that if confronted with another PHEIC, the organization would be more able than in 2009 to support sustained deployment of existing staff. This observation has been borne out by the current regional and global responses to the current Ebola outbreak in West Africa.
Recognizing that WHO’s limited financial resources greatly hinder its ability to mobilize in a pandemic, Fineberg reported on the review committee’s recommendation that hundreds of millions of dollars be made available to the
organization to support these efforts. Such funds, he suggested, could be authorized through the World Bank or International Monetary Fund as a line of credit conditioned upon the declaration of a public health emergency of international concern.
When asked whether the world was better prepared to respond to a pandemic today than in 2009, Fineberg answered yes, for such reasons as the advent of PIP agreements, and some (but not enough) progress toward implementation of the IHR (WHO, 2008). Nevertheless, the world could do far more to prepare but does not. Why?
“It is very difficult to invest in a possibility when you’ve got compelling alternative immediate demands,” Fineberg observed. When people are actually dying of other diseases, it is difficult to shift limited resources away from those toward a possible disaster, he said. Thus, he concluded, the case for pandemic preparedness is a case for insurance, requiring an investment that not every country is ready or able to make.
OVERVIEW OF EMERGING VIRAL DISEASES
An early session of the workshop was devoted to four presentations that provide context for the subsequent discussion of the emerging MERS-CoV and influenza A viruses by exploring the ecology and immunology of emerging viral diseases; the political and social conditions that have facilitated their emergence, which in turn are affected and shaped by the consequences of infectious disease; and the research response to viral disease emergence.
Emerging Diseases in Wildlife
Once studied only for their role in regulating wildlife populations, infectious diseases of wildlife have recently gained attention as potential threats to domestic animal and human health, according to speaker Jonathan Sleeman, of the U.S. Geological Survey (USGS) National Wildlife Health Center. Indeed, three-quarters of known emerging infectious diseases are zoonotic, of which the majority are of wildlife origin, he noted, and disease agents infecting wildlife are twice as likely to become EIDs as those without wildlife hosts (Taylor et al., 2001) (Dr. Sleeman’s contribution may be found on pages 248–262 in Appendix A).
Previously described global trends favoring disease emergence apply equally to diseases of wildlife, particularly ecological changes such as intensified farming, alterations in landscape and land use, human activity in formerly pristine areas, and climate change. These circumstances “are clearly leading to increased opportunities for spillover of pathogens from wildlife into domestic animal and human populations,” Sleeman said. Moreover, he added, while public and animal health initiatives in wealthy countries have prevented or controlled many emerging viral diseases, comparable infrastructure to address wildlife disease and
health issues barely exists. Likewise, the health and economic consequences of emerging diseases of wildlife such as white-nose syndrome in bats, colony collapse disorder in bees, chytridiomycosis in amphibians, canine distemper virus in wild and domestic animals, and Ebola virus, while surely significant, have been largely unrecognized, he observed.
- White-nose syndrome is an emergent disease of hibernating bats that has spread from the northeastern to the central United States at an alarming rate. Since the winter of 2007–2008, millions of insect-eating bats in 25 states and 5 Canadian provinces have died from this devastating disease. The disease is named for the white fungus Pseudogymnoascus destructans that infects skin of the muzzle, ears, and wings of hibernating bats (Blehert, 2012; Blehert et al., 2009, 2011; USGS, 2014).
- Colony collapse disorder (CCD) is a serious problem threatening the health of honey bees and the economic stability of commercial beekeeping and pollination operations in the United States. Despite a number of claims in the general and scientific media, a cause or causes of CCD have not been identified by researchers (USDA/ARS, 2014).
- Massive die-offs of amphibians are often caused by ranaviruses. USGS scientists have isolated ranaviruses associated with die-offs in more than 25 states involving more than 20 species of turtles and amphibians in mortality events ranging from one to thousands of individuals affected. Some events may involve a single species; others may involve multiple species. Frogs and salamanders in the same pond, for example, may die from ranaviral infections at the same time (USGS, 2013).
- Canine distemper virus (CDV) is the second most common cause of infectious disease death in domestic dogs and is a significant viral disease of global importance in common and endangered wild carnivores. It is a multihost pathogen with abundant wildlife reservoir species, such as raccoon dogs (Nyctereutes procyonoides). Identification of positive tiger CDV cases suggests wide distribution for the Arctic-like CDV strain that infects and kills Amur tigers (Seimon et al., 2013).
- Repeated outbreaks of Ebola have had a devastating impact on humans, chimpanzees, and gorillas in central Africa over the last decade. There are particular fears for western gorillas (Gorilla gorilla). Although all apes and chimpanzees are threatened, these gorillas have a habitat ranging over a particularly small area, with the majority of the population found in parts of Cameroon, Gabon, and Republic of Congo.15 It is estimated
_______________
15 Western lowland gorillas are endangered, but they remain far more common than their relatives, the mountain gorillas. They live in heavy rain forests, and it is difficult for scientists to accurately estimate how many survive in Angola, Cameroon, Central African Republic, Congo, the Democratic Republic of Congo, Equatorial Guinea, and Gabon. National Geographic, Western Lowland Gorilla Gorilla gorilla gorilla, http://animals.nationalgeographic.com/animals/mammals/lowland-gorilla (accessed November 12, 2014).
that one-third of the world’s gorilla population living under protection in national parks have died in the past 15 years from this disease (Animal Research Info, 2014).
It is clear that emerging viral diseases—including ranavirus in amphibians, canine distemper in tigers, and Ebola hemorrhagic fever in gorillas—have caused major wildlife population declines since the 1990s, according to Sleeman. Whereas in the 1970s, wildlife diseases investigated by the USGS tended to cause large but localized die-offs, since the 1990s, the agency has encountered an ever-increasing number of novel diseases that are hard to predict, spread rapidly over large geographic areas, impact multiple species, and cause dramatic population declines and even extinctions, he reported—and outbreak investigations involving collaboration among multiple sectors and agencies.
In his presentation to the workshop, Sleeman chose to highlight the role of wild animals in the emergence of avian influenza and the resurgence of bluetongue and epizootic hemorrhagic disease, the phenomenon of disease transmission at the human–primate interface, and the significance of bats as reservoirs for numerous emerging viruses—including SARS and MERS-CoV, Nipah and Hendra, Ebola and Marburg—that cause human disease.
Emerging Avian Influenzas
Wild birds—primarily waterfowl, shore birds, and gulls—are the natural reservoirs for avian influenza (AI) viruses, Sleeman observed. Recently emerging AI strains, including H5N1, H7N9, H10N8, and H5N8, have threatened the poultry industry as well as public health. He noted that many of these strains originated in Southeast Asia, where the intensification of farming practices—including the co-mingling of domestic and wild species—and the growth of live markets16 have fueled the spillover of viruses from wild to domestic birds, and then to humans.
The USGS genome studies have examined the genetic structure of AI viruses in Asia and North America, paying particular attention to continental edges in Alaska and along the eastern seaboard of Canada and Iceland, Sleeman stated. In such areas, the USGS found both Eurasian and North American strains, along with strains with mixed lineages—hot spots for the evolution of new viruses. “These are definitely areas of focus for surveillance,” he observed.
The agency is also keeping a close eye on unusual events such as the 2011–2012 die-off of harbor seals in New England due to an H3N8 AI virus, Sleeman noted. This virus proved similar to one that was circulating in wild North American waterfowl, he added, but its sequence suggested its recent adaptation
_______________
16 Variously known as live bird markets, live poultry markets, and wet markets, open marketplaces composed of stalls where live poultry (and often other live animals and fresh vegetables) are sold are found throughout China and many Southeast Asian countries. In this document, all such venues are denoted by the term “live market.”
to mammalian hosts—in contradiction to the widely accepted “mixing vessel hypothesis,”17 which defines a circuitous route of pandemic viral emergence from wild birds into domestic fowl, then pigs, before recombination with other mammalian viruses to create a pandemic viral strain. Instead, the H3N8 seal influenza virus appears to have jumped directly from wild birds into mammals through a yet-uncharacterized “direct pipeline.”
Bluetongue and Epizootic Hemorrhagic Disease
Epizootic hemorrhagic disease is an acute, infectious, often fatal viral disease of some wild ruminants. This infection is characterized by extensive hemorrhages and has been responsible for significant epizootics in deer in the northern United States and southern Canada. A similar hemorrhagic disease called bluetongue also occurs throughout the United States and Canada. The two diseases are antigenically different (Howerth et al., 2001).
Bluetongue (BT) is a noncontagious viral disease affecting domestic and wild ruminants (primarily sheep, cattle, goats, buffalo, antelope, deer, elk, and camels) that is transmitted by insects, especially biting midges of the Culicoides species. BT has a significant global distribution in regions where this insect vector is present, including Africa, Asia, Australia, Europe, North America, and several islands in the tropics and subtropics. The virus that causes BT is identified as a member of the Reoviridae family (OIE, 2014a).
Severe infections of domestic and wild ruminants by these similar viruses have resulted in dramatic die-offs among deer and livestock, Sleeman reported. BT has been recognized for decades but is recently resurging. “There has been a dramatic extension of the [geographic range of the] virus into Northern Europe, in particular the UK and Scandinavia,” he said. “Here in the United States we are seeing more severe, widespread outbreaks in wild ruminants. The disease has been found further north than it used to be, in states like Wisconsin and Michigan. It was found in New York for the first time several years ago.”
BT’s resurgence has been influenced by several drivers, Sleeman observed, including warmer temperatures in northern Europe that have allowed the disease vector to survive at ever-higher latitudes; higher summer and winter temperatures that have increased vector capacity and competence, causing more severe outbreaks; and novel viral serotypes that have appeared in North America, probably with the arrival of exotic game or illegally imported viremic cattle, he added.
_______________
17 Due to the segmented nature of the influenza virus genome (eight individual segments of RNA), influenza viruses can undergo genetic reassortment to produce new variant strains of virus. Pigs are hypothesized to serve as the “mixing vessels” in which two influenza viruses co-infect and undergo reassortment. Source: Influenza as a zoonotic disease; zoonotic swine influenza, http://www.vetmed.wisc.edu/pbs/zoonoses/influenza/swineflu.html (accessed February 19, 2015).
Diseases at the Human–Primate Interface
The best-known pandemic disease of zoonotic origin, HIV/AIDS, is widely believed to have emerged from the spillover of the simian immunodeficiency virus through trade in bushmeat, Sleeman noted (Hahn et al., 2000; Smith et al., 2012). The international trade in bushmeat is known to be extensive, he said, and although it is difficult to precisely estimate the size of this market, about 10 tons of bushmeat are illegally imported annually into countries as small as Switzerland (Falk et al., 2013). Partnering with several U.S. government agencies and nonprofit organizations, the USGS examined bushmeat confiscated at U.S. borders by the Fish and Wildlife Service and found several novel retroviruses and herpesviruses, Sleeman reported, demonstrating bushmeat’s potential as a pipeline for pathogen spread (Smith et al., 2012).
Human diseases can also spill over into great apes, Sleeman observed. “There is a lot of concern about the potential impact of human diseases on [the] Great apes, particularly in Africa,” he said, where many Great ape populations live near densely populated human settlements with high burdens of disease. Gorillas and chimpanzees are often habituated to tourists, further increasing their vulnerability to exposures to infection with human pathogens from ecotourists, Sleeman stated. Recent research suggests that chimp die-offs have resulted from such human pathogens as respiratory syncytial virus and metapneumovirus, he said (Kondgen et al., 2008). Sleeman further observed that in her book The Chimpanzees of Gombe primatologist Jane Goodall described an outbreak of flaccid paralysis in chimpanzees that occurred simultaneously with a local outbreak of polio in the human populations around this animal reserve (Goodall, 1986).
Bats as Reservoirs of Emerging Viruses
Bats, as noted previously, have been identified as actual or potential sources of several important emerging human viral diseases, yet, New World bat populations have been severely decimated by introduced diseases including white-nose syndrome, a fungal infection that has killed at least 6 to 7 million North American bats since 2007/2008 (Bat Conservation International, 2014), Sleeman observed. Perhaps, some have reasoned, there is something unique about bats’ biodiversity or immune system that allows them to harbor these viruses. In the case of whitenose syndrome, bats are vulnerable to fungal infection during hibernation, when their immune system is quiescent, and become ill when their awakening immune system hyper-responds to the pathogen, he said—a pattern that may underlie infection by other pathogens (Meteyer et al., 2012).
As illustrated in Figure WO-4, the 2007–2008 outbreaks of Marburg virus in Uganda were associated with caves. Caves were used by the local population for mining and also tourist attractions. African fruit bats were implicated as the reservoir for this filovirus. Bats were captured and sampled and peak viral
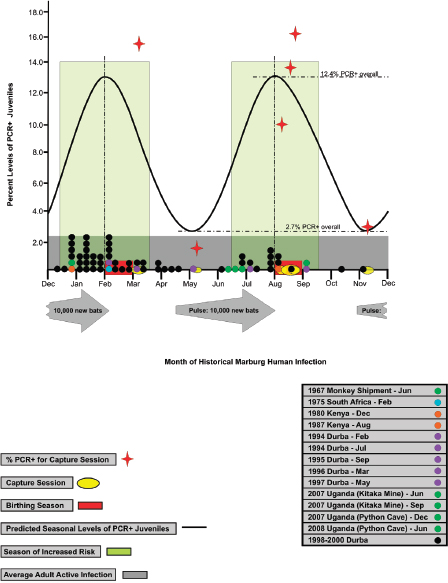
FIGURE WO-4 Increases in seasonal risk to human health. Historical spillover events (colored circles on x axis) compared to predicted seasonal levels of PCR+ juveniles (sinusoidal curve). The amplitude of the curve is based on average PCR+ juveniles experimentally determined during birthing (12.4%) and breeding (2.7%) seasons. Large light green vertical rectangles represent the proposed approximate 3-month seasons of increased risk based on the average level of juvenile infected bats at peak times of encompassing birthing (February and August) and breeding (May and November). Large gray arrows depict the twice yearly influx of newly autonomous juvenile bats born in the prior birthing season. The influx begins at the approximate time of the juvenile’s independence from their mothers.
SOURCE: Amman et al., 2012.
transmission occurred twice yearly during breeding seasons. Eighty-three percent of spillover events to humans occurred during that time (Amman et al., 2012).
When local populations were informed of the results of this investigation they responded by killing off all of the cave bats—perhaps an understandable yet tragic reaction of the local populations to this threat in their midst (Amman et al., 2014). This study also points to opportunities for human–bat coexistence by allowing mining to occur while restricting human access to the caves during the periods of peak viral shedding. Although the risk may still be considered too high, this case study illustrates the types of investigations that need to be conducted in order for indigenous wildlife and humans to coexist.
Disease Management Strategies
Interventions to reduce spillover and transmission risks offer one route to managing infectious diseases in wildlife, according to Sleeman. This includes temporal and spatial separation of humans and wildlife during high-risk periods as well as farm biosecurity, which researchers have identified as key to preventing AI outbreaks (Olson et al., 2014). Additional management strategies include improving food security in communities currently reliant on bushmeat and providing basic sanitation for human populations in close proximity to Great apes.
According to Sleeman, the larger task of preventing, preparing for, responding to, and recovering from emerging infectious diseases of wildlife origin will require the following elements:
- Basic epidemiology to respond to wildlife disease events through field investigations, laboratory diagnostics, and molecular techniques for pathogen discovery, such as deep sequencing.
- Surveillance: Can we start detecting pathogens in wildlife populations before they spill over into humans or domestic animals?
- Risk analysis to determine exposure pathways and identify hot spots for disease emergence in order to enable optimal allocation of resources.
- Monitoring: Robust existing Earth and climate monitoring systems, as well as public health monitoring, could be expanded to collect sorely needed wildlife health data, he noted. “There’s no real system of sharing that data or integrating it with public health data or domestic animal data, nationally or internationally,” he added, nor is there standard terminology for describing wildlife diseases.
- Modeling to increase understanding of the drivers of emerging diseases of wildlife origin and development of predictive models for decision support, especially Bayesian models that allow the incorporation of uncertainty into model selection.
- Tools for managing wildlife diseases, including vaccines of similar effectiveness to the oral rabies vaccine. The USGS is currently pursuing a
-
vaccine to control sylvatic plague in prairie dogs—a significant problem in the western United States (Abbott et al., 2012).
- Infrastructure, which is especially weak for wildlife health.
- Collaborative, cooperative partnerships to address health and environmental issues of mutual concern. In the United States, authority to manage a particular wildlife species or disease is not always clear, Sleeman observed, and that confusion results in delayed responses to disease threats and outbreaks. In the case of white-nose syndrome—an introduced fungal disease decimating New World bat populations—4 years elapsed between disease detection and the development of a national response plan.
The One Health approach18 provides a useful theoretical framework to address the interconnected health concerns of wild and domestic animals, humans, and the ecosystems they inhabit, Sleeman noted, but moving from this concept to implementation presents a major leadership challenge. In their recent study, Making One Health a Reality—Crossing Bureaucratic Boundaries, Sleeman and co-authors reviewed several case studies of projects involving multiple sectors in responses to such diseases as AI and anthrax in an attempt to identify factors that contributed to the success or failure of these projects (Rubin, 2014). They found that successful projects tended to include the following attributes:
- A sense of urgency and common purpose;
- A mandate or authority delegated to those who conduct the work;
- Oversight through an interagency steering committee or working group;
- A foundation of trust and a willingness to acknowledge all agencies’ concerns and perspectives;
- Mutually agreed-upon, science-based goals;
- Clearly defined roles and responsibilities; and
- Leadership rotation among all involved sectors.
On a more philosophical level, Sleeman asked a series of rhetorical questions of those who would lead the implementation of One Health:
- What are the core values of One Health?
- Is wildlife a threat to human health, or is it something that we value?
- Is this a classic team-building challenge?
- Do we have the individual leadership skills to make this successful?
- Do we have the ability to think outside the boundaries of our own agencies and our own perspectives?
_______________
18 One Health is the collaborative effort of multiple disciplines working locally, nationally, and globally to attain optimal health for people, animals, and our environment (AVMA, 2014).
- Do we have the ability to have influence when we have no real formal authority?
- Do we have the ability to compromise and reach consensus on issues?
The political, philosophical, and administrative challenges are every bit as daunting as the scientific hurdles that must be overcome in order to implement One Health, he concluded.
Eco-social Drivers of Viral Disease Emergence
Expanding on themes introduced by Fukuda and Sleeman, Dirk Pfeiffer of the Royal Veterinary College (UK) described six “global megatrends” that need to be considered when examining patterns of disease emergence (Anonymous, 2011):
- Rising living standards, as well as expectations for continued improvement in such amenities as food safety and quality will continue;
- Depletion of Earth’s limited supplies of natural mineral, energy, and water resources, which, in turn, affects food supplies;
- Increasing biodiversity loss;
- Movement of the balance of economic power toward the East and South;
- Lengthening human life span; and
- Expanding connectivity via communications and trade.
He also emphasized that it is important to appreciate the medium-level importance of the risk of pandemic disease and antimicrobial resistance within the broader context of perceived threats to human welfare, as shown in Figure WO-5—a perspective that politicians will adopt when prioritizing the allocation of limited resources, he noted (Anonymous, 2014) (Dr. Pfeiffer’s contributions may be found on pages 184–197, 197–209, 209–231, and 232–248 in Appendix A).
Briefly recapping points made by Fukuda and Sleeman, Pfeiffer noted that “eco-social changes”—phenomena such as urbanization, globalization, and land use changes—drive infectious disease emergence (see Figure WO-6). He paid particular attention to the intensification of agriculture in China, where a “food zone” in the northeast of 1,450 square kilometers19 is devoted purely to raising livestock and agricultural products, and noted the biosecurity challenges emerging from such an intensive production scenario. “There will be situations like that occurring around the world, because that often is being seen as the only mechanism of securing sufficient food supply for urban communities,” he
_______________
19 The China Jilin (Singapore) Modern Agricultural Cooperation Food Zone [in Jilin China] is an ambitious project covering 1,450 square kilometers, or 560 square miles, twice the area of Singapore. Source: http://www.nytimes.com/2010/09/28/business/global/28iht-rbofsing.html?_r=0 (accessed November 11, 2014).
FIGURE WO-5 The global risks landscape, 2014. Blue diamonds represent economic risk (e.g., fiscal crises, oil price shock, decline of the U.S. dollar). Orange diamonds represent geopolitical risks (e.g., state collapse, corruption, interstate conflict, terrorist attack). Green diamonds represent environmental risks (e.g., extreme weather events, climate change, natural catastrophes). Red diamonds represent societal risks (e.g., food crises, chronic diseases, antibiotic-resistant bacteria). Purple diamonds represent technological risks (e.g., cyber attacks, data fraud/theft). Survey respondents were asked to assess the likelihood and impact of the individual risks on a scale of 1 to 7, 1 representing a risk that is not likely to happen or have impact, and 7 a risk very likely to occur and with massive and devastating impacts.
SOURCE: Global Risks 2014, World Economic Forum, Switzerland, 2014.
FIGURE WO-6 Urbanization, globalization, and land use changes. (A) Percentage of urban population and agglomerations by size class, 2025. (B) Global pattern of meat production density (1961–2012). (C) Travel time to major cities. (D) Global forest cover loss and gain between 2000 and 2012.
SOURCE: Pfeiffer presentation, 2014; (A) http://esa.un.org/unpd/wup/Maps/CityGrowth/CityGrowth.aspx (accessed February 19, 2015); (B) data from FAOSTAT database; (C) http://www.newscientist.com/data/images/ns/cms/mg20227041.500/mg20227041.500-1_1000.jpg (accessed February 19, 2015); (D) Hansen et al., 2013. Science 342:850–853.
warned. Noting the expanding numbers of countries and communities dependent on “global value chains” for food and other goods, he observed that nonlinear development of increased numbers of linkages within and between these populations had resulted in the theoretically predicted “connectivity avalanche” that will allow rapid dispersal of infectious diseases as already observed, for example, for food-borne illness (Appel et al., 2012; Ercsey-Ravasz et al., 2012).
Pfeiffer also emphasized the need for the global community to adopt the concept of “ecosystem services,” which aims to recognize the vast benefits—including natural resources, food, climate regulation, and cultural riches—provided by the environment to human societies. Human use of natural resources threatens ecosystems worldwide, but such impacts are much greater in some places than in others, he noted (Haberl et al., 2007). As a consequence, some human populations are now exploiting distant ecosystem services to support their needs (Erb et al., 2009). The latter also contributes to increasing global connectivity.
Patterns of Emergence
The influence of eco-social change has played a role in many recent disease events, Pfeiffer emphasized. The rapid spread of foot-and-mouth disease throughout the United Kingdom in 2001, demonstrated the importance of livestock trade networks and within those particularly the role of markets as conduits for infectious disease spread. The recent expansion of viral diseases such as bluetongue and African swine fever20 into new geographic areas, and the emergence of novel viral pathogens, like Schmallenberg virus21 that was identified through an especially effective international collaboration, underscore the confluence of factors—including climate change, extreme weather events, transportation, and expanded international trade—that contribute to the expansion of these diseases into new hosts and environmental niches.
By contrast, Pfeiffer observed, most molecular clades of influenza A (H5N1) remain close to their apparent geographic origins in Southeast Asia and China—despite the availability of hosts such as migratory wild birds and exposure to global poultry and poultry-product trade networks (Pfeiffer et al., 2011). It
_______________
20 African swine fever (ASF) is a highly contagious tick-borne hemorrhagic disease of pigs, warthogs, European wild boar, and American wild pigs. With high virulence forms of the virus, ASF is characterized by high fever, loss of appetite, hemorrhages in the skin and internal organs, and death in 2–10 days on average. Mortality rates may be as high as 100 percent. ASF is caused by a DNA virus of the Asfarviridae family. Source: http://www.oie.int/fileadmin/Home/eng/Media_Center/docs/pdf/Disease_cards/ASF-en.pdf (accessed February 19, 2015).
21 In November 2011, scientists in Germany identified novel viral sequences in serum from cattle affected by a febrile syndrome that was reported during August–September 2011 in Germany and the Netherlands. Clinical signs included decreased milk production and diarrhea. The virus, named Schmallenberg virus (SBV), was isolated from blood of affected cattle, and similar clinical manifestations were observed in experimentally infected calves. In the Netherlands, SBV was detected retrospectively in serum from affected cattle in December 2011 (Reusken et al., 2012).
suggests that the characteristics of the eco-social systems in that part of the world provide more effective transmission opportunities than elsewhere. What factors underlie the apparent hot spot for AI evolution in Southeast Asia and China? “We are not absolutely certain about what the exact mechanisms are, despite all the research that has gone on,” he admitted. It is most likely the interaction between several factors, such as relatively intensive duck and chicken production traded across complex live bird marketing networks (Pfeiffer et al., 2013).
The Risk Governance Framework for Disease Management
The ecohealth or One Health approach to managing emerging viral diseases requires a systems perspective that takes into account the various drivers of emergence and places them within the context of institutions and societal goals, Pfeiffer explained. Thus, he said, traditional, linear, technocratic models of policy development solely based on translating scientific findings into policy as a “one way” street are being replaced by more interactive, cyclical models such as the “risk governance framework” depicted in Figure WO-7 (Pfeiffer, 2014). By identifying stakeholders and defining what is important to them, assessing risk with clear questions, using qualitative as well as quantitative analytical approaches, and evaluating the findings in the context of stakeholder priorities, policy makers using this framework can identify more effectively “what is to be done and how,” he said.
At the same time, Pfeiffer observed, researchers need to “embrace systems thinking and integrated research approaches . . . [and to] link biological and environmental [sciences] with [the] social sciences” in order to address the complex and dynamic impacts of global eco-social changes.
Studying Zoonoses in Their Natural Hosts
Bats, as illustrated by the flying fox in Figure WO-8, have been identified as the reservoirs of several recent emerging viral diseases (Calisher et al., 2006). As such, according to speaker John Lowenthal of the Commonwealth Scientific and Industrial Research Organisation (CSIRO) Biosecurity Flagship, bats deserve focused study (Dr. Lowenthal’s contribution may be found on pages 166–180 in Appendix A). Bats are the most abundant mammal on Earth. The members of the more than 1,200 bat species comprise about one-quarter of all mammals, he noted, yet many important questions about these mammals remain to be answered.
For example, Lowenthal observed, very little is known about bats’ response to disease. They are apparently asymptomatic carriers of viruses that threaten other animals and humans, including SARS, Hendra, rabies and other lyssaviruses, and Ebola, among more than 60 different viruses (and counting) identified to date, he said. Lowenthal remarked that it is only a matter of time until the next
FIGURE WO-7 Risk-governance framework.
SOURCE: Adapted from Renn, 2005.
novel virus emerges from bats—perhaps a new influenza A virus, as was recently detected in a bat species native to Central and South America (Tong et al., 2012).
Alternatives to Mouse Models
The containment of emerging infectious outbreaks is very challenging due to their unpredictable nature and the absence of effective medical countermeasures, such as vaccines and antivirals. The dearth of medical countermeasures is largely due to a lack of essential knowledge of the immune responses induced by zoonotic viruses, particularly the responses that are attributable to protection, Lowenthal noted. While mice have provided a useful and convenient model for understanding fundamental immune responses to infection—due to their ease of handling and rapid generation time—they may not always adequately model the behaviors of emerging infectious diseases in human hosts, he observed.
Mice “usually don’t accurately replicate the disease that is seen in the human,” Lowenthal explained. “They may allow viral replication, but a lot of the pathology and symptoms and responses are not directly relevant or related, and that creates a lot of problems. There are often differences in the symptoms of

FIGURE WO-8 Bats and emerging viruses. Bats are the most populous mammal—with more than 1,200 species representing approximately 25 percent of all classified mammal species—and are found in all regions of the world except for the North and South poles and some remote islands. Although they carry a number of zoonotic viruses without symptoms (e.g., SARS, Hendra, Nipah, and Ebola), little is known about their response to disease.
SOURCE: ©2014 EcoHealth Alliance/J. Epstein.
disease when you compare the disease in the natural host versus the transmission host versus what you see in the human,” he continued. For example, he observed, many zoonotic diseases “are asymptomatic and nonlethal in the natural host, but when they spill over into a transmission host or to a human, you can see potentially lethal effects and sometimes up to 100 percent mortality.”
Thus, nontraditional animal models—such as bats, chickens, and ferrets—are increasingly being used to study disease pathogenesis, host–pathogen relationships, and the nature of the immune responses to particular diseases, Lowenthal said. Studying immunology in nontraditional animal species might provide insights into improving the control of emerging diseases and suggest preparations for future pandemics or potential medical countermeasures against bioweapons, he added.
Ultimately, Lowenthal and fellow researchers question the use of animals other than the natural host in such studies, if that species or a close likeness is
available. For example, he noted, “Chickens have been used for a long time to study immune system development. They are a very good model for avian influenza. Bats are a host of very many different diseases. Ferrets are becoming the preferred model, and have been for a while now, for human influenza . . . [and] the woodchuck [serves] as a model for hepatitis B in humans.”
Chicken models of avian influenza Water birds, particularly ducks, are known to be the natural hosts of H5N1 influenza, which they generally carry without exhibiting symptoms, Lowenthal reported. “Infections in chickens and humans, on the other hand, can lead to very severe inflammation and very high levels of cytokine production (a phenomenon called the ‘cytokine storm’)”, he continued. “The big question is, is this aberrant immune response responsible for the high levels of mortality?”
To examine this question, one must first understand immunological differences among the species it infects. As illustrated in Figure WO-9, a duck infected with a highly pathogenic influenza strain is asymptomatic, with viral replication typically limited to the heart or to muscle tissue, coupled with a relatively low level of cytokine production, he said. The immune system has been shown to recognize the virus, but its response to it is mild. By contrast, he continued, the same virus causes a more severe reaction, involving the lung and possibly the brain, in intermediate hosts such as the pig, ferret, or mouse. Cytokine levels are elevated in these cases, but “nothing too serious,” he noted. In more susceptible hosts such as humans and chickens, however, infection is severe, spreads rapidly, and produces a cytokine storm (Bean et al., 2013).
The mechanisms underlying these differences could reveal novel approaches to mitigating influenza’s effects, Lowenthal observed. “It would be very useful to compare what is happening in terms of immunological responses in the natural host, which is resistant to the transmission host, and try to identify what the key differences and what the underlying differences are,” he explained. “If we can identify [the] factors that trigger an inappropriate inflammatory response that might inform us of therapeutic intervention strategies for human disease.” It has already been demonstrated that humans can be protected from infectious disease by treating transmission hosts to prevent them from spreading pathogens to humans, he reported, as was accomplished with an equine vaccine against Hendra virus that protected horses, and thus humans, from infection. (See Lowenthal original manuscript on pages 166–180.) This strategy could potentially be used to prevent human infection by other zoonotic viruses, he concluded.
Nontraditional Host Studies at the Australian Animal Health Laboratory
“There are lots of restraints and certain requirements in working with nontraditional animal species, particularly for highly pathogenic agents,” Lowenthal observed. Special high biocontainment facilities and trained staff are necessary, and a variety of animal husbandry and welfare issues relevant to the capture,
FIGURE WO-9 The host immune response to an infection influences the disease outcome. Infection with H5N1 influenza virus can cause very different disease outcomes in different reservoir and spillover host species. Waterfowl, such as wild ducks, are the natural host for this virus and develop a limited inflammatory response that is associated with low levels of cytokine expression. Intermediate hosts, including mice, pigs, and ferrets, are often used to study this infection and display mild to severe disease symptoms (depending on the H5N1 virus strain used) that are associated with increased levels of proinflammatory cytokines. By contrast, spillover hosts such as chickens and humans display a rapid and strong inflammatory response, often referred to as hypercytokinemia (or cytokine storm) and the infection becomes systemic, causing severe disease symptoms and high mortality rates.
SOURCE: Bean et al., 2013.
acclimatization, and breeding of wild animals must be addressed, he explained. In addition, immunological reagents for many nontraditional species are either scarce or nonexistent. Many of these hurdles have been overcome at the CSIRO Australian Animal Health Laboratory (AAHL), one of the few laboratories in the world that offers a wide range of different nontraditional animal species, including those of large animals, in expansive, sophisticated containment facilities, he said. “We have the whole farmyard covered,” he quipped.
In the AAHL’s Biosecure Immunology Laboratory, researchers can undertake comparative immunology across a spectrum of nontraditional species under biosafety level (BSL) 3 and 4 conditions,22 using state-of-the-art cell sorting and flow cytometry, Lowenthal stated. “We’re developing an immunological toolbox and cell lines for a number of these different species,” he continued; this will enable the use of high-throughput gene silencing to knock out individual immune genes, so as to measure their effects on the immune response to specific viruses in both natural and susceptible hosts. “We would love to be able to get a knockout bat or a knockout ferret,” he said. “I don’t think we’re too far away from doing that.”
Studying the immune response to a zoonotic pathogen in its natural reservoir species and comparing that with the response in spillover or transmission hosts will identify key processes and factors in disease susceptibility and transmission, Lowenthal stated. “Together with new technologies, such as gene knockout technology, we can identify new strategies to prevent and minimize the impact of emerging infectious diseases and enhance our pandemic preparedness,” he concluded.
_______________
22 BSL 3 is applicable to clinical, diagnostic, teaching, research, or production facilities where work is performed with indigenous or exotic agents that may cause serious or potentially lethal disease through the inhalation route of exposure. Laboratory personnel must receive specific training in handling pathogenic and potentially lethal agents, and must be supervised by scientists competent in handling infectious agents and associated procedures. All procedures involving the manipulation of infectious materials must be conducted within biological safety cabinets or other physical containment devices. A BSL-3 laboratory has special engineering and design features.
BSL 4 is required for work with dangerous and exotic agents that pose a high individual risk of aerosol-transmitted laboratory infections and life-threatening disease that is frequently fatal, for which there are no vaccines or treatments, or a related agent with unknown risk of transmission. Agents with a close or identical antigenic relationship to agents requiring BSL-4 containment must be handled at this level until sufficient data are obtained either to confirm continued work at this level, or redesignate the level. Laboratory staff must have specific and thorough training in handling extremely hazardous infectious agents. Laboratory staff must understand the primary and secondary containment functions of standard and special practices, containment equipment, and laboratory design characteristics. All laboratory staff and supervisors must be competent in handling agents and procedures requiring BSL-4 containment. The laboratory supervisor in accordance with institutional policies controls access to the laboratory. For further details of the BSL-3 and BSL-4 requirements please see the Biosafety in Microbiological and Biomedical Laboratories 5th Edition, http://www.cdc.gov/biosafety/publications/bmbl5 (accessed February 19, 2015). Source: HHS et al., 2009.
The NIAID Response to Emerging Viral Diseases
The National Institute of Allergy and Infectious Diseases (NIAID) funds a significant proportion of the nation’s efforts to address emerging and reemerging viral diseases, according to speaker Anthony Fauci, who directs the institute, and who described those efforts in his workshop presentation (Dr. Fauci’s contributions may be found on pages 133–136, 136–143, and 144–152 in Appendix A). NIAID undertakes a dual mandate unmatched by other National Institutes of Health (NIH) agencies, he noted: It must maintain and grow a portfolio of basic and applied research on EIDs, and rapidly respond to new and emerging threats.
Over the course of human history, extraordinary progress has been made in the control of infectious diseases, Fauci observed, noting the following key advances:
- The recognition that infections are caused by microbes;
- Improvements in sanitation, hygiene, and vector control;
- The discovery and implementation of antimicrobials;
- The development of vaccines and vaccination programs; and
- The development of diagnostics, enabling disease detection and monitoring.
With these breakthroughs, however, came a sense of complacency, illustrated by the following quote from the 1963 book The Evolution and Eradication of Infectious Diseases, by Aidan Cockburn:
We can look forward with confidence to a considerable degree of freedom from infectious diseases at a time not too far in the future. Indeed . . . it seems reasonable to anticipate that within some measurable time . . . all the major infections will have disappeared. (Cockburn, 1963)
This quote,23 Fauci said, also reflects “the extraordinary provincialism that we have in the developed world to have the temerity to say that we’re going to be free of infectious diseases when you have malaria, tuberculosis, and diseases that cause millions of deaths throughout the world.”
HIV: From Emerging Disease to Established Infection
NIAID had been studying emergent diseases for many years before HIV. Its efforts accelerated in 1981 with “the mother of all emerging and reemerging infectious diseases—HIV/AIDS,” Fauci recalled. “As devastating as this pandemic
_______________
23 About this quote, Fauci also noted that for many years, he and others had instead quoted U.S. Surgeon General William Stewart, who is claimed to have stated in 1967 or 1969, depending on the source, “It is time to close the book on infectious diseases, and declare the war against pestilence won” (Spellberg, 2008, p. 1). However, despite concerted efforts of Fauci’s scientific staff and many other individuals, including historians of public health, no primary source for this quote has been identified.
has been, it is really an extraordinary model for what happens when the public health and research communities mobilize with extraordinary resources to address an emerging public health threat.”
In the ensuing years, some 70 million people have been infected with HIV, resulting in 36 million deaths; more than 35 million people are currently living with the disease, Fauci reported. At the same time, collaboration between fundamental basic researchers, industry, clinical researchers in clinical trials, grantees, and contractors have produced more than 30 antiretroviral drugs that have transformed the lives of HIV-infected individuals, rendering an acute infectious disease that once killed an infected individual in less than a year into a chronic, manageable, illness. “Right now, if you go to any reasonable clinic in the developed world, and even in the developing world, and someone comes in who is 20-plus years old, recently infected with HIV, and you put them on triple combination,24 you could predict with accuracy and look them in the eye and honestly say, ‘If you take your medicine, you [are likely to] . . . live an additional 50 years,’” he said (Samji et al., 2013).
Implementing these interventions represents “another extraordinary accomplishment,” Fauci continued. Supported by the President’s Emergency Plan for AIDS Relief,25 the Global Fund,26 various philanthropies, and the governments of affected countries, deaths due to AIDS have declined by 30 percent since 2005, he said. Much of this improvement is due to a suite of nonvaccine preventive measures, including condom use, needle exchange, medical interventions to prevent mother-to-child transmission, circumcision, and prevention as treatment, he noted. Even so, he added, “It is far too soon for a victory lap. We have much to do in implementation, and also in discovery, such as with a vaccine or a cure.”
From SARS to MERS
The short-lived SARS pandemic, which consisted of approximately 8,000 cases resulting in 800 deaths before it was extinguished by “simple 19th-century public health measures,” introduced a new era of vaccine development, according to Fauci. Rapid sequencing platforms sped the characterization of SARS-CoV, showing that a safe and effective vaccine (which in the end was not needed) could be developed against a novel pathogen within a year of its discovery, he said.
“No sooner did we deal with one coronavirus . . . than we had another,” Fauci said, introducing the multipronged NIAID response to MERS, launched at a meeting in June 2013 (NIAID, 2013). The institute’s research response to the MERS-CoV is a rapid response capability similar to that applied to SARS, he explained, and focused on basic research; on developing animal models, vaccines,
_______________
24 Any antiretroviral regimen composed of three agents from at least two drug classes used to manage HIV infection, also known as highly active antiretroviral therapy (Henkel, 1998).
25 See http://www.pepfar.gov.
and therapeutics; and on identifying animal reservoirs. For example, he noted, a NIAID-supported animal model has been used to demonstrate that interferon-α and ribavirin are effective against MERS-CoV (Zhao et al., 2014)—a good start, he said, but “we likely need to do better than that.”
Similarly, NIAID supported the recent isolation of MERS-CoV from a large number of camels identical to viruses isolated from MERS patients—very much identical to the coronavirus of MERS, Fauci noted. However, it is still unclear whether camels have infected humans or vice versa, he added—a question NIAID researchers continue to investigate.
Reemerging Viruses
Among the many reemerging or resurging viral diseases, Fauci identified dengue27—a leading cause of illness and death in the tropics and subtropics—as being particularly important. Cases have increased sharply over the past 60 years, and the disease reemerged in the Caribbean islands and southern Florida;28 locally acquired cases had been absent in Florida since 1934. NIAID’s research approach to dengue encompasses fundamental basic research, vector biology, sharing research resources with the scientific community, and pursuing vaccines, therapeutics, and diagnostics, he said.
Another reemerging vector-borne virus, chikungunya, is also transmitted by more than one species of Aedes mosquitoes. Outbreaks of this debilitating, usually nonfatal disease have significant public health impact, and there are as yet no licensed vaccines or specific treatments for it, Fauci observed. Since crossing the Atlantic Ocean in December 2013, chikungunya has spread to more than 10 countries in the Americas and produced more than 780,000 suspected and 15,000
_______________
27 Dengue is caused by any one of four related viruses transmitted by mosquitoes, which infect as many as 400 million people each year. There are not yet any vaccines to prevent infection with dengue virus; the most effective protective measures are those that avoid mosquito bites. Early recognition of infection and prompt supportive treatment can substantially lower the risk of medical complications and death. Dengue emerged as a worldwide problem beginning in the 1950s. To date it has rarely occurred in the continental United States, but it is endemic in Puerto Rico and in Latin America, Southeast Asia, and the Pacific islands. Source: http://www.cdc.gov/dengue (accessed September 4, 2014).
28 Between 1946 and 1980, there were no reported cases of dengue acquired in the continental United States, and, according to the CDC, there hasn’t been an outbreak in Florida since 1934. However, in 2009, the first locally acquired case of dengue in the continental United States (other than those associated with outbreaks on the Texas–Mexico border) was detected in Key West, Florida. This outbreak was followed by several additional local cases (CDC, 2010). By the end of 2009, 27 cases of dengue infection had been confirmed in Key West residents. As of the end of June 2010, an additional 12 cases of locally acquired dengue had been reported in Key West and surrounding areas (Preidt, 2010). According to Dr. Harold Margolis, chief of the dengue branch at the U.S. Centers for Disease Control and Prevention, “[t]hese cases (in Key West) represent the re-emergence of dengue fever in Florida and elsewhere in the United States after 75 years. These people had not traveled outside of Florida, so we need to determine if these cases are an isolated occurrence or if dengue has once again become endemic in the continental United States” (Preidt, 2010).
laboratory-confirmed cases (CDC, 2014; Morens and Fauci, 2014). NIAID has pursued vaccine development to address this threat, he reported, and in November 2013, it completed a phase I trial of a candidate vaccine that appears to be both safe and immunogenic (Chang et al., 2014).
Prospects for a Universal Influenza Vaccine
Influenza is both a reemerging, due to its seasonality, and an emerging infectious disease, because novel influenza viruses with pandemic potential have arisen several times in the past century, Fauci observed. Human cases of AI H5N1 were first detected in 1997. Further spread was halted at the time through the mass slaughter of poultry in Hong Kong. Since 2003, the disease has smoldered in China when NIAID began tracking cases. Similarly, transmission of AI H7N9 continues within the live markets of China following detection of the first human cases in March 2013.
A series of challenges complicates efforts to address the threat of influenza, Fauci noted:
- Neither infection nor vaccination results in lifelong immunity.
- Seasonal strains inevitably exhibit genetic drift, necessitating a “timetable” approach to vaccine development.
- Seasonal strains for vaccine production must be predicted well in advance, providing a “best guess” rather than a precise match,
- The annual cost of preparing seasonal influenza vaccines is $2–$4 billion.
- Vaccines cannot be stockpiled for years ahead of time.
- Pandemic emergence is an ongoing threat.
Given these concerns, he asked, “Should we put on a full-court press to develop a universal influenza vaccine?”
Such a possibility has only recently become viable, thanks to advances in deep sequencing, structural biology, and crystallography, Fauci said. All of these technologies are being brought to bear in order to design antibodies that target the shared stem region of otherwise diverse influenza hemagglutinin molecules, as illustrated in Figure WO-10. “If you get an antibody to the stem, the antibody can find the stem, but the stem is not exposed enough to induce a very good antibody,” he explained. Researchers have now discovered multiple ways to “show” the stem to the immune system, thereby inducing potent antibodies against it—an advance Fauci called “the first good step towards developing a universal influenza vaccine.”
In this effort, as in all of its work on infectious diseases, NIAID focuses on three main goals, Fauci said:
- Supporting fundamental basic research;
- Producing resources to advance research; and
- Translating basic findings into clinical research with the ultimate goal of developing diagnostics, therapeutics, and vaccines.
In the case of emerging and reemerging infectious diseases, the microbes by their nature ensure that this will be “a perpetual and never-ending challenge,” he concluded.
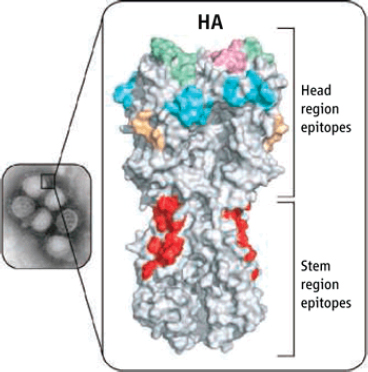
FIGURE WO-10 Antibody recognition. Most antibodies against influenza A virus (inset shows the 2009 H1N1 strain) bind to the highly variable part of the hemagglutinin (HA) glycoprotein at the surface of the virus particle (head region). In the H1 subtype, these antibodies recognize four major sites (Sa, Sb, Ca, and Cb are shown in green, pink, cyan, and yellow, respectively). The HA structure of the 2009 H1N1 virus is shown (PDB code 3LZG). Antibodies that neutralize multiple strains both within a virus subtype and from different subtypes bind to a highly conserved region (red) in the stem region of HA.
SOURCE: CDC (Inset); D. Ekiert, I. Wilson in Doms, 2010.
EMERGENCE OF MERS-COV
At the time of the workshop, in mid-March 2014 (and also at the time of this publication, nearly 1 year later) many more questions had been posed than answered about MERS-CoV in the 2 years since its initial identification as the cause of a novel viral respiratory disease syndrome.29 Globally, 1,026 laboratory-confirmed cases of infection with MERS-CoV including at least 376 related deaths have officially been reported to WHO as of February 23, 2015. About two-thirds of these cases are male, and the median age is 49 years old (9 months to 94 years old) (WHO, 2014f).
A β-coronavirus, MERS-CoV is a member of the large viral family that includes the SARS coronavirus as well as viruses that cause the common cold (WHO, 2014b). MERS-CoV appears to be circulating widely throughout the Arabian Peninsula, where all primary cases to date apparently became infected. While some secondary cases of MERS—including several large hospital outbreaks30—have arisen, the virus does not appear to be readily transmissible (WHO, 2014b). Intensive screening of MERS-CoV contacts revealed very few instances of household transmission (WHO, 2014d). Secondary cases tend to present with a milder disease than primary cases, and many of the recently reported secondary cases have been mild, or were people whose tests were positive for MERS-CoV but were asymptomatic.
According to WHO, a typical MERS patient presents with fever, cough, and shortness of breath, and is often found to have pneumonia. Some patients also experience gastrointestinal symptoms, including diarrhea. Severe illness can cause respiratory failure that requires mechanical ventilation and support in an intensive care unit. Some patients have had organ failure, especially of the kidneys, or septic shock. Nearly one-third of patients with laboratory-confirmed MERS-CoV have died. The virus appears to cause more severe disease in people with weakened immune systems, older people, and those with chronic comorbidities including diabetes, cancer, and chronic lung disease (WHO, 2014b).
How people become infected with MERS-CoV has yet to be determined. As is discussed below, strains of MERS-CoV that match human strains and antibodies to MERS-CoV have been isolated from camels in Africa and the Middle East, and strong similarity has found between viruses isolated from humans, camels, and bats (Haagmans et al., 2013; Memish et al., 2013; WHO, 2014b). Although camels are suspected to be the primary source of infection for humans, the routes of direct or indirect transmission remain unknown and investigations are ongoing.
_______________
29 A syndrome, in medicine and psychology, is the collection of signs and symptoms that are observed in, and characteristic of, a single condition.
30 In May 2014, WHO reported that the number of laboratory-confirmed MERS-CoV infections had risen sharply since mid-March, largely due to health care–associated outbreaks that occurred in Saudi Arabia and in the United Arab Emirates (WHO, 2014d).
“Further epidemiological investigations are urgently needed to confirm or refute these hypotheses,” according to WHO (2014d).
The five workshop presentations, summarized below, add considerable and intriguing detail to the current state of knowledge about MERS-CoV. They also reveal a multitude of pressing questions to be pursued, such as this list compiled by speaker Linda Saif, of Ohio State University:
- How was MERS-CoV transmitted to humans?
- What circumstances promote interspecies transmission of the virus?
- Are bats the only persistent animal reservoir for MERS-CoV?
- What is the role of camels: intermediate host or reservoir community?
- Are there subclinical MERS-CoV infections in humans?
- Why is there enhanced disease severity in people with comorbidities and the elderly?
- What is the role of cofactors and treatments (e.g., co-infections, antibiotics, corticosteroids) in enhancing the severity of MERS in people with comorbidities?
- Are there superspreading events as per SARS?
- What is the role of nonrespiratory viral shedding routes (feces, urine) in the transmission and pathogenesis of MERS-CoV (in both humans and camels)?
Human Coronavirus Emergence and Cross-Species Adaptation
Ralph Baric, of the University of North Carolina, opened the workshop session with a general description of coronavirus structure, as illustrated in Figure WO-11. Focusing on the spike (S) glycoprotein, which is embedded in the lipid bilayer surrounding the nucleocapsid, he noted that it mediates viral binding to host receptors and encodes critical determinants for cross-species transmission. The S glycoproteins of SARS-CoV and of another human coronavirus, known as HCoV-NL63, bind angiotensin I converting enzyme 2 (ACE2) on the surface of host cells for docking and entry; their more distant relative MERS-CoV recognizes instead dipeptidyl peptidase 4 (DPP4). “The spike glycoprotein is the major target for vaccine development because it encodes neutralizing epitopes, as well as T-cell epitopes, and it encodes the major component of protective immunity,” he explained.
MERS-CoV is the sixth animal coronavirus to have emerged in humans, according to Baric. Two other human coronaviruses, the closely related31 HCoVOC43 and HCoV-HKU1 strains, likely originated in mice and cattle, he said. Baric went on to observe that SARS-CoV, along with two additional closely related strains, HCoV-NL63 and HCoV-229E, are believed to be of bat origin, and
_______________
31 Based on spike protein sequence.
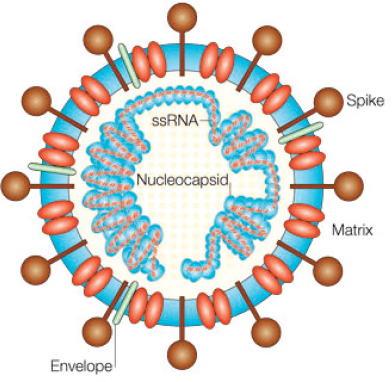
FIGURE WO-11 SARS-CoV virion—ssRNA, single-stranded RNA.
SOURCE: Dandekar and Perlman, 2005.
the closest known relatives to MERS-CoV are two viruses that have been isolated from bats. The emergence of MERS-CoV continues an accelerating pattern of cross-species transmission and emergence among coronaviruses: while the first human coronavirus appears to have emerged from bats more than 500 years ago, the vast majority of human strains either emerged suddenly (SARS-CoV, MERS-CoV) or were identified within the past 12 years (HCoV-NL63, HCoV HKU1), according to Baric (Graham et al., 2013; Huynh et al., 2012). In parallel, a number of new animal Nidovirales emerged over the past 30+ years, including porcine epidemic diarrhea virus, porcine respiratory coronavirus, porcine reproductive and respiratory disease virus, and bovine respiratory coronavirus (Graham et al., 2013), providing additional support for the hypothesis that cross-species transmission events are frequent among the Coronaviridae.
Mechanisms of Interspecies Transmission
Two distinct mechanisms enable the interspecies transmission (also known as “spillover”) of coronaviruses, Baric explained. The first is the ability of some
coronavirus S glycoproteins to bind analogous receptor proteins (receptor orthologs) in species other than their primary host (Bolles et al., 2011). These viruses, which include SARS-CoV and MERS-CoV, are capable of replicating in multiple hosts, typically clusters of species. Other coronaviruses with an even broader host range possess mutations in the S glycoprotein gene that render them (a) capable of recognizing receptor orthologs, (b) “preprogrammed” to fuse a variety of host cell proteins (Graham and Baric, 2010), or (c) easily mutatable, in the course of cell culture, to recognize heparin sulfate as a receptor for docking and entry to host cells, which vastly extends their host range (de Haan et al., 2005). These latter two mechanisms of host range expansion have only been identified using in vitro models, although the potential exists for similar mutants to emerge naturally in nature. Finally, several coronaviruses can use sugar moieties as receptors or co-receptors for entry, providing an alternative strategy for rapid trans-species movement (Li, 2013).
Many believe that the 2003 SARS epidemic resulted from the emergence of a bat-like coronavirus that also recognized the ACE2 receptor from civets, Baric recalled. Then, through a second rare mutational step, the civet-adapted virus acquired the ability to use human ACE2 for docking and entry. The resulting human-adapted strain was then thought to have circulated back and forth between civets and humans to mediate the expanding outbreak. The civet as an amplifying host in the open markets was clearly associated with the expanding epidemic. However, he continued, another, perhaps more likely, explanation for the initial emergence event was that bat SARS-like coronaviruses are naturally capable of recognizing ACE2 receptors from multiple species, including humans, civets, and a subset of other species (Graham et al., 2013). Once these generalists infect across species, additional mutations arose that permitted efficient cross-transmission between humans and civets as well as more efficient human-to-human transmission.
This scenario raises the prospect that additional coronaviruses will follow this path to emergence in humans—or in the reverse direction. “Can bat SARS-like coronaviruses use human ACE2 for docking and entry into human cells?” Baric wondered. Conversely, he asked, can human coronaviruses, such as epidemic SARS-CoV and HCoV-NL63, recognize bat ACE2 molecules? Interestingly, HCoV-NL63 is capable of replicating efficiently in select bat cell lines from North America raising the possibility of cross-species movement between human strains and bat species (Huynh et al., 2012). Researchers recently identified a cluster of SARS-like viruses in Chinese horseshoe bats, including two that were more than 99 percent homologous across the majority of their genome sequences with SARS-CoV, but only 90 to 95 percent homologous within their S glycoprotein sequence (Ge et al., 2013). “Our results provide the strongest evidence to date that Chinese horseshoe bats are natural reservoirs of SARS-CoV, and that intermediate hosts may not be necessary for direct human infection by some bat SL-CoVs [SARS-like coronaviruses],” the authors wrote.
Ge and coworkers (2013) were able to isolate one of these SL-CoVs and show that it could bind to human, civet, and bat ACE2 molecules. Baric and coworkers then replaced S protein in a molecular clone of SARS-CoV with S glycoproteins synthesized from two bat SL-CoV sequences and found that the resulting hybrid viruses could also replicate efficiently in cultured primary human airway epithelial lung cells to high titer (unpublished observation). “In essence, these two bat coronavirus spike glycoproteins—that, as far as we can tell, have never circulated through human populations—allow for extremely efficient replication in one of the primary targets for SARS coronavirus replication in humans,” he observed. “Both of these [viruses] are, in essence, poised to emerge in human populations.” There are 1,200 to 1,300 different bat species, each of which encodes its own versions of the major coronavirus receptor-proteins, including aminopeptidase N, ACE2, and DPP4—and each host species can support an estimated 7 to 10 different coronaviruses, he continued. All of this adds up, he concluded, to “a heck of a lot of epidemic potential.”
Routes to Emergence
Investigators in Baric’s lab sought to characterize bat coronaviruses in North America by surveying about 500 different bat species. One of the viruses, when isolated and sequenced, proved to be the closest known relative of HCoV-NL63 (Huynh et al., 2012). They also determined that both HCoV-NL63 and, less efficiently, SARS-CoV and a civet coronavirus, HCSC6103, were capable of infecting and replicating in these North American primary bat lung cells, and therefore probably recognize ACE2-like molecules from a range of mammalian hosts. “These observations support the hypothesis that human coronaviruses are capable of establishing zoonotic–reverse zoonotic transmission cycles that may allow some coronaviruses to readily circulate and exchange genetic material between strains found in bats and other mammals, including humans,” the authors concluded (Huynh et al., 2012).
In what Baric termed the “classic model” of viral emergence, zoonotic RNA viruses—which have high error frequencies—produce a “quasispecies” or a random swarm of heterologous mutants and some encode mutations that enable host range expansion when another appropriate warm-blooded host (such as a human) comes into contact with their primary host species. If more mutations arise within the new host that increase transmission efficiency within that species, a disease outbreak may be more likely. However, the possibility that some zoonotic coronaviruses are “programmed” to infect other species, as previously described and as suggested by the work of Huynh et al. (2012) and Ge et al. (2013), streamlines the route to viral emergence. “You don’t need a random mutation,” Baric observed. “They can immediately jump into different species, like humans, and move back and forth—after which, of course, [an additional few mutation(s)] might be required . . . to [establish] severe disease and transmission and pandemic potential.”
In addition to error-prone transcription typical of RNA viruses, coronaviruses encode two unique genetic factors that further promote variation, Baric noted. The first is a replication strategy that produces high rates of RNA recombination during mixed infections. “If you infect the cell with two different but closely related coronaviruses, up to one-third of the progeny that come out of those cells may be recombinants containing genome material from both parents,” he stated. The second variation-enhancing feature is an enzymatic proofreading activity called ExoN, which functions as a novel RNA fidelity proof-reading complex that is unique to coronaviruses (Denison et al., 2011; Graham et al., 2012). Baric speculated that this proof-reading function was probably under tight regulatory control, allowing for fluctuations in fidelity. Thus, the newly emerged virus could adapt quickly by relaxing fidelity control and then tightening fidelity control to stabilize these adaptive mutations in the new host species.
Host age has also been identified as a factor in regulating the cross-species transmission of coronaviruses, Baric added. In mice infected with SARS-CoV, Baric noted that the virus will replicate efficiently but not cause disease. A mouse-adapted virus—with mutations in the S and membrane (M) glycoproteins and the viral replicase resembling those that arose in humans during the 2003 SARS epidemic—may be created by passaging32 the virus multiple times through mice. Baric explained that this adaptation process requires 15–25 passages at 2-day intervals in young mice, which then develop mild alveolitis33 and bronchiolitis.34 In 1-year-old mice, only three to four passages are required for an adapted virus with S protein mutations to induce an acute, lethal, respiratory distress syndrome and end-stage lung disease (unpublished observation). About 6–9 or 1 mutation is needed for increased virulence in young and aged animals, respectively. This pattern, according to Baric, strongly resembles the progress of SARS and MERS in humans, suggesting that host age not only influences pathogenesis, but could also enhance animal-to-human cross-species transmission. It is possible that sufficient human-to-human and animal-to-human transmission events have occurred in the Middle East to model the role of aging in MERS-CoV transmission.
Characterizing MERS-CoV
Given the pandemic potential of coronaviruses, the emergence of a novel human virus, MERS-CoV, within a decade of the SARS epidemic is not surprising. Like SARS, MERS is a β-coronavirus that belongs to a phylogenetic group that includes a large number of bat viruses. That MERS-CoV can infect Pipistrellus bats and camels, as well as humans—via DPP4—raises an important question
_______________
32 Transferring some or all cells from a previous culture to a fresh growth medium. Subculture is used to prolong the life and/or expand the member of cells or microorganisms in culture. Source: cell passage; www.ruf.rice.edu (accessed February 26, 2015).
33 An inflammation of the alveoli of the lungs caused by the inhalation of an allergen.
34 An acute viral infection of the small air passages of the lungs called the bronchioles.
for vaccine development, Baric noted: Would a vaccine that was effective against MERS-CoV protect against other viruses that recognized DPP4? It was recently reported by two groups that another group 2c β-coronavirus, BtCoV HKU4, can also use human and bat DPP4 as receptors for docking and entry, but that efficient entry into human cells is limited by the availability of downstream S glycoprotein proteolytic processing, which is needed for virion fusion and entry (Wang et al., 2014; Yang et al., 2014).
To examine the antigenic variation among coronaviruses, investigators in Baric’s laboratory expressed S glycoproteins from the three known group 2c coronaviruses and most of the reported group 2b coronaviruses. Antisera against MERS-CoV S glycoprotein were capable of neutralizing two different human MERS-CoV strains. However, antisera against the group 2c MERS-like bat viruses HKU4 and HKU5 could not neutralize MERS-CoV or SARS-CoV. Likewise, antisera against closely related group 2b bat coronaviruses could not neutralize SARS-CoV or MERS-CoV (Agnihothram et al., 2014). These data led the investigators to conclude that sufficient “antigenic space” exists within the group 2b and 2c gene clusters to allow for three or more antigenically unique coronaviruses to emerge, he said. It is, therefore, not surprising that the previously mentioned pair of novel SARS-like bat viruses isolated by Ge et al. (2013) evade vaccines and immunotherapeutics that were developed against SARS-CoV, especially in highly vulnerable aged animals, he added.
In another effort to characterize MERS-CoV, Baric and coworkers synthesized a full-length cDNA clone of the coronavirus and used it to reconstitute virus that functioned similarly to the wild-type isolate (Scobey et al., 2013). Using this recombinant virus, tagged with a fluorescent protein, they demonstrated that MERS-CoV replicates preferentially in differentiated primary lung cells, like nonciliated bronchial epithelial cells, type II pneumocytes, and endothelial cells, and, therefore, shows much broader tissue tropism than SARS-CoV.
Unfortunately, Baric observed, MERS-CoV does not replicate in mice, ferrets, or guinea pigs, all of which are frequently used as small animal models for immunological studies. As he and his collaborators discovered, this incompatibility results from differences in the receptor interface between MERS-CoV and DPP4 among these species, which ultimately hinge on 1 or 2 amino acid differences and the presence of a glycosylation site in the small animal DPP4 interface sequence (Cockrell et al., 2014). These seemingly subtle distinctions mean that it will be difficult (but not impossible) to make a mouse-adapted strain of MERS-CoV, he said, and only slightly less difficult to use the guinea pig instead.
To get around this obstacle, Zhao and coworkers (2014) made an adenovirus gene therapy vector that encoded the human DPP4 receptor, transduced the lung of mice, then infected with MERS coronavirus, which yielded a viral replication model without serious disease. Taking a different approach, Baric and coworkers created a surrogate model for MERS-CoV that replicates in mice (Zhao et al., 2014). To do this, they synthesized the full-length genome of the MERS-like bat coronavirus HKU5—that can replicate in human cells, but cannot spread between
them—and then replaced its S glycoprotein with that of SARS-CoV. This recombinant virus was able to replicate well in the same tissues as SARS-CoV, he noted. The recombinant virus also produces severe disease, to which older animals are much more vulnerable. He anticipated using this surrogate MERS-CoV to test both therapeutics and vaccines targeting non-S glycoprotein antigens.
This is important, he explained, because NIH spent nearly $40 million on two killed SARS vaccines early in the epidemic that induced a strong immune response that protected in young and to a lesser extent in aged animals. Unfortunately, a Th2 immune response to the nucleocapsid protein enhanced immune pathology and eosinophilia, which may result in enhanced disease in some vaccines. These types of immune complications are often revealed in a robust animal model that recapitulates the human disease phenotypes. Curiously, Baric added, “when you do this experiment using the HKU5 challenge virus with animals vaccinated against either the HKU5 or the MERS nucleocapsid protein, there’s no increase in the number of eosinophils.” Perhaps, then, the pathology associated with the SARS vaccines might not recur with MERS-based vaccines, he suggested.
Pathogenic Potential of Coronaviruses
“Coronaviruses do two novel things to innate sensing and signaling programs to promote their disease potential,” Baric stated, reflecting on comparisons of human lung cell responses to SARS- and MERS-CoV, and to H1N1 and H5N1 influenza viruses. After infecting airway epithelial cells with these viruses, the researchers monitored interferon-stimulated genes that are important in establishing cell-intrinsic immunity and antiviral defense and discovered that approximately 150 interferon-stimulated genes (ISGs) are quickly turned on in human airway cells. Most of these ISGs are also strongly induced within 3 to 7 hours in airway cells treated with low-pathology H1N1 influenza virus, he added, whereas high-pathology H5N1 influenza turns on only a subset of these ISGs, and turns off many others. This suggests that H5N1 “has some additional trick to down-regulate cell-intrinsic immune responses to allow for more efficient replication,” he speculated.
Even more intriguing, “If you infect [airway] cells with SARS, for the first 12 to 24 hours, you see no measurable ISG response,” Baric observed; some ISGs are activated at 24 hours, and most, but not all, follow. Meanwhile, however, SARS reaches peak titers between 24 and 30 hours, “so by the time the cell-intrinsic defense mechanism gets turned on, coronaviruses are done with the cell,” he concluded. “MERS does the same thing. It has this huge delay in cell intrinsic immune recognition and ISG induction. It also downregulates a subset of these ISGs, just like H5N1, so that they never get turned on after infection. The mechanism underlying this response is probably epigenetically regulated,” he added, and it allows both the SARS-CoV and high-pathology H5N1 influenza virus to manipulate host cell intrinsic response, thereby increasing disease severity.
Lessons from Animal Coronaviruses
Both α- and β-coronaviruses are known to infect a range of species, and some—such as the β-coronavirus MERS-CoV—appear to be host range mutants that evolved through interspecies transmission and adaptation, as Baric described. Known species of the other two genera, gamma- and deltacoronaviruses, are largely avian viruses. Saif provided further context for understanding MERS-CoV through her discussion of coronaviruses known to cause disease in domestic livestock.
The α-coronaviruses—transmissible gastroenteritis epidemic virus (TGEV) and porcine epidemic diarrhea virus (PEDV), along with the feline infectious peritonitis virus (FIPV) and the canine coronavirus (CCoV)—appear to have evolved from a common ancestor (Le Poder, 2011), Saif noted. Such closely related viruses may have altered cell or tissue tropisms, enabling interspecies recombination events that drive genomic modification (e.g., of the spike glycoprotein gene). As summarized in Table WO-2, coronavirus variants have arisen through diverse mutational routes, which suggests that they have multiple ways of adapting to infect new species or tissues, she added.
Emerging Porcine Coronaviruses
Saif described three emerging α-coronaviruses of swine: TGEV, PEDV, and porcine respiratory coronavirus (PRCV) (see Table WO-3). TGEV and PEDV circulate in U.S. herds, causing intestinal infections and high mortality in seronegative piglets. PRCV, a spike glycoprotein gene deletion mutant of TGEV, instead infects the upper and lower respiratory tract, where it causes an atypical pneumonia that resembles SARS.
PEDV, TGEV, and PRCV all share the same aminopeptidase N receptor, but she explained that the loss of a sialic acid-binding spike region in PRCV apparently disabled it from binding to mucins associated with binding of TGEV strains in the gut. In laboratory studies, PRCV was found to induce partial immunity to TGEV, but when PRCV circulates as an endemic among swine populations, it induces repeat infections and the development of widespread herd immunity to TGEV, she noted. Although both are α-coronaviruses, PEDV and TGEV do not induce cross-neutralizing antibodies and do not cross-protect.
In April 2013, a virulent PEDV strain emerged in the United States as a highly fatal diarrheal disease in baby pigs, as illustrated in Figure WO-12. According to Saif, this epidemic is ongoing and still spreading among U.S. swine herds. Based on available genome sequence data, the virus has continued to evolve, including variants with insertions and deletions in the spike glycoprotein gene (S INDEL strains) that have been associated with milder disease (Vlasova et al., 2014), she reported. It remains to be determined whether these attenuated variants were introduced into the United States along with virulent PEDV, or if their mutations arose here.
| Resulting CoV/host | Suspected original CoV/host | Genomic modification | References | |
| TGEV/pig | CCoV-II/dog | ORF3 insertion | Decaro et al., 2007 | |
| CCoV-II/dog | TGEV/pig | Recombination in the 5′ end of the spike gene | Decaro et al., 2009 | |
| CCoV-II/dog | CCoV-I/dog and unknown CoV | Recombinant spike gene | Lorusso et al., unpublished | |
| FIPV/cat | FCoV/cat and CCoV/dog | Substitutions in M and ORF7b genes and FCoV-CCoV recombinations in spike and pol genes | Brown et al., 2009 | |
| PRCV/pig | TGEV/pig | 621-681-nt deletion in the 5′ end of the spike gene; deletions in ORF3 | Wesley et al., 1991 | |
| HCoV-OC43/human | BCoV/cow | 290-nt deletion (corresponding to the absence of BCoV nsp 4.9 kDa and nsp 4.8 kDa) | Vijgen et al., 2005 | |
| HECV-4408/human | BCoV/cow | Not known | ||
| GiCoV/giraffe | BCoV/cow | Deletion in the S1 subunit (amino acid 543-547) of the spike protein | Hasoksuz et al., 2007 | |
| SARS-CoV/human | Bat and civet SARS-CoV/ horseshoe bat and civet cats | 29-nt deletion in ORF8 and substitutions in spike gene and ORF3 | Lau et al., 2005 | |
SOURCE: Vlasova and Saif, 2013.
This is but one of a list of unresolved questions associated with the emergence of PEDV in swine, Saif observed. Also unknown are the following questions:
- What is the host reservoir from which PEDV emerged in European swine in the 1970s (a bat virus is the closest relative)?
- Why did PEDV outbreaks cease in Europe in the late 1990s without implementation of immunization against the virus, much as SARS disappeared from China while its bat host remains? Did both viruses emerge, and then disappear, from secondary hosts to which they were not well adapted?
TABLE WO-3 Emerging Porcine Coronaviruses—Target Tissues and Diseases
| Genus | Virus | Host | Disease/Infection Site | Year | ||
| Respiratory | Enteric | Other | ||||
| Alpha | TGEV | Pig | x | x | 1946 | |
| PRCV | Pig | x | 1989 | |||
| PEDV | Pig | x | 2013 | |||
| PEDV S | Pig | x (mild?) | 2013 | |||
| Beta | HEV | Pig | x | CNS | 1962 | |
| Delta | PDCoV | Pig | x? | 2014 | ||
NOTE: CNS, central nervous system.
SOURCE: Saif Presentation, 2014.
- Why did a more virulent form of PEDV emerge in China in 2010—did the use of live, partially attenuated vaccines in swine select for this variant?
- What is the origin of PEDV in U.S. swine? Was it imported from China, as sequence similarities suggest?
- Will spike-variant S INDEL PEDV strains associated with milder disease moderate PEDV’s impact here as well?
In February 2014, porcine delta-coronavirus was first identified as a cause of diarrheal disease in U.S. swine (Li et al., 2014a). Outbreaks in herds infected with this virus resembled those associated with PEDV and TGEV, but were less severe, Saif said. It remains to be determined whether this virus actually causes diarrhea in swine, she noted. This association is of interest because it suggests an expanded disease potential among δ-coronaviruses, which previously had been confined mainly to avian species. In this case, she observed, a virus of the same species that infects sparrows also infects swine and another mammals, including the Asian leopard cat, signaling possible spillover events.
Evidence of Interspecies Transmission
Over the past two decades, Saif’s group has amassed evidence showing that some (but not all) coronaviruses have broad host ranges; this body of research is illustrated in Figure WO-13. Early experiments involved inoculating immunologically naïve germfree calves with enteric coronaviruses isolated from disease outbreaks in captive wild ruminants such as sambar, white-tailed deer, and waterbuck, as well as a human enteric coronavirus isolate. Bovine β-coronaviruses share sequence identity with coronaviruses from these and other animal species. “We put all these [viruses] into our calves, and all the calves got diarrhea, they

FIGURE WO-12 Emerging CoVs in U.S. swine (2013 to present).
SOURCE: Saif presentation, 2014.
all shed the virus, and they all seroconverted with neutralizing antibodies to bovine coronavirus,” Saif reported (Han et al., 2006 Tsunemitsu and Saif, 1995; Tsunemitsu et al., 1995). “This was early evidence that coronavirus from wild ruminants or humans can experimentally infect young calves.” Such studies were among the earliest to challenge the dogma that coronaviruses are highly species-specific by demonstrating the potential for coronaviruses to cause interspecies infections.
Both SARS- and MERS-CoV have proven to be promiscuous with regard to host, Saif noted. SARS-CoV has been found either naturally or experimentally to infect humans, civet cats, raccoon dogs, horseshoe bats, swine, nonhuman primates, ferrets, cats, mice, guinea pigs, and hamsters. MERS-CoV can infect humans, bats, camels, and rhesus macaques, as well as monkey and pig cells in culture.
Among circumstances that favor spillover at interfaces between wildlife, domestic animals, and humans, Saif expressed particular concern regarding fecal contamination of animal food sources, against which few protections exist. For example, grain destined for livestock feed is often stored in open areas, where many species of birds can readily eat and defecate on it, as shown in Figure WO-14. “We talk about food safety from the human perspective,” she observed, “but maybe we should talk about food safety for animals, too.”
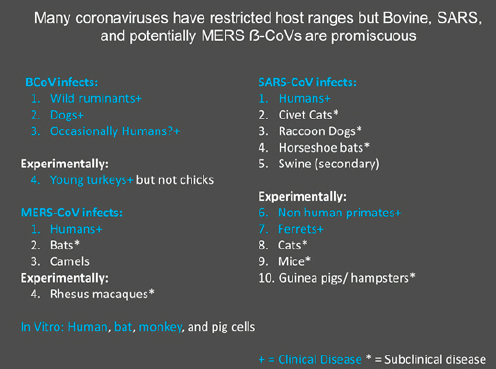
FIGURE WO-13 Coronaviruses with broad host ranges.
SOURCE: Saif presentation, 2014.

FIGURE WO-14 Birds consume and contaminate livestock feed.
SOURCE: USDA, 2010.
A One Health Challenge
Discovering whether bats are the sole animal reservoir for SARS-CoV or whether it also persists in an intermediate host or community reservoir is an important step in determining the likelihood that the virus will reemerge in humans, according to Saif. Equally pressing is the need to identify the host reservoir of MERS, as she and several workshop speakers noted throughout this workshop. Efforts to pursue this question are discussed in the section, “Ecology and Animal Origins of MERS-CoV.” More generally, Saif noted, two key potential reservoir animals for coronaviruses—birds (for δ- and γ-coronaviruses) and bats (for α- and β-coronaviruses)—include migratory populations that congregate in high densities, often as multiple species—conditions that further favor the interspecies viral transmission of coronaviruses, which are already genetically predisposed to adapt to new hosts.
The emergence of SARS, MERS, and coronavirus diseases of domestic animals such as those Saif described, combined with accumulating knowledge of the potential for interspecies transmission among coronaviruses, suggests that coronavirus spillover presents an ongoing threat to animal and human health. It will therefore be important to survey diverse coronavirus strains from wild and domestic animals and to study their pathogenesis in the natural host, she advised. Such a goal exemplifies the One Health approach to addressing zoonotic diseases, and one best pursued by multidisciplinary teams that combine the expertise and efforts of medical and veterinary scientists.
Tracing the Origins of MERS-CoV
Soon after the first MERS case was recognized and the novel virus identified, the EcoHealth Alliance in partnership with the Center for Infection and Immunity at Columbia University, and the Kingdom of Saudi Arabia’s Ministry of Health, became involved in efforts to identify animal reservoirs of MERS-CoV. Presentations by researchers Jonathan Epstein and Kevin Olival described the multipronged approach taken by the team that involved simultaneous epidemiological, immunological, ecological, and evolutionary investigations (Drs. Epstein and Olival’s contribution may be found on pages 119–133 in Appendix A).
Epidemiology and Immunology
“Early on, as the genetic code was being analyzed for this new virus and there were linkages being made to other coronaviruses—namely, bat coronaviruses that were linked to [bats] Pipistrellus and Tylonycteris species from Hong Kong—we also had some insight into the potential bat reservoirs based on some
work we were doing under the USAID-funded PREDICT program,”35 Epstein noted. To catalog viral diversity among bats, rodents, and primates that inhabit so-called hot spots for infectious disease emergence (Jones et al., 2008), PREDICT researchers created a genomics library that included novel bat coronaviruses, some of which proved to be closely related to MERS-CoV.
The team began its work in Bisha, where the first human MERS case was identified. The index case was a 60-year-old man who died from respiratory disease, and who had no reported history of animal contact or underlying disease, Epstein said. To better understand how this patient might have been exposed to and infected by this virus, they met with and interviewed his family and toured his several properties, where they found domestic animals, including pet camels. While few people recognized that there were bats in the vicinity, the researchers observed several in flight on a single evening and ultimately discovered bats roosting in abandoned buildings in downtown Bisha, as well as a colony of about 500 bats just outside of town. There they set up a mobile laboratory and collected samples of saliva, feces, urine, and blood from seven different bat species to search for MERS-CoV, which they found in the Egyptian tomb bat, Taphozous perforatus—depicted in Figure WO-15 (Memish et al., 2013).
At the same time, several research groups investigating MERS-CoV in camels and dromedaries found indications of the widespread presence of MERS-CoV and closely related viruses (Haagmans et al., 2013; Hemida et al., 2013; Meyer et al., 2014; Perera et al., 2013; Reusken et al., 2013). Other livestock species have been sampled, including sheep and goats, but to date all have proven negative for both serology and molecular evidence for MERS-CoV infection, Epstein stated (Alagaili et al., 2014).
When Columbia University and EcoHealth Alliance, in collaboration with scientists from King Saud University, conducted a survey of camels throughout Saudi Arabia for evidence of MERS-CoV exposure, including archived and fresh camel blood samples dating back to 1992, and nasal and rectal swabs, they found PCR-positive camels across Saudi Arabia, as well as antibodies from past infection (Alagaili et al., 2014). About 35 percent of juvenile camels and 15 percent of adult camels were PCR-positive for the virus, he reported. The majority of these positives were detected in nasal swabs from camels in western Saudi Arabia, near Taif and Jeddah. They detected four different clades of MERS-CoV, suggesting that diverse strains of the virus are circulating among camels, either as a result of camel movement or of multiple spillover events from a reservoir host (Alagaili et al., 2014).
_______________
35 The U.S. Agency for International Development (USAID) funds an Emerging Pandemic Threats program composed of four projects: PREDICT, RESPOND, IDENTIFY, and PREVENT. PREDICT project partners, including the EcoHealth Alliance, conduct research to identify novel infectious diseases that could become a threat to human health, focusing on wildlife species that inhabit geographic hot spots for infectious disease emergence. Source: http://www.ecohealthalliance.org/programs/28-predict_program (accessed February 19, 2015).

FIGURE WO-15 Egyptian tomb bat. The Egyptian tomb bat (Taphozous perforatus) is a species of sac-winged bat in the family Emballonuridae (Mickleburgh et al., 2004). It is a medium- to large-sized bat with a mass of approximately 30 g (1.1 oz) (Monadjem et al., 2010). It is an aerial insectivore, foraging in open space (Monadjem et al., 2010). Based on individuals captured in Ethiopia, it is thought to feed predominantly on Lepidoptera, but is also known to feed on Isoptera, Coleoptera, and Orthoptera (Skinner and Chimimba, 2005). Viral RNA matching MERS-CoV was found in an Egyptian tomb bat near the victim’s home in Saudi Arabia.
SOURCE: Photo courtesy of Jonathan Epstein, EcoHealth Alliance © 2014.
Data on camel imports and exports compiled by the FAO reveal that Saudi Arabia is predominantly an importer of camels, mainly since 1993, when an urban development boom may have increased demand, Epstein said. Although this date coincides with their earliest serological finding of MERS-CoV antibodies in camels, he added, “that’s not to say that there wasn’t MERS prior to 1992.” More importantly, Epstein noted, researchers should be looking for MERS-CoV in camels and humans in those countries that export the majority of camels to Saudi Arabia: Somalia, Oman, United Arab Emirates (UAE), Qatar, Djibouti, and Sudan (Oman, UAE, and Qatar have reported human cases). For example, he said, about 232,000 camels were imported by Saudi Arabia in 2005. Assuming an infection rate among camels of 25 percent (reflecting their PCR survey results), about 58,000 infected camels entered the country that year.
Epstein hypothesized that a relatively small proportion of the male population of Saudi Arabia, which totals 13 million, that includes camel herders, owners, and traders, would have the most frequent contact with camels and therefore the highest risk of coming into contact with an infected animal. And because camel
importation and evidence for camel infection predates 2012 by at least 20 years, human MERS infections have likely occurred in Saudi Arabia (and elsewhere) prior to 2012. He cautioned, however, that additional epidemiological data were still needed to determine whether and how camel-to-human transmission of MERS-CoV actually occurs. In addition, further studies of MERS-CoV in bats are needed to support the initial finding that Egyptian tomb bats are a reservoir, and to determine whether MERS coronavirus is circulating in other bats or other wildlife species, and whether transmission from bats to camels, or to humans, occurs.
Ecological and Evolutionary Approaches
Olival described ecological and evolutionary approaches to understanding the frequency, timing, and geographic “footprint” of MERS-CoV transmission among wildlife, livestock, and humans, and thereby predicting future spillover events. He noted that EcoHealth Alliance had conducted similar work to understand the emergence of Nipah virus in Malaysia (Epstein et al., 2006)—a threat he likened to MERS-CoV as another bat-borne viral disease likely driven to emerge by human-precipitated ecological changes. In the case of Nipah in Malaysia, bats shed the virus into pigpens through their waste products along with partially chewed fruit that pigs subsequently consume. Olival wondered, What comparable interactions might take place between bats and camels—or bats and humans—that could result in spillovers of MERS-CoV from their animal host(s) to humans?
Bats cohabit with humans far more often and more easily than we appreciate, Olival observed. While detailed ecological surveys are needed to better describe bat–livestock and bat–human interfaces, a few detailed initial investigations would provide significant preliminary data, he said. Thus he and coworkers mapped bat species richness—and by association, coronavirus richness—in Saudi Arabia. They also examined viral diversity within a single bat species, in which they detected 7 to 10 different coronaviruses (Memish et al., 2013). On the basis of these results, they estimate that in some regions of Saudi Arabia, hundreds of coronaviruses could be circulating in bat communities, he concluded.
Bats in Western Europe have been shown to shed virus seasonally, coincident with periods of birthing and lactation, Olival reported (Drexler et al., 2010). Presumably this holds true elsewhere, he said, but this premise should be tested through longitudinal studies in bat host populations for MERS-CoV.
Phylogenetic studies of MERS-CoV isolates suggest that either camels transmitted the virus to humans through multiple spillover events, or that viral diversity is being maintained in both camel and human populations, Olival said. His own analysis suggests generally strong cophylogeny between β-coronavirus species and their bat hosts, but there are some cases of crossover. He and coworkers believe that T. perforatus is the reservoir host for MERS-CoV, but because
bat-to-bat spillover of the virus may have occurred, he advised investigation of other bat species is needed. “There are probably a lot of [MERS-CoV] related coronaviruses out there in bat populations, and if you believe my cophylogeny, these include the genus Taphozous,” he added.
As illustrated in Figure WO-16, bat species richness varies across the landscape of the Arabian Penninsula. Some bat “hot spots” occur in pockets across the known geographic range of human MERS-CoV cases, as well as in Africa and South Asia, Olival reported. Fourteen species of Taphozous are found across a broad swath of Africa, South Asia, and Australia—including a substantial portion of the geographic range for camels. The import and export of camels between countries could further have facilitated localized spillover. Therefore, he concluded, it is entirely possible that MERS-CoV emerged outside of the Middle East.
Given this possibility, surveillance for MERS-CoV virus should not be confined solely to the Middle East, Olival advised. “We need more global surveillance in both livestock and presumptive wildlife reservoirs,” he insisted, and noted that EcoHealth Alliance is pursuing active viral surveillance in bats on a global scale. Moreover, he said, the demonstrated ability of MERS-CoV to replicate in cell lines from multiple species (Eckerle et al., 2014) suggests that “we should be casting a wider net when we do our animal surveillance.” Returning to the human experience with MERS-CoV, Olival urged workshop participants to consider its ecological context. Why did the disease first become noticeable in the Middle East? Did it truly emerge there? The answers to these questions likely involve ecology, he suggested.
MERS Epidemiology and Pandemic Potential
Early Epidemiology of a Novel Disease
Trish Perl of Johns Hopkins University began her discussion with a detailed account of the index case mentioned by Epstein: a 60-year-old man admitted to the Dr. Soliman Fakeeh Hospital in Jeddah, Saudi Arabia, in June 2012 (Zaki et al., 2012) (Dr. Perl’s contribution may be found on pages 181–184 in Appendix A). He had suffered fever, productive cough, and shortness of breath for the prior week. He had no history of cardiopulmonary or renal disease, took no regular medications, and did not smoke. Treated initially as a case of influenza, he received the antiviral medication oseltamivir, as well as antibiotics and antifungals to treat apparent opportunistic infections. His condition continued to deteriorate, and he subsequently suffered renal failure and died within 2 weeks.
The virologist at this hospital, Ali Moh Zaki, is “one of the heroes in this particular story,” according to Perl. Zaki applied tracheal aspirate from the index patient to two monkey cell lines, recognized that they became infected with a coronavirus, and sent samples of the virus to the Erasmus Medical Center in the
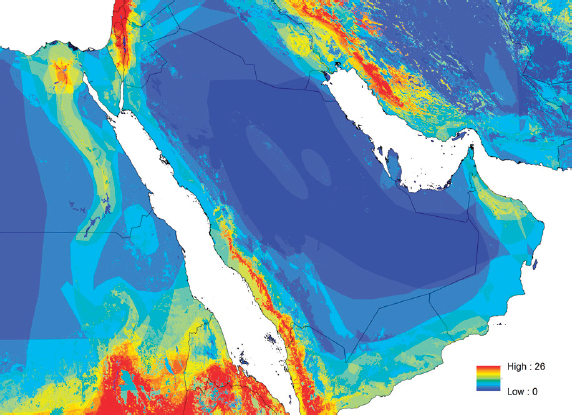
FIGURE WO-16 Arabian bat species richness. While some areas have low bat richness and bat densities, some areas are very rich in bat diversity. Good locations to target for understanding the baseline of viruses “zoonotic pool” in these hosts. Some Arabian bat hot spots include Southwest KSA and Yemen; Iraq and Iran; Jordan and Israel; Egypt along the Nile; and parts of Sudan, Eritrea, and Ethiopia.
SOURCE: Permission provided by EcoHealth Alliance, 2014.
Netherlands for sequencing, where its novelty and phylogeny was verified, she explained. Perl noted that the discovery, announced on ProMED36—another hero in this story—alerted clinicians in Qatar’s National Health Service who were treating a patient with similar symptoms; this patient was later identified as the second case. Thirteen close contacts of this case with mild self-limiting respiratory illnesses were tested for the virus, but none were found to be infected with it, she reported. Within 2 weeks, the novel virus was identified in a patient in the United Kingdom who had recently returned from travel in the Middle East.
An earlier MERS case cluster was recognized to have occurred in Jordan in April 2012, based on retrospective testing of specimens from two deceased patients in a hospital intensive care unit and their contacts (Hijawi et al., 2013). Most of those found to be positive for the virus were health care workers, much as had occurred a decade earlier during the SARS epidemic, Perl pointed out. She then described an intrahospital MERS case cluster also reminiscent of SARS, which occurred in several Al Hasa facilities in April 2013.
A rural governorate comprising about one million people, Al Hasa is located in eastern Saudi Arabia. “Initially, it appeared that this cluster was located in two dialysis units and several of the intensive care units in this hospital,” Perl recalled. “We went in to do chart review and investigate the hospital outbreak.” The initial case was recognized on April 8, 2013, by an infection control practitioner investigating multiple deaths from pneumonia in one hospital. As a result, infection control measures were put in place, and expanded about 1 week later. Once effective measures were put in place in all three local institutions, she said, no additional MERS cases were reported. A total of 21 confirmed and 2 probable cases were acquired by person-to-person transmission in dialysis units, intensive care units, or in-patient units. Among 217 household contacts and more than 200 health care worker contacts, MERS-CoV infection developed in only 5 family members and 2 health care workers (Assiri et al., 2013). There were additional health care workers who had febrile illnesses at that time who were not tested and are now suspected to have had MERS as well, she added.
Using the epidemiological information they collected, Perl and coworkers traced the path of infection through multiple contacts, as shown in Figure WO-17. One patient clearly transmitted the virus to multiple people in various settings, she stated, although she was reluctant to call him a “superspreader.” “Were these people more vulnerable, for some reason?” she wondered. “We don’t have a case-control study to tell us. But there was something about this patient.”
Perl’s team of investigators was also able to estimate the incubation period for MERS—5.2 days—and the serial interval (the time between successive cases
_______________
36 ProMED—the Program for Monitoring Emerging Diseases—is an Internet-based reporting system dedicated to the rapid global dissemination of information on outbreaks of infectious diseases and acute exposures to toxins that affect human health, including those in animals and in plants grown for food or animal feed.
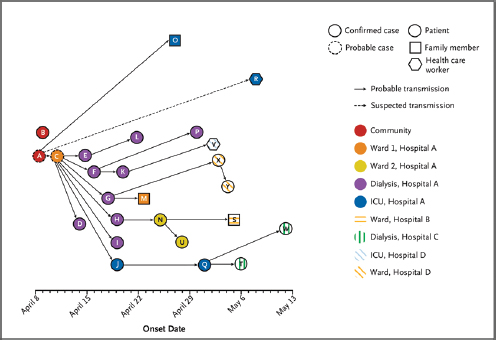
FIGURE WO-17 Transmission map of outbreak of MERS-CoV infection. All confirmed cases and the two probable cases linked to transmission events are shown. Putative transmissions are indicated, as well as the date of onset of illness and the settings. The letters within the symbols are the patient identifiers.
SOURCE: Assiri et al., 2014.
in a chain of transmission) at about 7.6 days (Assiri et al., 2013). Both the incubation period and the serial interval closely resembled those for SARS (4 days and 8.6 days, respectively), Perl reported.
Genetic mapping of Al Hasa and other MERS-CoV isolates was consistent with the epidemiological findings, suggesting that the Al Hasa cases were closely related and distinct from cases from other locations, Perl said (Cotten et al., 2013). Based on genetic information on the Al Hasa strains, minor changes were made to the transmission map and to the incubation and serial interval estimates, which were initially derived from epidemiological findings, she noted.
Assessing Pandemic Potential
Using data from the Al Hasa outbreak, Perl and coworkers estimated the reproductive number (the average number of secondary infections attributable to a single infectious individual in a susceptible population, or R0) for MERS to be about 0.6. Other estimates have ranged from 0.69 (Breban et al., 2013) to up to 1.3 (Cauchemez et al., 2014); she noted for comparison that the R0 for SARS
in the early days of that epidemic was calculated to be 0.8. Because the MERS estimates are based on scant information, and therefore uncertain, she suggested that a more salient current indicator of MERS’ pandemic potential is the relatively small sizes of case clusters, and the fact that person-to-person transmission has so far successfully been controlled using standard interventions against infectious diseases. Analysis of the epidemic curve and of genetic sequences of various MERS isolates “indicates a slowly growing epidemic either in humans or in an animal reservoir,” she asserted.
Analysis of epidemiological parameters by Cauchemez and coworkers (2014) suggests that sustained transmission of MERS could be possible if the characteristics of the current animal and human environments remain relatively stable, Perl noted. Thus, where infection or animal control measures are lacking, there is significant potential for self-sustaining MERS transmission, she concluded. Treatments for the disease are needed, and clinical communities should collaborate in order to systematically assess the use of agents such as interferon or ribavirin that have shown some promise against MERS, she advised. “I don’t think any of us want to be caught like we were with SARS, where we were giving people steroids,” she said, referring to their use, which remains controversial in the absence of assessment by clinical trial (Gomersall, 2004). Similarly, she observed, diagnostic strategies for MERS have not been established, although deep and multiple sampling appears crucial.
Perl also urged efforts to identify factors favoring MERS transmission, which appears primarily to occur person to person or animal to person. “There [are] some data suggesting that this organism survives in the environment better than influenza,” she noted; therefore, fomite transmission may be an important route of exposure (van Doremalen et al., 2013). “Analysis of individual time course of transmissibility could really help us in determining and prioritizing interventions,” she continued. Much might be learned about seasonality of transmission by comparing epidemic curves of virus shedding in bats and humans. Finally, she cautioned, asymptomatic MERS transmission has been very poorly characterized. “The fact that we think that there are asymptomatics out there is going to be an albatross in terms of control measures, if we don’t figure that out,” she declared.
MERS’ pandemic potential should not be discounted, Perl concluded—and it should prompt the creation of better theoretical models (e.g., through the synthesis of genetic and incidence data) to guide ongoing research, as well as the sharing of data on a global scale in order to expedite the development of appropriate interventions.
Mass Gatherings and the International Spread of MERS
Proclaim the pilgrimage to all people. They will come to you on foot and every kind of swift mount, emerging from every deep mountain pass.
—Qur’an Chapter 22, verse 27
With this passage from Qur’an, Kamran Khan of the University of Toronto reminded workshop participants that the ancient ritual of pilgrimage—and particularly that of Muslims’ required journey to Mecca known as the Hajj—brings vast numbers of people from disparate locations together in close proximity, after which they return to their communities. As the world’s population grew, and particularly as modern transportation developed, the “swift mount” of choice shifted from camels to airplanes, which transport increasing numbers of pilgrims from around the globe to Mecca (see Figure WO-18). In 2012, about three million pilgrims performed the Hajj, about 60 percent of whom came from outside Saudi Arabia.
Although the Hajj passed in 2012 and 2013 without triggering the international spread of MERS, this and the Umrah—another annual pilgrimage to Mecca that peaks during Ramadan—are annual events, “so we’re going to be facing this issue again,” Khan cautioned. “We really can’t get too complacent, unless, of course, MERS disappears in the next 4 months or so.” And although the number of pilgrims performing Hajj declined in 2013, likely due to the threat of MERS, that trajectory is certain to continue upward, he insisted.
Mass Gatherings and Infectious Disease
Khan compared the global transportation network created by commercial air travel to an organism, with passengers flowing through its “arteries” creating patterns akin to physiological states (see Figure WO-19).
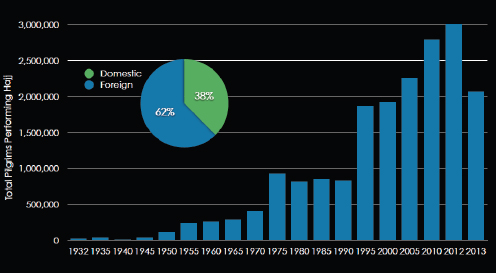
FIGURE WO-18 Pilgrims performing Hajj—1932 to 2013.
SOURCE: Khan presentation, 2014.
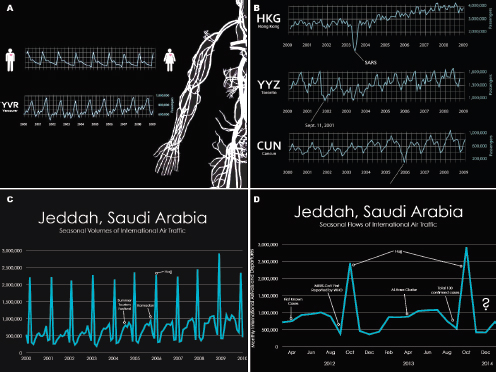
FIGURE WO-19 Seasonal passenger and air traffic volume. (A) Seasonal air passenger volume in Vancouver, Canada, from 2000 to 2009. (B) Seasonal air passenger volume in Hong Kong; Toronto, Canada; and Cancun, Mexico. Major public health and extreme weather events can be seen to have an impact on travel. (C) Seasonal volumes of international air traffic in Jeddah, Saudi Arabia. (D) Seasonal flows of international air traffic by month from April 2012 to December 2014.
SOURCE: Khan presentation, 2014.
“Each one of these particular flight lines, in Figure WO-19 (a) has its physiology, its own pattern. Understanding that pattern is particularly important since this is a major conduit for the international spread of infectious diseases. As illustrated in Figure WO-19 (b) this system can get sick, almost like an individual can, from things like viruses, where we can see a change in the normal physiologic pattern, if you will, things like terrorism even affecting a city like Toronto, which wasn’t directly affected by the attacks of September 11, 2001, and even things like natural disasters, Hurricane Katrina having a big impact on global population movements. There is some evidence of how behavior can be changed negatively, where travel can decrease. The opposite may also occur in association with mass gatherings. As depicted in Figure WO-19 (c), the seasonal pattern of aircraft going into Jeddah, the city that is closest to Mecca where pilgrims tend to arrive prior to going by road for an hour or so over to Mecca, shows a very, very large
spike in terms of population movements into this particular city.”37 Turning to Figure WO-19 (d), Khan reported, “If we look at some of the events that occurred [in 2012], this is where MERS CoV is first being reported . . . some of the [9 or 10] initial cases were thought to occur, back in April.”
As has occurred in response to other health threats and disasters (e.g., the terrorist attacks of September 11, 2001, the SARS epidemic, and Hurricane Katrina), air passenger transit through Saudi Arabia declined in 2013, as awareness of MERS began to spread globally. But this temporary disruption occurs against the backdrop of globalization, the context that enables mass gatherings to amplify and disperse disease worldwide, Khan pointed out. This can occur when infected individuals travel to a mass gathering, where they transmit disease to other international travelers who then introduce the disease back to their home countries upon their return, or when—as is possible in the case of MERS—travelers to a mass gathering site expedite the international spread of a formerly local epidemic. By their nature, pilgrimages such as the Hajj involve crowded conditions, Khan observed, further facilitating the spread of communicable diseases.
To anticipate and prepare for the possible spread of MERS following a future Hajj, Khan and coworkers sought clues to where exported cases would likely appear, and how quickly specific nations will be able to detect, identify, and effectively respond to such an event. By gathering and integrating voluminous and detailed information on the geographic origins of pilgrims, on air traffic patterns in the Arabian Peninsula, and on the public health capacity of nations to which infected pilgrims might return (see Figure WO-20), they learned that nearly two-thirds of air travelers leaving the Arabian Peninsula during this period returned to low-income or lower-middle-income countries, he reported (Khan et al., 2013). This finding does not augur well for the timely detection of and response to international MERS transmission, he noted. Indeed, it suggests that the Hajj presents a greater challenge to global disease control than the Olympic Games, which attracts wealthier travelers with better access to health care.
Pandemic Risk and Epidemiologic Blind Spots
Why did MERS not spread internationally after the 2013 Hajj? “There may be a variety of reasons why,” Khan explained: chance, enhanced infection control measures undertaken within Saudi Arabia in response to MERS, or the reduction by one million in the number of pilgrims in Mecca as compared with previous years. But given such containment of MERS, it is surprising that international
_______________
37 Hajj/October; there is an earlier bump in population due to the summer tourism festival . . . every year in Jeddah. It is mainly a domestic event, but there are people traveling into Saudi Arabia from the neighboring countries. . . . Right after that, you have this period where Ramadan is occurring and you have large numbers of pilgrims performing Umrah [lesser pilgrimage] . . . large spike here is the Hajj, . . . [with] very, very large numbers of people coming into the country in a short period of time and performing this particular event.
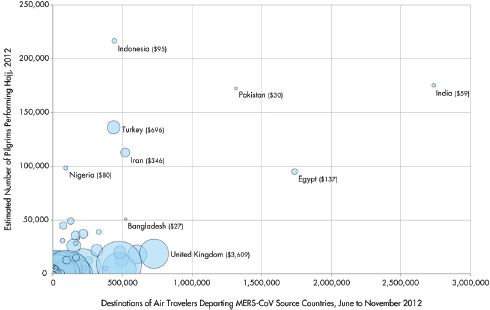
FIGURE WO-20 Country-level destinations of air travelers departing MERS-CoV source countries,* origins of Hajj pilgrims,† and health care expenditures per capita.‡
* Final destinations of air travelers departing Saudi Arabia, Jordan, Qatar, and the United Arab Emirates via commercial flights between June and November 2012.
† Estimated for 2012.
‡ Sizes of the circles are proportionate with health care expenditures per capita as estimated by the World Bank, 2011.
SOURCE: Khan et al., 2013.
cases arose prior to the 2013 Hajj in Western Europe and North Africa. Meanwhile, no imported cases were reported in travelers from South Asia and Afghanistan, Pakistan, India, Nepal, and Bangladesh, which together comprise almost 30 percent of international air passengers out of the Arabian Peninsula, and about 25 percent of Hajj pilgrims, Khan stated. “It’s possible, certainly with the spectrum of an illness ranging from subclinical to minimally clinical, that there were undetected cases that may have moved into those [more likely] areas,” he pointed out.
Additional countries vulnerable to MERS spread include Egypt and Indonesia, Khan said. “Cairo has the strongest ties of any city in the world to [Saudi Arabia]. . . . About 10 percent of all the international air traffic winds up in Egypt, and about 5.5 percent of all the pilgrims.” However, he added, “We didn’t observe any imported cases of MERS there either.” Similarly, no cases have been reported in Indonesia, the world’s most populous Muslim country and home to more than 12 percent of all Hajj pilgrims, he said. Conversely, MERS cases were introduced through travel to the United Kingdom, France, Italy, and Tunisia, which together
account for about 7 percent of all the international air travelers and 2 percent of all the Hajj pilgrims, he noted.
Even with the most sophisticated mathematical modeling, according to Khan, it remains difficult to predict the international spread of an infectious disease resulting from a mass gathering. Factors including the amount and activity of the disease, the volume of travel leaving the particular region, and public health measures within the country complicate patterns of disease transmission, he noted. Nevertheless, Khan added, in both the SARS and H1N1 influenza epidemics, the association between geographic and temporal patterns of global travel and those of disease spread were clear. Thus, it is fair to wonder whether the unexpected distribution of international MERS cases results from actual transmission, or from “epidemiologic blind spots,” where cases have gone undetected in resource-limited settings.
Clearly, many more questions have been raised about MERS than have been answered and, as Khan observed, their pursuit is crucial to the mitigation and prevention of this and other emerging infectious diseases.
MERS in Context
In the discussion that concluded this session, the topic of immunization strategy led to an exchange that illustrates how difficult it remains to gauge the threat MERS presents, and therefore to address it appropriately. Saif raised the possibility that if camels were determined to be an important or intermediate host for MERS-CoV, it might make sense to vaccinate them (once a vaccine is developed) in order to break the chain of transmission to humans.
William Karesh of the EcoHealth Alliance noted that despite abundant opportunities for camel-to-human transmission of MERS-CoV in Saudi Arabia, there had been very few human infections:
If you go to Saudi Arabia . . . and you go to an abattoir there where they are slaughtering camels . . . [you’ll see that] these animals are bled out on the floor, and somebody is standing there with a hose, spraying. All this blood is being aerosolized. No one has a mask or a glove on. . . . [Yet] in Egypt, where they tested abattoir workers, all are seronegative, but the camels are seropositive. . . . There are hundreds of thousands of camels [imported] to Saudi Arabia annually. . . . Most of those are being slaughtered. . . . And we have only had 40 or 50 or 60 people that seem to be primary cases.
Given these odds, he wondered, why is MERS-CoV so feared? “We’re thinking maybe we should put $40 million38 into vaccine development for camels, and 55,000 people die of rabies every year?” he asked. “We have a great rabies vaccine that you could give to dogs and prevent human rabies. But we can’t seem to
_______________
38 The cost of developing a SARS vaccine, which was never used.
muster that together, and tens of thousands of people are dying every year, when we’re thinking about a MERS vaccine for camels,” he said.
Perl noted that a case-control study and additional “shoe-leather epi”39 would go a long way toward defining the threat posed by MERS-CoV without costing $40 million. Baric observed that four human coronaviruses “have solved all the problems in terms of transmission and disease,” he said, and thus have considerable pandemic potential. Despite the adoption of robust approaches to prevent the spread of infectious diseases, novel α-coronaviruses, including PEDV, TGEV, and PRCV are able to circumvent those transmission barriers and produce significant disease in swine.
“To some extent, the SARS epidemic was an example of the public health success story that prevented an expanding outbreak,” Baric noted. “But what if the virus had evolved mutations that allowed it to transmit 36 hours earlier or 24 hours earlier, before symptomatic disease? It would have been a very different story. The window is actually quite small. With MERS,” Baric continued, “we have a virus that has a 40 percent mortality rate. We have a virus family that is able to solve fundamental problems in transmission and to produce high mortality rates, both in animals and in humans.” He argued, therefore, that “another equally good question is: would we be responsible stewards if we did nothing? I think the answer is, we should do something.”
Saif emphasized that coronaviruses cause devastating diseases in livestock. Millions of pigs have died in Asian outbreaks, and they have also occurred in the United States, where “they have not been able to keep this PEDV coronavirus out, even with the most stringent high-security measures,” she warned. “It is a concern.”
EMERGENCE OF INFLUENZA A VIRUSES IN ASIA
Avian Influenzas A (H7N9) and A (H5N1)
Ruben Donis, of the CDC,40 provided an introduction to two important type A avian influenzas that recently emerged in humans: H7N9 (in 2013) and H5N1 (in 1997). Neither virus has yet adapted sufficiently to humans to support sustained person-to-person transmission, he noted. “Who knows if they ever will?” he added. Even so, he said, “We need to be prepared for that.”
_______________
39 The term shoe-leather epidemiology is often synonymous with field epidemiology or intervention epidemiology. All three terms imply investigations initiated in response to urgent public health problems and for which the investigative team does much of its work in the field (i.e., outside the office or laboratory) (Koo and Thacker, 2010).
40 Donis presented for Robert Webster, of St. Jude Research Hospital.
H7N9 in Context
Concern about the severity and pandemic potential of H7N9 has prompted comparisons with its predecessor, H5N1. That virus, which is highly pathogenic in birds, first infected humans during a poultry outbreak in Hong Kong in 1997 (WHO, 2014a). It was controlled, but later reemerged in 2003, spreading from Asia to Europe and Africa. Since then, it has caused more than 650 human cases, of which nearly 400 were fatal. H5N1 has become endemic in poultry in some countries, where it has had a severe economic impact.
For each of the 22 human cases of H5N1 reported between September 2013 and March 2014, H7N9 caused more than 10 cases in China alone, Donis reported. As Fukuda noted, most people infected with H7N9 have had direct contact with poultry through urban live bird markets. By contrast, most H5N1 cases have been linked to domestic “backyard” poultry rearing. “This difference in exposure settings we think is more a reflection of the circulation of the virus in different parts of the chain,” he said. “I don’t think that there is a fundamental difference in the way that these viruses are transmitted to people.”
In response to the 1997 emergence of H5N1, Hong Kong closed its live bird markets temporarily, which drastically reduced infections in both humans and poultry. However, as Donis pointed out, this could only be a temporary solution, given the importance of such markets to the economy and culture of this and many other Asian countries.41 After a series of less stringent solutions failed to suppress transmission, the authorities required bird market vendors to slaughter all birds at the end of each day and disinfect the premises, which halted the previously simmering outbreak.
Chinese officials also temporarily closed live markets in response to each of two waves of human H7N9 cases, in April 2013 and February 2014, resulting in a drastic drop in case incidence, Donis reported.
Origins of H7N9
H7N9 is a novel, reassortant influenza A virus, and genetic analyses have provided the clearest clues to its origins. Phylogenetic analyses of 100 closely related sequences for each viral gene suggest that H7N9 is derived from at least four viral strains with distinct origins: “duck origin for HA, duck (probably also wild bird) origin for NA, and at least two H9N2 chicken viruses for the internal genes” (see Figure WO-21) (Cohen, 2013; Liu et al., 2013). Researchers hypothesize that the H7N9 NA gene originated in viruses carried by wild birds, and that wild ducks probably transferred the viruses to domesticated ducks. Later, H7N9 began to circulate with low pathogenicity in chicken populations (Liu et al., 2013).
_______________
41 Donis observed that by closing live markets for extended periods, “you could potentially make the [transmission] problem worse by creating alternative opaque channels of distribution. . . . At least if you have open channels you can regulate, you can inspect, you can promote better biosecurity.”
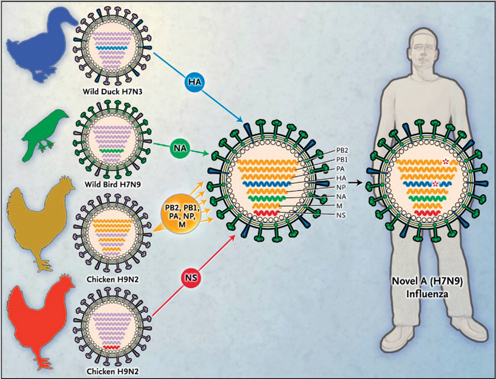
FIGURE WO-21 Origin of the novel avian influenza A H7N9 virus. On the basis of published sequences and phylogenetic analyses, it has been hypothesized that the novel avian H7N9 influenza virus is a reassortant virus containing gene segments derived from four separate avian influenza viruses, including two different wild-bird viruses contributing the H7 hemagglutinin (HA) (closest match to wild-duck virus) and N9 neuraminidase (NA) (closest match to a wild-bird isolate) gene segments, and two different domestic poultry–derived H9N2 viruses contributing the other six “internal” genes (polymerase PB2, PB1, and PA genes), the nucleoprotein (NP) gene, and the matrix (M) and nonstructural (NS) genes. The avian origin of each of the eight H7N9 gene segments is coded by color.
SOURCE: Morens et al., 2013. ©2013 Massachusetts Medical Society. Reprinted with permission.
The amino acid composition of the H7N9 cleavage site resembles that of human seasonal influenzas and renders it less severe in birds, which develop asymptomatic infections. By contrast, the multi-basic cleavage site of H5N1 is recognized by proteases present in all tissues of its avian host, facilitating systemic, often fatal infection. Poultry die-offs may signal outbreaks that threaten human populations. Lacking this “warning system,” H7N9 outbreaks in humans have served as a sentinel for detection of the virus in birds, Donis observed.
When H7N9 is detected in China, consumers tend quickly to avoid buying poultry, according to Donis. “This causes tremendous economic loss to the
industry, to the whole value chain,” he said. So far, the Chinese economy has lost about $16 billion as a result of H7N9 emergence, he reported. “H7N9 today is low pathogenic in birds, but it could change,” Donis observed. “These subtypes of the hemagglutinin, the H5 and the H7, are the only two ones that we know have consistently shifted from low pathogenic phenotypes to the high pathogenic phenotype upon multiple rounds of replication in chickens,” he stated.
Transmission
When human influenza viruses are inoculated into ferrets that are placed in close proximity to—but not in direct contact with—naïve animals, transmission occurs through respiratory droplets, and infection is successful nearly 100 percent of the time, Donis stated. By contrast, H7N9 does not often transmit via droplets, and H5N1 never does under normal circumstances. This happens because H7N9 and H5N1 do not efficiently recognize receptors in the upper respiratory tract. H7N9 has evolved to recognize so-called partially human-like receptors present in the upper airway, nasal turbinates, and trachea. The two mutations responsible for this ability arose, separately, in the 1957 (H2N2) and 1968 (H3N2) pandemic influenza strains, Donis observed. This change is completely unprecedented for the H7 subtypes, he said, and has never been detected in chickens—a situation he deemed “a reason for concern.”
Ecology
Aquatic and gallinaceous42 birds (poultry) are reservoir species for the H5N1 virus, which has been transmitted into and dispersed by wildlife into pigs and to other mammalian species. A few human-to-human transmission events have been reported, but none were sustained. By comparison, H7N9 appears to be restricted to its reservoir in gallinaceous birds, occasionally infecting aquatic birds, and, rarely, humans. “If H7N9 becomes transmissible in wild migratory birds, it will likely spread to many countries,” Donis warned, as did H5N1 after it became adapted to wildlife hosts. And if H7N9 reached the frozen lakes of Siberia, a major breeding ground for migratory birds, that would be even worse, he added. The virus could be widely dispersed, and it would be difficult to eradicate from this frigid environment, where infectious virus can persist. To date, Donis reported, neither H5N1 nor H7N9 persistence has been detected in this location, which is being closely monitored.
_______________
42 Gallinaceous birds, or galliforms, belong to an order (Galliformes) of heavy-bodied ground-feeding birds that includes the turkey, grouse, chicken, New and Old World quail, ptarmigan, partridge, and pheasant.
Influenza A (H7N9) in Poultry
As Donis made clear, the low pathogenicity of H7N9 in birds—in contrast to its severe consequences for humans—creates significant challenges for detecting the virus and preventing human exposure. Speaker David Swayne, of the U.S. Department of Agriculture, further explored this question by describing the distribution and behavior of the virus among chickens and other species sold as poultry in China’s live markets, which have been strongly associated with human cases of H7N9 influenza (Dr. Swayne’s contribution may be found on pages 263–284 in Appendix A).
Poultry Production in China
Today, amid a period of rapid agricultural modernization, China’s poultry production system comprises two equal and independent tracks that serve the industrial market and live markets, Swayne explained. Industrial producers raise breeds collectively known as white chickens. These are birds that grow rapidly and/or readily produce eggs. Farmers who supply live markets raise “yellow” chickens, which grow more slowly but are preferred by customers. Live markets also feature other bird species: ducks and geese, quail and other gallinaceous birds, pigeons, and captive-reared wild waterfowl, he noted.
The Chinese industrial chicken meat production system is highly integrated, Swayne said: production companies control the supply of chicks, feed, veterinary care, and processing. Contract growers’ farms must meet the company’s biosecurity standards, as well as stringent government standards and high consumer expectations. Meat production cycles are short and farms are large (generally 20,000 to 200,000 chickens). Most industrial chickens are processed and purchased as fresh or frozen meat by urban grocery stores and chain restaurants, which largely serve China’s young, two-career families.
The chicken production system for live markets is increasingly integrated, as its companies consolidate their resources, according to Swayne. Thus, he noted, farmers in this system may buy chicks from a broker, feed from a feed mill, and medicines from another source—or all of those things from a single source. While most growers are independent, some are under contract, as in the industrial system; in either case, chickens are raised indoors. There still are chickens in the village setting that are raised for local consumption, he added, but those that are transported to live markets are raised on larger farms. Figure WO-22 is a schematic of the production system supplying retail live markets via wholesale markets; large cities in China often have several live markets.
Regulatory and consumer oversight of the live market production system is lax in comparison to industrial production, Swayne observed. Yellow chickens could be raised at high density and may be moved over long distances to the live markets, he noted. Yellow chicken producers use large amounts of vaccines and antibiotics, he said. Largely due to the conditions under which chickens are
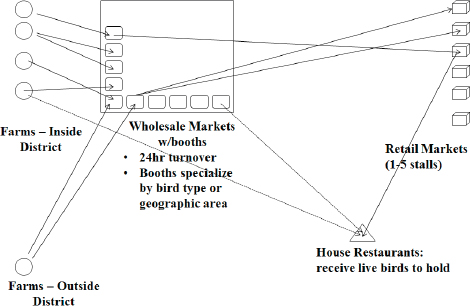
FIGURE WO-22 Live poultry market system.
SOURCE: Swayne presentation, 2014.
raised, he continued, biosecurity is significantly lower in this sector than in industrial production. In addition, the chickens live three times longer before being consumed as compared to industrial white chickens.
Response to H7N9 Emergence in Humans
Soon after the first human cases of H7N9 were reported, China quickly undertook active surveillance for the virus by expanding its existing H5N1 surveillance program. In 2003, more than 1 million serum and swab samples were taken from poultry; only 83 were determined to be positive for the virus, Swayne reported. However, because the H5N1 program was directed toward industrial chicken production, he noted, the samples were skewed to that population, and so did not accurately represent viral prevalence in the live poultry markets or farms that supply them. Based on the first 24 human cases that could be traced back to specific live markets, 22 markets had H7N9-positive birds or environments, he said—suggesting that the contaminated environment, and not individual birds, was the major driver of H7N9 persistence and transmission. In response to this finding, half a million birds were culled from live markets, the markets were closed, and movement restrictions were placed on poultry. The markets reopened within 3 months under new sanitary standards and monitoring practices.
Subsequent investigations determined that live bird markets were heavily contaminated with H7N9, and that they had no protocol for daily closure to allow cleaning and disinfection, Swayne said. In fact, he and coworkers suggested that environmental samples better predicted the presence of H7N9 in a given market than swab samples from individual birds. The actual poultry species that served as the source of the virus that infected humans remains unknown, but, based on sampling and field epidemiology, it has been strongly associated with yellow chickens—and the fact that no new human infections arose during the temporary closure of live markets further supports yellow chickens in live markets as the source, he observed.
Between Two Waves
In addition to the range expansion of H7N9 cases between the 2013 and 2014 waves mentioned by Fukuda, Swayne noted that cases also spread beyond live market exposures to include poultry workers and farmers in the second wave. The results of poultry testing also reflected geographic range expansion between the two waves, from a few provinces to a large region of the country. “We don’t know where the affected farms are, but it appears that the locations of affected farms have spread, and we have [affected] farms in more distant provinces than we had last year, which means the problem is growing and not declining,” he said. Thus, while transmission control measures directed at live markets have proven effective in limiting human cases in the short term, the more challenging and equally necessary work of identifying viral reservoir populations remains to be realized.
Pathogenesis in Poultry
To better understand the environmental context of human H7N9 cases, Swayne and coworkers sought to answer the following questions about avian infection (Pantin-Jackwood et al., 2014):
- What bird species are susceptible to infection by H7N9? By inoculating various poultry species across a range of viral titers, the researchers produced infection in all species tested.
- Is clinical disease associated with infection in birds? The only evidence of infection was reduced weight gain.
- Do infected poultry shed large amounts of virus? Chickens and quail were found to shed large amounts of virus for up to 11 days following inoculation. Muscovy duck had similar shedding behaviors but represent only a small percentage of birds in live markets, which are dominated by chickens (and to a lesser extent, quail).
- Does the virus become systemic in birds, thereby facilitating food-borne transmission to humans? H7N9 is generally limited to the upper
respiratory tract with little evidence of systemic spread. In all bird species tested, virus was detected at much higher levels in oral swabs, i.e., respiratory tract replication, than cloacal swabs, i.e., digestive tract replication.
Eradication Strategies
Experience with the H5N1 influenza A virus provides a basis for developing eradication strategies for H7N9, Swayne observed. When he and coworkers compared the effectiveness in eradicating H5N1 on a national basis, against a range of economic indicators, they found no statistical association between rapid eradication and wealth alone, he reported (Pavade et al., 2011). They did, however, find that countries belonging to the Organisation for Economic Co-operation and Development—which are not only wealthy, but also maintain strong principles of governance—had shorter and significantly fewer avian outbreaks, quicker eradication times, and lower mortality rates, he added. Further analysis by the OIE (see the section “The OIE Perspective”) found that countries with “core competencies”43 in national, provincial, and local veterinary services and practice were most effective in limiting H5N1 spread. “Without strong veterinary services you cannot control and eradicate [zoonotic] diseases,” Swayne concluded.
These strategies appear all the more important given the evolutionary history of H5N1 and H7N9, as Swayne briefly explained during the discussion that concluded his workshop presentation. Both viruses acquired their internal genes from influenza A (H9N2), a low-pathogenicity virus found throughout Asia and the Middle East, he said. Epidemiological surveys of poultry in China frequently find H9N2 infections in as many as 10 percent of the birds tested. H9N2 is endemic among poultry raised in many countries, often in conditions that expose them to wild birds, he added. Additional reassortant viruses could easily continue to evolve and emerge much as H5N1 and H7N9 did. As long as H9N2 moves freely through the environment, “We are going to continually have this emerging disease issue in humans from avian influenza virus, because we still have the donors out there, the donors of the internal genes,” he explained.
Influenza A (H7N9) in Humans
While not the first influenza A virus of the subtype H7 to emerge causing illness in humans, H7N9 is the first of its subtype to cause an extensive number of infections in humans, according to speaker Dan Jernigan of the CDC. Other H7 infections had been confirmed in people who had direct contact with infected birds, often during outbreaks in poultry. The severe symptoms of H7N9 in
_______________
43 Veterinary core competencies include staffing of veterinarians and para-veterinarians, professional competencies and continuing education of the veterans, emergency funding, veterinary laboratory diagnosis, epidemiological surveillance, availability of veterinary medicines and biologicals, transparency, disease prevention, control, and eradication.
humans are also unusual, compared with previously reported H7 infections; these were generally mild, causing conjunctivitis and influenza-like illness (Belser et al., 2009). Although H7N9 caused severe illness in humans, it caused little to no disease in poultry. H7N9 is truly something new, he observed.
To illustrate this point, Jernigan described two recent H7 influenza outbreaks in humans. In 2003, an outbreak of highly pathogenic influenza H7N7 in the Netherlands resulted in the culling of more than 30 million birds (Fouchier et al., 2004). The virus was also detected in 86 people who had been directly exposed to infected poultry, and in three of their family members, he said. Among them, 91 percent had conjunctivitis, and one person—a 57-year-old veterinarian with underlying conditions—died. A 2012 outbreak of highly pathogenic H7N3 influenza in Mexico, in which at least 3.8 million birds were culled, resulted in two cases of conjunctivitis without fever or respiratory symptoms in persons exposed to poultry, he added (CDC, 2012).
On March 31, 2013, the China Health and Family Planning Commission notified WHO of three cases of human infection with influenza A H7N9 in Shanghai Municipality (CDC, 2013; WHO, 2013): an elderly man, 87 years old, who had visited poultry markets; a 27-year-old worker in a live market; and a 35-year-old housewife from Guangxi. All three suffered from respiratory tract infection that progressed to fatal pneumonia, he stated. As there was no coincident poultry die-off, the disease had not previously been recognized in poultry in the area where human cases emerged.
Jernigan identified some oft-cited factors in infectious disease emergence with particular relevance to H7N9: crowded conditions44 in the region where the first cases were reported and high-density areas within that region such as live poultry markets and airports; interconnectivity between these regions and the rest of the world via international travel and trade; and an expanding animal–human interface coincident with increasing meat consumption.
International Response
As shown in Figure WO-23, and as previously noted, two waves of human H7N9 cases had occurred at the time of the workshop. Because they occurred in a “post-SARS, post-H1N1 world,” Jernigan said, the response to H7N9 was much swifter and more effective than in past crises such as SARS. The CDC has a long history of public health collaboration with China at multiple levels, including cooperative agreements, laboratory training, and supporting the Chinese National Influenza Center, and the WHO Collaborating Centre for Reference and Research on Influenza established in 2010, he noted. The preparation in China allowed for
_______________
44 This region includes about 575 million people, which is 45 percent of the population of China and 8 percent of the population of the world. Approximately 131 million people, 241 million domestic chickens, and millions of ducks and pigs live within 50 kilometers of the first 60 H7N9 cases, according to Jernigan.
FIGURE WO-23 Number of confirmed human H7N9 cases by week as of July 14, 2014.
SOURCE: WHO, 2014c.
a rapid response to H7N9 emergence. At the global level, WHO’s GISRS played an important role in international influenza coordination, and virus sharing was organized under a global agreement known as the Pandemic Influenza Preparedness Framework. The CDC developed, manufactured, and oversaw the global distribution of H7N9 PCR testing kits and developed an H7N9 candidate vaccine virus, now completing clinical trials.
Three different surveillance systems, in place when H7N9 emerged, contributed to monitoring the virus in China. The Chinese National Influenza-Like Illness Surveillance Network of more than 900 sentinel facilities submitted about 60,000 swabs from laboratories and influenza-like illness patients throughout the country between March and April 2013, of which only six samples from known affected provinces were found to be positive for the virus by PCR (Xu et al., 2013). Specimens collected by the Severe Acute Respiratory Infection and Pneumonia of Unknown Etiology (PUE) surveillance systems were also screened for H7N9 (Xiang et al., 2013); PUE surveillance was the most common means by which cases were identified, Jernigan said.
Epidemiology
At the time of this writing, as at the time of the workshop, no human cases of H7N9 influenza had been reported outside China. “It’s surprising that we have not seen more of these [exported cases],” Jernigan remarked. “I would have expected to have a couple of more of these than we’ve actually seen.”
Peak H7N9 case numbers have coincided with those of seasonal influenza. At the time of this writing, confirmed cases and deaths from H7N9 in China during the most recent wave—from late 2013 through early spring 2014—are shown in Figure WO-24. The focus of disease has moved southward, and now affects Guizhou and Guangxi provinces on the border with Vietnam, Jernigan noted. This is troubling, he said, because Vietnam does not have China’s resources for controlling viral transmission.
Using geospatial mapping coupled with surveillance data to develop risk maps for H7N9 spread in Asia, Fuller and coworkers (2014) identified northern Vietnam as a likely site of disease emergence. “That’s an area where we do need to spend a lot of focus and effort,” Jernigan insisted. He noted that the CDC has worked with the government of Vietnam to improve symptomatic surveillance for humans along the border with China, and also to conduct environmental testing in live poultry markets in order to detect the presence of H7N9. However, he added, H7N9 may have already arrived in Vietnam, as illegal poultry movement between China and Vietnam is common.
At the time of the workshop, Jernigan reported that nearly 400 cases of H7N9 had been reported and eight confirmed clusters had been identified. Since then, as compiled in Table WO-4, 98 percent of confirmed cases were hospitalized, with a 32 percent case fatality. “This is very different than the previous H7 cases in humans—something that is very substantial and has to be addressed,” he observed.
About 70 percent of H7N9 cases to date are males, most of whom live in urban areas and have had direct contact with poultry in live markets, according to Jernigan. Among 139 cases and more than 2,600 contact evaluations they reviewed from the first epidemic wave, Li and coworkers discovered four family disease clusters in which person-to-person transmission could not be ruled out; however, among the large numbers of close contacts, only 28 developed respiratory symptoms, and none tested positive (by PCR) for the virus (Li et al., 2014b). Commenting on this demonstrated lack of efficient and sustained person-to-person transmission of H7N9, Jernigan remarked that the possibility of an emerging pandemic is “clearly something that we have to follow closely.”
The incubation period for H7N9 influenza has been calculated at 3.3 days (Yu et al., 2014), as compared with about 2 days for seasonal influenza, Jernigan reported. As the case contact study suggests, the reproductive number (R0) for H7N9 is quite low: 0.1, as calculated by Chowell et al. (2014). This compares with novel influenzas as a whole (R0 = 0.34) and seasonal flu (R0 = 1.28).
Among the first 111 cases, 61 percent had an underlying condition (Gao et al., 2013). In the previously mentioned study of 139 cases, about 73 percent had
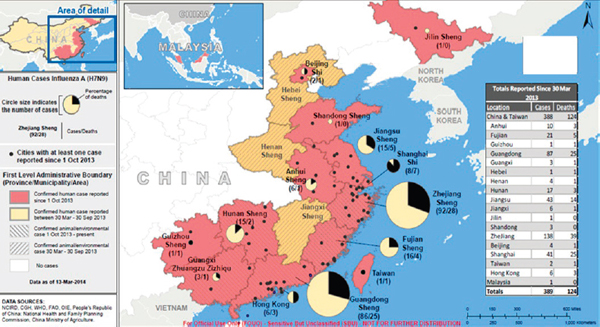
FIGURE WO-24 Confirmed cases and deaths from avian influenza A (H7N9) from October 1, 2013, to mid-March 2014.
SOURCE: Jernigan presentation, 2014.
TABLE WO-4 Avian Influenza A (H7N9) Update: March 17, 2014
| Cumulative counts by report date | 30 Mar–30 Sept, 2013 | 1 Oct 2013–Present | Total | |
| Countries affected | China | China, Malaysia | China, Malaysia | |
| Number of confirmed casesa | 135 | 257 | 392 | |
| Number of confirmed cases hospitalized | 131 | 256 | 387 (98%) | |
| Number of fatal confirmed cases | 45 | 80 | 125 (32%) | |
| Cases of confirmed human-to-human transmissionb | 0 | 0 | 0 | |
| Number of confirmed clustersc | 5 | 3 | 1 | |
| Number of asymptomatic infections | 1 | 0 | 1 | |
a Confirmed cases include persons with laboratory confirmation of H7N9 infection through report from China CDC or Provincial CDC.
b Represents transmission between confirmed cases.
c Confirmed clusters are two or more confirmed cases of H7N9 that are close contacts of one another.
SOURCE: Jernigan presentation, CDC, 2014.
underlying disease (Li et al., 2014b). Hypertension was by far the most common, followed by diabetes. Jernigan described the typical H7N9 influenza case—a member of the target group for intervention—as an older man with chronic heart disease that frequents live markets. Nearly all confirmed H7N9 cases analyzed by Gao and colleagues (2013) developed pneumonia; 71 percent developed acute respiratory distress syndrome (Murray et al., 2012), and 76 percent were admitted to intensive care. “These are tremendously high numbers, showing a very severe infection,” Jernigan remarked. At illness onset, 71 percent of cases presented with cough, which was higher than among human cases of H5N1, he noted (Cowling et al., 2013). The median age of H7N9 influenza cases in the first wave was calculated at 61 years, as compared with 26 years for H5N1 (Cowling et al., 2013; Li et al., 2014b). Clearly, Jernigan concluded, the epidemiology of H7N9 differs markedly from that of H5N1, its likeliest comparator.
Is H7N9 New?
Exploring the possibility that H7N9 is not a novel virus, but merely newly recognized, several studies have produced somewhat conflicting results, Jernigan
observed (Bai et al., 2013; Lebarbenchon et al., 2013; Yang et al., 2013), but overall, the preponderance of data lead to the conclusion that H7N9 has emerged relatively recently and is not merely an artifact of improved detection. Moreover, he added, phylogenetic studies suggest little to no dissemination of H7N9 by waterfowl, as was alluded to previously. It is, therefore, not surprising that serologic studies conducted by the CDC demonstrate that the U.S. population lacks cross-reactive antibodies to H7N9.
Role and Repercussions of Live Markets
Of the 139 cases described by Li and coworkers, more than 80 percent had exposure to animals, which, in nearly every case, included chickens (Li et al., 2014b). In 65 percent of those instances, that exposure came through visiting a poultry market, “So this [poultry markets] is the risk factor of interest,” Jernigan emphasized.
In addition to the significant toll H7N9 influenza has already exacted on human life and health and the attendant costs of medical care, the H7N9 influenza epidemic has also led to major economic losses to the poultry industry in China, Jernigan reported (Qi et al., 2014; Wu and Gao, 2013). The figures associated with these economic effects vary wildly, he noted, but their implications are clear. The poultry industry losses amounted to $1.24 billion in 10 affected provinces and $0.59 billion in 8 nonaffected adjacent provinces (Qi et al., 2014). Economic loss associated with live poultry market closures was in excess of $8 billion in one report (Wu and Gao, 2013).
The closure of live markets dramatically reduced the mean daily number of H7N9 infections in four cities, according to a study by Yu and co-authors (2014). An editorial that accompanied this publication in Lancet also credited the media with informing people about how to avoid infection, Jernigan said. Live market closures have been sporadic in many areas, he noted. “Honestly,” he said, “it has been very difficult to find out exactly what places have been closed and for how long. . . . Clearly there are places that have been implementing measures.” On the other hand, he noted, some traders in Shanghai reportedly sidestepped live market closures by selling poultry online.
What’s Next?
Is H7N9 here to stay? Recalling their recent experience with the H3N2 swine influenza virus, which has caused several human infections since August 2011, Jernigan observed that he and his colleagues at the CDC expected the H3N2 epidemic to last several years. Instead, “We had huge numbers of cases in 2012, and then very few last year. Is that going to happen for H7N9?” Jernigan continued. “I don’t think that’s the case at all.”
“All of the factors really point towards this being an intransigent problem that we will have a hard time detecting, and so it may be a new H5N1 that will be harder to monitor,” Jernigan surmised. He, therefore, argued for improvements in the active surveillance of live markets for the detection of influenza A viruses, “because it’s hard to know exactly where the H7 is, and that may help us to know what to do with human health measures.” Meanwhile, he added, surveillance for H7N9 in humans will unfortunately be a sentinel for animal disease.
A vaccine has been developed against H7N9 and may be stockpiled, Jernigan stated. “The Chinese government is also supporting a vaccine being developed,” he reported, which may become available to the CDC as well. Vaccination of animals may also become an option, albeit a controversial one, he noted.
Ultimately, response to H7N9 must be coordinated on a global basis, Jernigan said, echoing remarks by several other speakers. “We at the CDC will maintain our stocks of diagnostic reagents and other things to help manufacturers, and we will maintain readiness ourselves in working very closely with the Chinese government,” he stated.
INTERNATIONAL AND DOMESTIC RESPONSES TO EMERGING VIRAL DISEASES
A series of workshop presentations described two distinct approaches to addressing the threat posed by emerging viral diseases: first, efforts to control, mitigate, and study recent and ongoing epidemics caused by influenza A (H5N1 and H7N9) and SARS- and MERS-CoV; second, research directed toward predicting the pandemic potential of viruses such as MERS-CoV and H7N9 that are identified during the early stages of their emergence into human populations.
H7N9 Emergence: The Big Picture
The OIE Perspective
Responding to the H7N9 epidemic, the World Organisation for Animal Health (OIE)45 offered assistance to China, and upon being invited to do so, coordinated with the FAO46 and WHO to help that country create a national strategy to address the crisis, according to speaker Alex Thiermann, of the OIE. Several problems hindered this process, he noted: live bird markets were closed too late and infected birds were not sampled on time; a lack of coordination among the many national laboratories involved with the response; and limited sharing of reagents, which were of variable quality.
_______________
45 World Organisation for Animal Health.
46 Food and Agriculture Organization of the United Nations.
These difficulties could arise anywhere, Thiermann emphasized. To improve participation by most countries in detecting and reporting these zoonotic disease outbreaks, countries must have the proper animal health infrastructure, he noted. The epidemiological characteristics of the next pandemic may be entirely different from those of outbreaks of the past, so we must be prepared to detect and respond to any emerging pathogen. To ensure that this happens, every country must have adequate capacity for disease detection and control—an underlying principle of the IHR, and also of the OIE international animal health standards for veterinary services. From the point of view of the OIE, the most effective route to reduce the burden of known and emerging diseases is through assistance and capacity building activities, provided by the OIE, at member country’s request, Thiermann explained. His presentation focused on an evaluation tool developed by the OIE to evaluate and strengthen national veterinary services in their ability to comply with the OIE standards (OIE, 2014c).
Evaluating the performance of veterinary services The OIE Tool for the Evaluation of Performance of Veterinary Services (PVS Pathway) is a continuous process intended to assist countries in evaluating the performance of their veterinary services against 47 critical elements; identifying gaps and weaknesses in their ability to comply with OIE international standards; and to determine a path to improvement and sustainable efficiency, Thiermann stated (OIE, 2014c). Figure WO-25 diagrams this process, which he compared to the course of diagnosing and treating illness. PVS also supports the implementation of international standards, as is the case under the IHR for public health; however, he noted, unlike the WHO, the OIE provides assistance, training and resources to countries to conduct the PVS and follow-up activities, funded through the OIE Animal Health and Welfare Fund.
Thiermann briefly described each step in the PVS Pathway: evaluation; gap analysis; capacity building activities on legislation, laboratories, etc., as well as, follow-up evaluation missions.
Evaluation assesses four categories of critical competencies based on the OIE standards: human, physical, and financial resources; technical capability and legislative authority; interaction with interested parties; and market access. The Evaluation help to raise awareness and to improve the understanding across sectors of the requirements for the effective functioning of the veterinary services, this step results in the creation of a detailed, reliable document for analysis by national authorities. As of June 2014, 129 of the OIE’s 178 member countries had requested PVS evaluations, of which 117 evaluations were completed, 86 reports have been finalized, and 19 are publicly available.
Gap analysis facilitates the identification of priorities for strategic action to address the gaps and improve compliance of veterinary services with the OIE
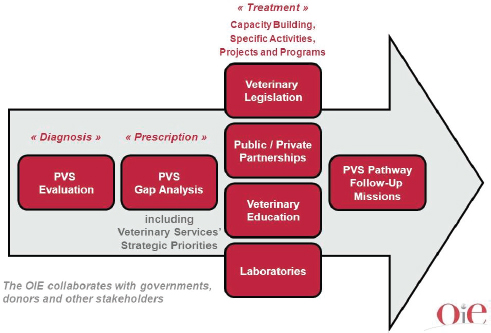
FIGURE WO-25 The PVS Pathway.
SOURCE: OIE, 2014b; Courtesy of the World Organisation for Animal Health. Available at: http://www.oie.int/en/support-to-oie-members/pvs-pathway (accessed June 12, 2014).
international standards and pursue national goals over the next 5 years. In June 2014, the OIE had received 95, and completed 73, requests for gap analyses. This generated 42 completed reports, 11 of which are publicly available.
Capacity building supports the improvements of identified needs through gap analysis. The PVS Pathway Laboratory Mission also helps national veterinary services identify and allocate appropriate resources to the various areas, including the national animal health laboratory system. The missions on Legislation Support offer advice in examining and modernizing national laws and regulations pertaining to veterinary services.
Follow-up missions are conducted every three to five years to measure progress toward the implementation of the PVS-defined strategy to improve compliance with OIE standards.
Many large countries, including China, have not yet participated in the PVS evaluation process, Thiermann reported. However, the OIE, at China’s request, trained 1,000 Chinese veterinarians to become familiar with the PVS Pathway, and apply the same concept at a national level. The European Union is also opting
for internal evaluations, he said. “I’ve been part of discussions between the US, Canada, New Zealand, and Australia on this subject,” he added.
In the discussion that followed Thiermann’s prepared remarks, he noted that countries derive trade benefits from engaging in the PVS process, which produces trusted impartial evaluations by the OIE trained experts. In addition, these evaluations can assist member countries in the process by which the OIE officially recognises countries free of certain animal diseases, creating additional incentives for participation. In order to maintain disease-free status, countries must conduct and share the results of their ongoing disease surveillance activities. This, he noted, allows member countries to focus on diseases and routes to eradication of national importance, providing incentives to maintain a surveillance network that could eventually detect emerging infectious diseases.
The OIE and WHO As global institutions responsible for animal and human health, intergovernmental standards, and strengthening infectious disease surveillance, detection, reporting and response capacity, the OIE and WHO have great potential to work synergistically to advance a One Health agenda, Thiermann stated. As previously noted, the OIE has much greater leverage in influencing member states to comply with its standards than does WHO to enforce the IHR. Now, with support from the World Bank, the OIE and WHO are examining the possible harmonization of national animal and public health capacities for assessing zoonotic disease response, he announced.
Three countries are serving as pilots for the OIE-WHO harmonization effort: Costa Rica, Kenya, and Thailand. The OIE and WHO have been mapping shared outcomes and critical elements, and the OIE provides resources such as costing tools to WHO and encouraging collaboration between animal and human health sectors, Thiermann said. They also plan to implement a joint OIE-WHO workshop at a regional level.
The OIE is also conducting research aimed at determining the cost of controlling or preventing specific diseases, and eventually hopes to demonstrate that this can be best accomplished through collaboration between public health and veterinary services, Thiermann reported. “They don’t have to be merged into a common agency,” he added. “The issue is they need to learn how to work together.” He also noted that the potential for creating joint animal and public health laboratory facilities had been discussed, and countries such as Canada provide good examples for such synergistic arrangements.
The USAID Perspective
Dennis Carroll, director of USAID’s Pandemic Influenza and Other Emerging Threats Unit, discussed research in countries neighboring China that may be at risk for introduction of the H7N9 influenza A virus. As he began his presentation, he noted that it was often said about SARS: “If only we had known what
we could have done to have disrupted and prevented this situation from becoming a global situation.” Ten years later, having detected H7N9 at a much earlier stage—while the disease is still zoonotic, and also geographically limited—we are faced with the challenge of deciding what to do, he observed.
Efforts over the past decade to study the periodic emergence of new H5N1 clades and subclades and their spread within Asia have been very useful in considering how H7N9 may spread geographically, Carroll noted. Thanks to these studies, the role of value chains and the marketing dynamics that move poultry (and disease) throughout Asia is better understood, and the resulting routes and flows that spread disease have been mapped, he explained. Analysis of farm-to-market dynamics—as illustrated in Figure WO-26—reveal how a virus in Shanghai might spread to Guangdong, and from there to Guangxi, and onward to the border of north Vietnam. While not perfect, such “first-order” understanding of these routes is viewed by USAID as an opportunity to target surveillance to monitor for the presence and plan interventions to control the spread of H7N9.
FIGURE WO-26 Live bird market (LBM) networks in Guangxi, Yunnan, and Hunan. Representation of the two-mode “market–source node” network of poultry movement in southern China according to the k-core value. The k-core is a network parameter that measures the centrality of a node within a network. Some LBMs have a higher k-core than others, especially in Hunan and Guangxi provinces, where some LBMs displayed a maximum k-core value of 4 and could play a greater role in highly pathogenic avian influenza virus (HPAIV) maintenance.
SOURCE: Martin et al., 2011.
Implications of early detection Currently, with H7N9 still largely contained within animal reservoirs and limited in geographic distribution, we may be able to disrupt its further spread, and potentially preempt its emergence as a virus with efficient human to human transmission, Carroll stated—something that has never been done before. However, he cautioned, pursuing this goal demands strategies and approaches that fully exploit early detection in ways that do not unnecessarily trigger an emergency response which results in the disruption of ongoing public health and veterinary services. As advances in technology increasingly allow zoonotic pathogens to be detected at a stage before they can move efficiently from person to person, we need to make an effort to distinguish these circumstances from emergencies, Carroll observed. That, he noted, will require coordination and cooperation between ministries of health and agriculture.
Exploiting early detection USAID’s strategy for H7N9 is to attempt to disrupt its spread at the point of introduction, while human disease prevalence is low and the affected poultry population is limited, Carroll stated. Since May 2013, the agency has partnered with FAO, WHO, and the CDC, and eight countries in Asia (see Figure WO-27) to establish capacities for early detection of the virus in both poultry and human, and rapid control of the virus at the point of introduction, he reported. At the same time, as part of its support for operations led by FAO and WHO in China, USAID established H7N9 surveillance activities in border provinces such as Guangxi and Yunnan.
Based primarily on knowledge of regional poultry trade dynamics, the eight Asian countries were categorized in terms of the risk for H7N9 introduction, Carroll explained. In high-risk areas, USAID supports surveillance of live bird markets and human populations. In all countries, the agency is supporting the strengthening clinical care practices for H7N9; disseminating communications to educate political leaders, market owners, traders, and consumers about the virus;47 and supporting the development of a disease-control “tool kit” of interventions (live market closures, cleaning, depopulation, movement control) to contain the virus should it be detected. The agency also cohosted a series of planning and review sessions in China, Myanmar, Rome, and Thailand that brought together representatives from Ministries of Health and Agriculture with technical experts, with the goal of educating ministries on H7N9, and how to use that knowledge to create preparedness plans and recognize needs; these functions continued at subsequent national planning sessions.
_______________
47 The communications package, targeting local authorities as well as high-risk groups in the general public, is in part intended to garner support for possible market closures or culling of apparently healthy but infected birds, Carroll explained—and thereby, reduce the chance that if such methods are used, they will not result in market shocks or the unmonitored movement of poultry (and virus). The importance of creating a supportive environment for disease control is a lesson learned from the global experience with H5N1 influenza, he noted.
FIGURE WO-27 Stratifying risk.
SOURCE: Carroll presentation, 2014. Adapted from FAO.
As previously noted, low-pathogenicity H7N9 infection is difficult to detect in all but humans. Thus, Carroll said, it is important for Ministries of Health to coordinate their monitoring for human infection with the Ministries of Agriculture, as knowledge of poultry production would inform determinations of vulnerable “points of entry” for H7N9 into their countries. Guidance documents were distributed to encourage standardized sample collection and diagnostic approaches, along with a group of recommended contingency control measures, should the virus be detected.
Operations As previously illustrated in Figure WO-27, USAID ranked the three countries sharing a border and direct commercial trade with China—Laos, Myanmar, and Vietnam—as having a “high risk” for H7N9 introduction, and five others— Bangladesh, Bhutan, Cambodia, Indonesia, and Nepal—as having a “moderate risk.” USAID support in each country reflects its relative risk.
High-risk entry points have been identified (and illustrated in Figure WO-28) within Laos and Myanmar where joint planning between the Ministry of Health and Ministry of Agriculture has focused on surveillance of live bird markets, along with influenza-like illness and severe acute respiratory illness surveillance in people living near market sites, as previously described by Jernigan. “This is a work in progress,” Carroll observed. In Laos, poultry and human surveillance has been aligned in the most high-risk provinces, and in several locations in

FIGURE WO-28 Ongoing surveillance for H7N9: Laos and Myanmar. Joint planning between ministries of health and ministries of agriculture has increased coordinated monitoring for H7N9 in targeted live bird markets and adjacent health facilities.
SOURCE: Carroll presentation, 2014. Adapted from FAO.
Myanmar, “but they’re still working out the details of further alignment within both of these countries,” he reported. In Vietnam, more than 20,000 samples obtained from 70 live bird markets in 12 northern provinces were all negative, according to Carroll. Surveillance is also ongoing in live bird markets deemed high risk for H7N9 introduction in Bangladesh, Bhutan, Cambodia, Indonesia, and Nepal, he said.
In China, analysis of initial data on market closure strategies does not clearly demonstrate that it is effective in breaking the chain of transmission, Carroll reported. USAID and FAO plan to gather additional evidence on the effects of market closures and cleanings on influenza rates over the long term. More generally, he noted, the emergence of H7N9 provides an opportunity to evaluate the feasibility of infectious disease disruption through a combination of early detection and rapid control measures instituted during the “introductory phase” of emergence.
Disruption as a long-term strategy Can we develop strategies that exploit the earlier and earlier identification of emergent zoonoses? Can appropriate interventions be developed to minimize the spread of such a pathogen, thereby disrupting its ability to adapt to humans? If so, Carroll said, this would represent a long-term strategy for disease control, particularly in areas of Asia where multiple strains of avian influenza viruses are cocirculating (e.g., H5N1, -N2, and -N8; H6N1; H7N1; H9N2; and H10N8—in addition to H7N9). Southeast Asia is “a rich cauldron for new viral emergence,” he observed; those viruses that infect poultry will move along regional value chains. This presents the possibility of “normalizing” strategies for early detection and control of emergent viruses through existing surveillance platforms and interventions focused on live animal markets, he suggested.
Even so, Carroll emphasized, disrupting the adaptation of emergent viruses to human hosts is not the same as preempting the emergence of zoonotic threats. Strategies for disruption are not a solution; they are a response to problems that arise from current systems of livestock production and marketing. These factors continue to raise the risk for pandemic influenza and other pathogens that can only be reduced through measures that truly improve biosecurity on farms and in markets, he concluded.
β-CoV Challenges in Health Care Facilities
Having played a central role in responding to and describing the SARS outbreak in Toronto, speaker Allison McGeer, of Mt. Sinai Hospital, confessed to finding her task of health care issues associated with emerging viruses “a little bit depressing”—not because of the memories it rekindled, but because there has been relatively little progress on these issues in the ensuing years (Raboud et al., 2010) (Dr. McGeer’s contribution may be found on pages 181–184 in Appendix A). To illustrate this point, she described the scenario of one of the
last smallpox outbreaks in the United Kingdom, which occurred in Bradford, England, in 1962. In much the same way that smallpox spread to 13 contacts of the index case within a hospital, coronaviruses and other emerging diseases could spark a nosocomial outbreak.
Several things about emerging diseases spell trouble for hospitals, according to McGeer: in the case of smallpox, an infectious period that extended late into the disease (not common among bacterial or viral pathogens); an unrecognized disease; open waiting areas and emergency department bays, along with multibed rooms; lack of standard practices adequate to control the transmission of infectious disease; and hospitals that are inadequate to contain spread of communicable pathogens. “The only thing that has changed in the last 50 years is that we have eradicated smallpox, and we have better vaccination programs at a hospital level,” she observed.
SARS vs. MERS
Unlike MERS, SARS spread rapidly and widely, McGeer recalled. More than three-quarters of SARS cases in Hong Kong and in Toronto were hospital associated, she reported; about a third of those were health care workers, and the remaining two-thirds were visitors and patients in the hospital. The fatality rate of MERS at first appeared much higher than that of SARS, she said, “but in fact this difference is driven almost entirely by the difference in infected populations.” About 40 percent of people who were infected in SARS outbreaks outside of China were health care workers, between the ages of 25 and 40, without underlying illness, she explained. By contrast, most people who have been infected with MERS have been hospital patients with primary infections, who are much older and more likely to have underlying illness. In Toronto, for example, the case fatality rate in 60-year-olds was 54 percent, and in patients with nosocomial infections, it was 50 percent. “That looks a lot like MERS,” she observed.
Indeed, she continued, “The longer we’ve been watching MERS evolve, the more closely it resembles SARS.” Both are primarily pulmonary diseases, she noted, which have shown similar times from onset of symptoms to hospitalization (4 days for MERS; 3 for SARS), as well as incubation times (5.2 days for MERS; 4.6 for SARS) and serial intervals (7.6 days for MERS; 8.4 for SARS), she reported. With its slightly longer incubation period and slightly shorter serial interval, MERS patients can transmit the disease earlier in their infection than could SARS patients, she observed. “What saved us in SARS was that people were not infectious until they were really sick in the hospital,” she said. That does not appear to be true of MERS, and if that is the case, it is a significant difference, she concluded.
While primary MERS cases had been predominantly male, health care–associated cases were 80 percent female, McGeer reported. This percentage is roughly equivalent to the gender ratio of that population, she stated—supporting
Fukuda’s earlier observation that the demographics of primary MERS cases largely reflect exposure, rather than specific vulnerability. Nevertheless, McGeer continued, “There are still some mysteries about what goes on [with MERS] in hospitals.” In the Al-Musa Hospital outbreak, previously described by Perl, about 100 health care workers were thought to have been exposed to the virus, including 43 patients on dialysis, of whom 17 were confirmed probable cases. However, she added, of 18 full-time staff in that unit, only 1 presented with fever for 2 days and was not tested for MERS. The other health care workers remained apparently healthy—a dramatically different outcome compared with what happened during SARS. “I don’t yet have serology to know whether there was a substantial number of asymptomatic infections [among hospital staff at Al-Musa],” she acknowledged. However, she added, it is mystifying that so much transmission occurred there between patients without the development of illness among staff members.
It is also notable that MERS does not resemble other viral respiratory diseases. Influenza, for example, produces more cases of mild illness, fewer people with severe disease, and a much lower case fatality rate, McGeer said. This behavior resembles meningococcal meningitis, she observed: “You either don’t get sick, or you get really sick and you have a high case fatality rate.” Clearly, she concluded, this is “different from what we’re used to seeing, and because of that, significant.”
The current case fatality rate of MERS in Saudi Arabia of 41 percent suggests that it either causes a more severe disease in young healthy people than SARS, or that only a small fraction of infections in health care workers are being detected, according to McGeer. “Either way, it’s bad news, because either we’re looking at a disease that has a 10 percent case fatality rate in health care workers . . . or we’re looking at a disease that is much more transmissible to health care workers than we’re recognizing in Saudi Arabia,” she said.
McGeer noted two important differences between SARS and MERS: the length of the period of infectiousness, and the reproductive ratio (R0). The number of secondary cases from every index case of SARS ranged from 2.2 to 3.9 in various locations, she stated. Even in the outbreak in Al Hasa, the R0 for MERS was found to be 0.5, and it lies between 0.4 and 0.6 for other MERS cases to date, she reported. However, she added, “It doesn’t seem like there is necessarily going to be a big jump for MERS to become more transmissible and more like SARS.”
MERS in Health Care
There are several reasons to be concerned about the impact of coronaviruses such as SARS and MERS in health care settings, according to McGeer, including the fact that health care–-associated cases represent a significant proportion of disease with these coronaviruses (WHO MERS-CoV Research Group, 2013); their high case fatality rates; and the inherent difficulties involved in diagnosis and prevention of transmission.
On its face, preventing coronavirus transmission is simple, McGeer observed: put people in private rooms and wear barriers when you take care of them—that is, follow so-called droplet contact precautions. But how can you tell you are dealing with SARS or MERS? “With coronavirus, as [with] other viral respiratory illnesses . . . you are dreaming in Technicolor if you think you can distinguish one cause of respiratory illness from another,” she quipped. “It cannot be done, and in fact many older people who have viral respiratory illnesses present with complications of those illnesses rather than the illnesses themselves, so what seems like something that is really simple turns out to be really difficult.” These circumstances raise the following important challenges for health care institutions.
Evaluating basic practices Results from several studies analyzing health care worker protection during the SARS epidemic suggest that basic practices such as hand hygiene are helpful in controlling the spread of coronaviruses, McGeer concluded. However, research is ongoing to identify factors that influence how viruses are transmitted in health care, as well as what can be done to control it, she said. Past efforts toward this goal have to some extent improved our ability to prevent infectious disease transmission, but much remains to be done.
Infection control education during the SARS outbreak was associated with a reduction in the risk of infection, McGeer reported. “The better we train our health care workers, the better we as health care workers understand how to implement prevention, the better off we will be,” she declared. She also acknowledged, however, that it is hard to persuade health care workers to change. “It took us 2 months in the middle of [the SARS] outbreak to persuade health care workers that they needed to be adhering to precautions against infection,” she recalled. “So you can imagine how hard it is to do in a much lower risk situation” such as MERS. “It will require a revolution in the provision of care in our hospitals to manage the kind of change that we need [in order to] to protect people from emerging viruses,” she predicted.
Changing the built environment “If we didn’t have open bays in our emergency department, we would not have had the SARS outbreak in Toronto, and that would have saved us about $1.2 billion,” McGeer stated. “We could have built closed rooms in every emergency department in the country for that price.” Efforts to change any hospital’s built environment to reduce infectious disease transmission will require careful, appropriate analysis of cost effectiveness, she added.
Recognizing disease Thanks to ongoing progress in point-of-care diagnostics, patients with coronavirus infections such as SARS and MERS may someday be rapidly identified, McGeer predicted. “Recognition of disease is a critical element of managing these cases, and our abilities to do that is within reach now,” she
said. “We need to accelerate our ability to diagnose disease and, in particular, diagnose communicable disease in hospitals.”
Therapeutic prospects β-coronavirus diseases are “begging for therapy,” McGeer observed, and with MERS, as with SARS, it is likely to be difficult—if not impossible—to develop new drugs in short periods of time, let alone discover them. An old methodology, convalescent plasma therapy—which apparently reduced mortality when used during the 1918 influenza pandemic—might be worth investigating as a stopgap measure, she suggested (Hung et al., 2013; Luke et al., 2006). “It seems to me a really important, if perhaps relatively small, intervention that might help in the future emergence of disease in hospitals,” she concluded.
Health care workers’ expectations “Health care workers in truth are at very low risk of occupational disease or injury in any circumstance, but we have become accustomed to thinking that we are safe,” McGeer observed. Indeed, coronavirus infections are only some of the risks faced by health care workers, which also include higher rates of influenza and antimicrobial resistance, she reported. Moreover, she added, occupational risk associated with emerging infections is not limited to health care workers—but the nature of these additional occupations, the risks involved, and how to mitigate them, remain largely to be determined.
Predicting Pandemic Potential of Zoonotic Influenza Viruses
While the emergence of a pandemic strain of H5N1 or H7N9 influenza appears unlikely to happen, it could be disastrous if it did. Since the emergence of H5N1 in Hong Kong in 1997, the question of how easily this virus could evolve to transmit readily among humans has preoccupied many researchers and policy makers; now it is being asked about H7N9 as well. “Many of the best flu labs in the world for over 10 years were working on trying to figure out whether or not such viruses could go airborne among mammals,” observed speaker Derek Smith, of Cambridge University.
In 2012, after two groups of scientists separately showed that H5N1 viruses could be genetically engineered through so-called gain-of-function experiments (Herfst et al., 2012; Imai et al., 2012), Smith and coworkers demonstrated that the likelihood of these changes occurring naturally was sufficient to present a “potentially serious threat” (Russell et al., 2012). In his presentation to the workshop, Smith discussed the state of research and policy on zoonotic threats in light of these discoveries.
Predicting Transmissibility of Influenza Viruses in the Ferret Model System
“It’s absolutely clear what we should do next,” Smith argued: test naturally occurring influenza viruses to see if they possess the functional equivalent of the
substitutions determined by experiment to confer transmissibility between mammals; choose those that are closest to making this transition and test them for their ability to transmit between mammalian animal models (e.g., ferrets); and then experimentally determine which substitutions make this possible. Using this method, he and coworkers discovered a 2006 German isolate of H5N1 that he deemed “closest to transmissible” (Herfst et al., 2012).
“There is an enormous amount that we can know about what the emergence potential of this particular virus is,” Smith stated. Moreover, researchers can gain information from such discoveries to refine predictions about which viruses are more likely to transmit, or to require the fewest adaptations to transmit, and continue to test those predictions, he said. “We can learn this for H5 based on what we know, and we can apply it to other influenza threats as well,” he added.
However, Smith continued, such experiments are not being conducted in a systematic way, for reasons that are understandable. A comprehensive program to discover preemergent viral threats would require a major commitment of resources, he noted, and it would constitute “dual-use research of concern” as its results could be misused to pose a biologic threat to public health and/or national security.48 As predicted a decade ago in the influential report Biotechnology Research in an Age of Terrorism (NRC, 2004), the need for biosecurity has had a chilling effect on efforts to identify preemergent influenza viruses, he observed.
Addressing Dual-Use Concerns
For many of the experiments Smith and colleagues have conceived to explore influenza transmission it is not easy to determine whether the risk they pose for dual use outweighs their potential benefit, he said. A robust, consensus process needs to be developed that involves both scientists and national security experts in making such decisions, he argued—and soon; otherwise, scientists will simply stop doing work that supports such decisions for lack of funding and trained personnel. “If we don’t come together on this . . . we do run the risk that we will lose . . . the scientific partners because it’s just too hard to do work in the area. What’s really critical is that the people on the science [and health care] side, and the people on the national security side, need to be around the same table, because neither . . . are experts in the other domain,” he insisted.
Both do, however, understand the concept of risk, and this should be the basis for their deliberations, according to Smith. While acknowledging that “it’s very easy to overestimate risks, and it’s very easy to underestimate risks,” he suggested
_______________
48 Dual-use research of concern is life sciences research that, based on current understanding, can be reasonably anticipated to provide knowledge, information, products, or technologies that could be directly misapplied to pose a significant threat with broad potential consequences to public health and safety, agricultural crops and other plants, animals, the environment, materiel, or national security. Source: http://osp.od.nih.gov/office-biotechnology-activities/biosecurity/dual-use-research-concern (accessed June 12, 2014).
that these extremes could be balanced in much the same way as an actuary sets the price of insurance.
On the other hand, Smith added, these decisions should not only reflect careful estimates of risk, but also human judgments that should be applied in a quantitative, transparent way whenever possible—perhaps guided by peer review. “If these decisions are taken in a way where this normal scientific process can’t apply to at least some of the calculations, then I think we really do run the risk of driving the scientists away,” he warned. Fortunately, he said, efforts are under way to better connect academic research with national security efforts.
“We know so little about H7N9 compared to what we could know about it,” Smith observed. Barring introduction of a universal influenza vaccine—as previously described by Fauci, and which would obviate the need to predict pandemic potential—there is no alternative to gain-of-function experimentation, he insisted.
A Pandemic Risk Assessment Framework for Animal Influenza Viruses
Speaking this time about one of his own CDC projects, Donis reminded workshop participants that real-time PCR diagnosis of influenza, which first came into widespread use in U.S. hospitals and laboratories in 2009, vastly increased detection and comprehension of many influenza subtypes, most notably zoonotic strains. Expressing hope that this technology, now becoming common in Europe, will eventually gain a foothold in Asia as well, he observed, “The more we use these molecular diagnostic tools, the more we’re going to find what novel virus is causing sporadic infections” such as human cases of H5N1 and H7N9, he said.
A Basis for Comparison
But this expanding catalog of novel influenza viruses presents a challenge: how to identify those likeliest to develop the capacity for human-to-human transmission? “We have a number of viruses that are being detected in zoonotic infections, and we have to have a mechanism to understand their relative importance,” Donis stated. To meet this need, he and coworkers have developed the Influenza Risk Assessment Tool (IRAT) to identify, define, and assess risk associated with a specific viral subtype relative to others, providing actionable information to risk management programs (Trock et al., 2012). As such, it could be considered an instrument of the “risk governance” model advocated by Pfeiffer (see the section “The Risk Governance Framework for Disease Management”).
A risk-scoring algorithm informed by expert observation, Donis explained that the IRAT was designed to answer two key questions: What is the risk of a given virus emerging as a pandemic? And, if it does so, how severe would the pandemic be? IRAT’s developers determined that three categories of factors contributed to pandemic risk: the properties of the virus, the attributes of the population, and the ecology and epidemiology of the virus. Each category contains several risk elements. For example, “transmission in animal models”
is an element of the category, “properties of the virus”; “disease severity” is an “attribute of the population”; and “global distribution” is an element of “ecology and epidemiology.” Each risk element is defined precisely in terms of what constitutes low, moderate, or high risk, he said, and each risk level is assigned a numerical score. A subject-matter expert—such as a researcher knowledgeable about a particular virus—assigns the various scores for that virus, which incorporate both range and confidence level.
Once calculated, the total scores for several viruses can then be compared to each other, in order to answer the two key questions: the relative risk of emergence, and of high public health impact, Donis said. However, the risk elements composing those scores must first be weighted to reflect the question being posed, as different factors favor pandemic potential and severity of disease, he noted.
Using IRAT
The IRAT offers researchers and policy makers a consistent approach to evaluate risk, Donis observed; it reduces bias in comparisons among viruses and documents information used in decision making. It is a tool that is useful not only for the comparisons it facilitates, but for the facility by which it allows information to be shared. IRAT is also readily modifiable, he pointed out.
“The use of the IRAT has also been humbling in some ways, because many times we have very little data to perform a score,” Donis acknowledged. “Most of the viruses of concern are those that we know very little about, and especially when there are a large number of cases. H7N9 was a perfect example of that,” he observed, noting Jernigan’s description of the onset of severe human disease from a virus that formerly was associated with conjunctivitis. “Now, it’s a totally different virus,” he observed. When they attempted to compute an IRAT score for H7N9, Donis and coworkers found only a few H7N9 sequences in the database. “We know a lot about Eurasian H7s, but this is a totally different beast. This has H9N2 internal genes. So you’re faced with a lot of gaps, huge gaps of knowledge,” he explained. “I think many times we’re forced to compare apples with pineapples.”
Next Steps
The more IRAT is used, the more useful it will become, according to Donis. He hopes to put it in the hands of anyone who can benefit from it, and particularly WHO, in order to support GISRS.49 “Please go and find novel viruses and score them and put the information on the table for everybody to discuss,” he urged, because that will create incentive for even greater exploration and information sharing.
_______________
49 See http://www.who.int/influenza/gisrs_laboratory/en (accessed June 12, 2014).
In this way, IRAT could foster a global effort in sample collection and analysis extending to the creation of standardized methods for virus assessment such as transmission studies or measures of human population immunity, Donis observed. “It is hoped this will lead us to better databases, better data, better reporting, and ultimately, to better public health,” he concluded.
Abbott, R. C., J. E. Osorio, C. M. Bunck, and T. E. Rocke. 2012. Sylvatic plague vaccine: A new tool for conservation of threatened and endangered species? EcoHealth 9(3):243-250.
Agnihothram, S., R. Gopal, B. L. Yount, Jr., E. F. Donaldson, V. D. Menachery, R. L. Graham, T. D. Scobey, L. E. Gralinski, M. R. Denison, M. Zambon, and R. S. Baric. 2014. Evaluation of serologic and antigenic relationships between Middle Eastern respiratory syndrome coronavirus and other coronaviruses to develop vaccine platforms for the rapid response to emerging coronaviruses. Journal of Infectious Diseases 209(7):995-1006.
Alagaili, A. N., T. Briese, N. Mishra, V. Kapoor, S. C. Sameroff, P. D. Burbelo, E. de Wit, V. J. Munster, L. E. Hensley, I. S. Zalmout, A. Kapoor, J. H. Epstein, W. B. Karesh, P. Daszak, O. B. Mohammed, and W. I. Lipkin. 2014. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio 5(2):e00884-14.
Amman, B. R., S. A. Carroll, Z. D. Reed, T. K. Sealy, S. Balinandi, R. Swanepoel, A. Kemp, B. R. Erickson, J. A. Comer, S. Campbell, D. L. Cannon, M. L. Khristova, P. Atimnedi, C. D. Paddock, R. J. Kent Crockett, T. D. Flietstra, K. L. Warfield, R. Unfer, E. Katongole-Mbidde, R. Downing, J. W. Tappero, S. R. Zaki, P. E. Rollin, T. G. Ksiazek, S. T. Nichol, and J. S. Towner. 2012. Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathogens 8(10):e1002877.
Amman, B. R., L. Nyakarahuka, A. K. McElroy, K. A. Dodd, T. K. Sealy, A. J. Schuh, T. R. Shoemaker, S. Balinandi, P. Atimnedi, W. Kaboyo, S. T. Nichol, and J. S. Towner. 2014. Marburgvirus resurgence in Kitaka Mine bat population after extermination attempts, Uganda. Emerging Infectious Diseases 20(10):1761-1764.
Animal Research Info. 2014. Ebola vaccine & western gorillas. http://www.animalresearch.info/en/listing/114/ebola-vaccine-western-gorillas/%23ebolavirus (accessed September 4, 2014).
Anonymous. 2011. The European environment—state and outlook 2010: Assessment of global megatrends. Copenhagen, Denmark, European Environment Agency. 128 pp.
Anonymous. 2014. Global risks 2014. Geneva, Switzerland: World Economic Forum. 59 pp.
Appel, B., G.-F. Böl, M. Greiner, M. Lahrssen-Wiederholt, and A. Hensel. 2012. EHEC Outbreak 2011—Investigation of the outbreak along the food chain. Berlin, Germany: Federal Institute for Risk Assessment.
Assiri, A., A. McGeer, T. M. Perl, C. S. Price, A. A. Al Rabeeah, D. A. Cummings, Z. N. Alabdullatif, M. Assad, A. Almulhim, H. Makhdoom, H. Madani, R. Alhakeem, J. A. Al-Tawfiq, M. Cotten, S. J. Watson, P. Kellam, A. I. Zumla, and Z. A. Memish. 2013. Hospital outbreak of Middle East respiratory syndrome coronavirus. New England Journal of Medicine 369(5):407-416.
AVMA (American Veterinary Medical Association). 2008. One Health: A new professional imperative. Schaumburg, IL: AVMA.
AVMA. 2014. One Health—What is One Health? https://www.avma.org/KB/Resources/Reference/Pages/One-Health94.aspx (accessed September 4, 2014).
Bai, T., J. Zhou, and Y. Shu. 2013. Serologic study for influenza A (H7N9) among high-risk groups in China. New England Journal of Medicine 368(24):2339-2340.
Bat Conservation International. 2014. White-nose syndrome: A crisis for America’s bats. http://www.batcon.org/pdfs/whitenose/WNS_FAQjuly14.pdf (accessed September 4, 2014).
Bean, A. G. D., M. L. Baker, C. R. Stewart, C. Cowled, C. Deffrasnes, L.-F. Wang, and J. W. Lowenthal. 2013. Studying immunity to zoonotic diseases in the natural host—keeping it real. Nature Reviews: Immunology 13(12):851-861.
Belser, J. A., C. B. Bridges, J. M. Katz, and T. M. Tumpey. 2009. Past, present, and possible future human infection with influenza virus A subtype H7. Emerging Infectious Diseases 15(6):859-865.
Blehert, D. S. 2012. Fungal disease and the developing story of bat white-nose syndrome. PLoS Pathogens 8(7):e1002779.
Blehert, D. S., A. C. Hicks, M. Behr, C. U. Meteyer, B. M. Berlowski-Zier, E. L. Buckles, J. T. Coleman, S. R. Darling, A. Gargas, and R. Niver. 2009. Bat white-nose syndrome: An emerging fungal pathogen? Science 323(5911):227.
Blehert, D. S., J. M. Lorch, A. E. Ballmann, P. M. Cryan, and C. U. Meteyer. 2011. Bat white-nose syndrome in North America. Microbe 6(6):267-273.
Bolles, M., E. Donaldson, and R. Baric. 2011. SARS-CoV and emergent coronaviruses: Viral determinants of interspecies transmission. Current Opinion in Virology 1(6):624-634.
Breban, R., J. Riou, and A. Fontanet. 2013. Interhuman transmissibility of Middle East respiratory syndrome coronavirus: Estimation of pandemic risk. Lancet 382(9893):694-699.
Brown, M. A., J. L. Troyer, J. Pecon-Slattery, M. E. Roelke, and S. J. O’Brien. 2009. Genetics and pathogenesis of feline infectious peritonitis virus. Emerging Infectious Diseases 15(9):1445.
Calisher, C. H., J. E. Childs, H. E. Field, K. V. Holmes, and T. Schountz. 2006. Bats: Important reservoir hosts of emerging viruses. Clinical Microbiology Reviews 19(3):531-545.
Carver, S., A. Bestall, A. Jardine, and R. S. Ostfeld. 2009. Influence of hosts on the ecology of arboviral transmission: Potential mechanisms influencing dengue, Murray Valley encephalitis, and Ross River virus in Australia. Vector Borne and Zoonotic Diseases 9(1):51-64.
Cauchemez, S., C. Fraser, M. D. Van Kerkhove, C. A. Donnelly, S. Riley, A. Rambaut, V. Enouf, S. van der Werf, and N. M. Ferguson. 2014. Middle East respiratory syndrome coronavirus: Quantification of the extent of the epidemic, surveillance biases, and transmissibility. Lancet Infectious Diseases 14(1):50-56.
CDC (Centers for Disease Control and Prevention). 2010. Report suggests nearly 5 percent exposed to dengue virus in Key West. http://www.cdc.gov/media/pressrel/2010/r100713.htm (accessed September 8, 2014).
CDC. 2012. Notes from the field: Highly pathogenic avian influenza A (H7N3) virus infection in two poultry workers—Jalisco, Mexico, July 2012. Morbidity and Mortality Weekly Report 61(36):726-727.
CDC. 2013. Emergence of avian influenza A (H7N9) virus causing severe human illness—China, February-April 2013. Morbidity and Mortality Weekly Report 62(18):366-371.
CDC. 2014. Notes from the field: Chikungunya virus spreads in the Americas—Caribbean and South America, 2013-2014. Morbidity and Mortality Weekly Report 63(22):500-501.
CDC. 2015. 2014 Ebola outbreak inWest Africa—Case counts. http://www.cdc.gov/vhf/ebola/outbreaks/2014-west-africa/case-counts.html (accessed February 25, 2015).
Chang, L. J., K. A. Dowd, F. H. Mendoza, J. G. Saunders, S. Sitar, S. H. Plummer, G. Yamshchikov, U. N. Sarwar, Z. Hu, M. E. Enama, R. T. Bailer, R. A. Koup, R. M. Schwartz, W. Akahata, G. J. Nabel, J. R. Mascola, T. C. Pierson, B. S. Graham, and J. E. Ledgerwood. 2014. Safety and tolerability of chikungunya virus-like particle vaccine in healthy adults: A phase 1 dose-escalation trial. Lancet 384(9959):2046-2052.
Chowell, G., L. Simonsen, S. Towers, M. A. Miller, and C. Viboud. 2014. Transmission potential of influenza A/H7N9, February to May 2013, China. BMC Medicine 11:214.
Clayton, B. A., L. F. Wang, and G. A. Marsh. 2013. Henipaviruses: An updated review focusing on the pteropid reservoir and features of transmission. Zoonoses and Public Health 60(1):69-83.
Cockburn, A. 1963. The evolution and eradication of infectious diseases. Baltimore: Johns Hopkins Press.
Cockrell, A. S., K. M. Peck, B. L. Yount, S. S. Agnihothram, T. Scobey, N. R. Curnes, R. S. Baric, and M. T. Heise. 2014. Mouse dipeptidyl peptidase 4 is not a functional receptor for Middle East respiratory syndrome coronavirus infection. Journal of Virology 88(9):5195-5199.
Cohen, J. 2013. New flu virus in China worries and confuses. Science 340(6129):129-130.
Cotten, M., S. J. Watson, P. Kellam, A. A. Al-Rabeeah, H. Q. Makhdoom, A. Assiri, J. A. Al-Tawfiq, R. F. Alhakeem, H. Madani, F. A. AlRabiah, S. Al Hajjar, W. N. Al-nassir, A. Albarrak, H. Flemban, H. H. Balkhy, S. Alsubaie, A. L. Palser, A. Gall, R. Bashford-Rogers, A. Rambaut, A. I. Zumla, and Z. A. Memish. 2013. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: A descriptive genomic study. Lancet 382(9909):1993-2002.
Cowling, B. J., L. Jin, E. H. Lau, Q. Liao, P. Wu, H. Jiang, T. K. Tsang, J. Zheng, V. J. Fang, and Z. Chang. 2013. Comparative epidemiology of human infections with avian influenza A H7N9 and H5N1 viruses in China: A population-based study of laboratory-confirmed cases. Lancet 382(9887):129-137.
Dandekar, A. A., and S. Perlman. 2005. Immunopathogenesis of coronavirus infections: Implications for SARS. Nature Reviews: Immunology 5(12):917-927.
Dawood, F. S., A. D. Iuliano, C. Reed, M. I. Meltzer, D. K. Shay, P. Y. Cheng, D. Bandaranayake, R. F. Breiman, W. A. Brooks, P. Buchy, D. R. Feikin, K. B. Fowler, A. Gordon, N. T. Hien, P. Horby, Q. S. Huang, M. A. Katz, A. Krishnan, R. Lal, J. M. Montgomery, K. Molbak, R. Pebody, A. M. Presanis, H. Razuri, A. Steens, Y. O. Tinoco, J. Wallinga, H. Yu, S. Vong, J. Bresee, and M. A. Widdowson. 2012. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: A modelling study. Lancet Infectious Diseases 12(9):687-695.
de Haan, C. A., Z. Li, E. te Lintelo, B. J. Bosch, B. J. Haijema, and P. J. Rottier. 2005. Murine coronavirus with an extended host range uses heparan sulfate as an entry receptor. Journal of Virology 79(22):14451-14456.
Decaro, N., C. Desario, D. D. Addie, V. Martella, M. J. Vieira, G. Elia, A. Zicola, C. Davis, G. Thompson, E. Thiry, U. Truyen, and C. Buonavoglia. 2007. The study molecular epidemiology of canine parvovirus, Europe. Emerging Infectious Diseases 13(8):1222-1224.
Decaro, N., M. Viviana, M. Campolo, A. Lorusso, M. Camero, G. Elia, V. Martella, P. Cordioli, L. Enjuanes, and C. Buonavoglia. 2009. Recombinant Canine Coronaviruses Related to Transmissible Gastroenteritis Virus of Swine Are Circulating in Dogs. Journal of Virology 83(3):1532-1537.
Denison, M. R., R. L. Graham, E. F. Donaldson, L. D. Eckerle, and R. S. Baric. 2011. Coronaviruses: An RNA proofreading machine regulates replication fidelity and diversity. RNA Biology 8(2):270-279.
Doms, R. W. 2010. Prime, boost, and broaden. Science 329(5995):1021-1022.
Drexler, J. F., F. Gloza-Rausch, J. Glende, V. M. Corman, D. Muth, M. Goettsche, A. Seebens, M. Niedrig, S. Pfefferle, S. Yordanov, L. Zhelyazkov, U. Hermanns, P. Vallo, A. Lukashev, M. A. Muller, H. Deng, G. Herrler, and C. Drosten. 2010. Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. Journal of Virology 84(21):11336-11349.
Eckerle, I., V. M. Corman, M. A. Muller, M. Lenk, R. G. Ulrich, and C. Drosten. 2014. Replicative capacity of MERS coronavirus in livestock cell lines. Emerging Infectious Diseases 20(2):276-279.
Epstein, J. H., H. E. Field, S. Luby, J. R. Pulliam, and P. Daszak. 2006. Nipah virus: Impact, origins, and causes of emergence. Current Infectious Disease Reports 8(1):59-65.
Erb, K.-H., F. Krausmann, W. Lucht, and H. Haberl. 2009. Embodied HANPP: Mapping the spatial disconnect between global biomass production and consumption. Ecological Economics 69(2):328-334.
Ercsey-Ravasz, M., Z. Toroczkai, Z. Lakner, and J. Baranyi. 2012. Complexity of the international agro-food trade network and its impact on food safety. PLoS ONE 7(5):e37810.
Falk, H., S. Durr, R. Hauser, K. Wood, B. Tenger, M. Lortscher, and G. Schupbach-Regula. 2013. Illegal import of bushmeat and other meat products into Switzerland on commercial passenger flights. Revue Scientifique et Technique 32(3):727-739.
Fauci, A. S., and D. M. Morens. 2012. The perpetual challenge of infectious diseases. New England Journal of Medicine 366(5):454-461.
Fineberg, H. V. 2014. Pandemic preparedness and response—Lessons from the H1N1 influenza of 2009. New England Journal of Medicine 370(14):1335-1342.
Fouchier, R. A., P. M. Schneeberger, F. W. Rozendaal, J. M. Broekman, S. A. Kemink, V. Munster, T. Kuiken, G. F. Rimmelzwaan, M. Schutten, G. J. Van Doornum, G. Koch, A. Bosman, M. Koopmans, and A. D. Osterhaus. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proceedings of the National Academy of Sciences of the United States of America 101(5):1356-1361.
Fuller, T., F. Havers, C. Xu, L.-Q. Fang, W.-C. Cao, Y. Shu, M.-A. Widdowson, and T. B. Smith. 2014. Identifying areas with a high risk of human infection with the avian influenza A (H7N9) virus in East Asia. Journal of Infection 69(2):174-181.
Gao, H. N., H. Z. Lu, B. Cao, B. Du, H. Shang, J. H. Gan, S. H. Lu, Y. D. Yang, Q. Fang, Y. Z. Shen, X. M. Xi, Q. Gu, X. M. Zhou, H. P. Qu, Z. Yan, F. M. Li, W. Zhao, Z. C. Gao, G. F. Wang, L. X. Ruan, W. H. Wang, J. Ye, H. F. Cao, X. W. Li, W. H. Zhang, X. C. Fang, J. He, W. F. Liang, J. Xie, M. Zeng, X. Z. Wu, J. Li, Q. Xia, Z. C. Jin, Q. Chen, C. Tang, Z. Y. Zhang, B. M. Hou, Z. X. Feng, J. F. Sheng, N. S. Zhong, and L. J. Li. 2013. Clinical findings in 111 cases of influenza A (H7N9) virus infection. New England Journal of Medicine 368(24):2277-2285.
Ge, X. Y., J. L. Li, X. L. Yang, A. A. Chmura, G. Zhu, J. H. Epstein, J. K. Mazet, B. Hu, W. Zhang, C. Peng, Y. J. Zhang, C. M. Luo, B. Tan, N. Wang, Y. Zhu, G. Crameri, S. Y. Zhang, L. F. Wang, P. Daszak, and Z. L. Shi. 2013. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503(7477):535-538.
Gomersall, C. D. 2004. Pro/con clinical debate: Steroids are a key component in the treatment of SARS. Pro: Yes, steroids are a key component of the treatment regimen for SARS. Critical Care 8(2):105-107.
Goodall, J. 1986. The chimpanzees of Gombe. Cambridge, MA: Bellknap Press.
Gostin, L. O. 2004. Pandemic influenza: Public health preparedness for the next global health emergency. Journal of Law, Medicine & Ethics 32(4):565-573.
Gostin, L. O., D. Lucey, and A. Phelan. 2014. The Ebola epidemic: A global health emergency. Journal of the American Medical Association 312(11):1095-1096. doi: 10.1001/jama.2014.11176.
Graham, R. L., and R. S. Baric. 2010. Recombination, reservoirs, and the modular spike: Mechanisms of coronavirus cross-species transmission. Journal of Virology 84(7):3134-3146.
Graham, R. L., M. M. Becker, L. D. Eckerle, M. Bolles, M. R. Denison, and R. S. Baric. 2012. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nature Medicine 18(12):1820-1826.
Graham, R. L., E. F. Donaldson, and R. S. Baric. 2013. A decade after SARS: Strategies for controlling emerging coronaviruses. Nature Reviews Microbiology 11(12):836-848.
Haagmans, B. L., S. H. Al Dhahiry, C. B. Reusken, V. S. Raj, M. Galiano, R. Myers, G. J. Godeke, M. Jonges, E. Farag, A. Diab, H. Ghobashy, F. Alhajri, M. Al-Thani, S. A. Al-Marri, H. E. Al Romaihi, A. Al Khal, A. Bermingham, A. D. Osterhaus, M. M. AlHajri, and M. P. Koopmans. 2013. Middle East respiratory syndrome coronavirus in dromedary camels: An outbreak investigation. Lancet Infectious Diseases 14(2):140-145.
Haberl, H., K. H. Erb, F. Krausmann, V. Gaube, A. Bondeau, C. Plutzar, S. Gingrich, W. Lucht, and M. Fischer-Kowalski. 2007. Quantifying and mapping the human appropriation of net primary production in Earth’s terrestrial ecosystems. Proceedings of the National Academy of Sciences of the United States of America 104(31):12942-12947.
Hahn, B. H., G. M. Shaw, K. M. De, and P. M. Sharp. 2000. AIDS as a zoonosis: Scientific and public health implications. Science 287(5453):607-614.
Han, M. G., D. S. Cheon, X. Zhang, and L. J. Saif. 2006. Cross-protection against a human enteric coronavirus and a virulent bovine enteric coronavirus in gnotobiotic calves. Journal of Virology 80(24):12350-12356.
Hansen, M. C., P. V. Potapov, R. Moore, M. Hancher, S. A. Turubanova, A. Tyukavina, D. Thau, S. V. Stehman, S. J. Goetz, T. R. Loveland, A. Kommareddy, A. Egorov, L. Chini, C. O. Justice, and J. R. G. Townshend. 2013. High resolution global maps of 21st-century forest cover change. Science 342:850-853.
Hasoksuz, M., K. Alekseev, A. Vlasova, X. Zhang, D. Spiro, R. Halpin, S. Wang, E. Ghedin, and L. J. Saif. 2007. Biologic, antigenic, and full-length genomic characterization of a bovine-like coronavirus isolated from a giraffe. Journal of Virology 81:4981-4990.
Hatz, C. F., E. Kuenzli, and M. Funk. 2012. Rabies: Relevance, prevention, and management in travel medicine. Infectious Disease Clinics of North America 26(3):739-753.
Hemida, M. G., R. A. Perera, P. Wang, M. A. Alhammadi, L. Y. Siu, M. Li, L. L. Poon, L. Saif, A. Alnaeem, and M. Peiris. 2013. Middle East respiratory syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Euro Surveillance 18(50):20659.
Henkel, J. 1998. Attacking AIDS with a “cocktail” therapy? FDA Consumer 33(4):12-17.
Herfst, S., E. J. Schrauwen, M. Linster, S. Chutinimitkul, E. de Wit, V. J. Munster, E. M. Sorrell, T. M. Bestebroer, D. F. Burke, D. J. Smith, G. F. Rimmelzwaan, A. D. Osterhaus, and R. A. Fouchier. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336(6088):1534-1541.
HHS (U.S. Department of Health and Human Services), CDC, and NIH (National Institutes of Health). 2009. Section IV—Laboratory biosafety level criteria. Bethesda, MD: NIH.
Hijawi, B., M. Abdallat, A. Sayaydeh, S. Alqasrawi, A. Haddadin, N. Jaarour, S. Alsheikh, and T. Alsanouri. 2013. Novel coronavirus infections in Jordan, April 2012: Epidemiological findings from a retrospective investigation. Eastern Mediterranean Health Journal 19(Suppl 1):S12-S18.
Hoffmann, B., M. Scheuch, D. Hoper, R. Jungblut, M. Holsteg, H. Schirrmeier, M. Eschbaumer, K. V. Goller, K. Wernike, M. Fischer, A. Breithaupt, T. C. Mettenleiter, and M. Beer. 2012. Novel orthobunyavirus in cattle, Europe, 2011. Emerging Infectious Diseases 18(3):469-472.
Howerth, E. W., D. E. Stallknecht, and P. D. Kirkland. 2001. Bluetongue, epizootic hemorrhagic disease, and other orbivirus-related diseases. Infectious Diseases of Wild Mammals 3:77-97.
Hung, I. F., K. K. To, C. K. Lee, K. L. Lee, W. W. Yan, K. Chan, W. M. Chan, C. W. Ngai, K. I. Law, F. L. Chow, R. Liu, K. Y. Lai, C. C. Lau, S. H. Liu, K. H. Chan, C. K. Lin, and K. Y. Yuen. 2013. Hyperimmune IV immunoglobulin treatment: A multicenter double-blind randomized controlled trial for patients with severe 2009 influenza A (H1N1) infection. Chest 144(2):464-473.
Huynh, J., S. Li, B. Yount, A. Smith, L. Sturges, J. C. Olsen, J. Nagel, J. B. Johnson, S. Agnihothram, J. E. Gates, M. B. Frieman, R. S. Baric, and E. F. Donaldson. 2012. Evidence supporting a zoonotic origin of human coronavirus strain NL63. Journal of Virology 86(23):12816-12825.
Imai, M., T. Watanabe, M. Hatta, S. C. Das, M. Ozawa, K. Shinya, G. Zhong, A. Hanson, H. Katsura, S. Watanabe, C. Li, E. Kawakami, S. Yamada, M. Kiso, Y. Suzuki, E. A. Maher, G. Neumann, and Y. Kawaoka. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486(7403):420-428.
IOM (Institute of Medicine). 1992. Emerging infections: Microbial threats to health in the United States. Washington, DC: National Academy Press.
IOM. 2003. Microbial threats to health: Emergence, detection, and response. Washington, DC: The National Academies Press.
IOM. 2007. Ethical and legal considerations in mitigating pandemic disease: Workshop summary. Washington, DC: The National Academies Press
IOM. 2008. Global climate change and extreme weather events. Washington, DC: The National Academies Press.
IOM. 2010. Infectious disease movement in a borderless world. Washington, DC: The National Academies Press.
IOM. 2014. The influence of global environmental change on infectious disease dynamics. Washington, DC: The National Academies Press.
Jones, K. E., N. G. Patel, M. A. Levy, A. Storeygard, D. Balk, J. L. Gittleman, and P. Daszak. 2008. Global trends in emerging infectious diseases. Nature 451(7181):990-993.
Kay, B. H., A. M. Boyd, P. A. Ryan, and R. A. Hall. 2007. Mosquito feeding patterns and natural infection of vertebrates with Ross River and Barmah Forest viruses in Brisbane, Australia. American Journal of Tropical Medicine and Hygiene 76(3):417-423.
Khan, K., J. Sears, V. W. Hu, J. S. Brownstein, S. Hay, D. Kossowsky, R. Eckhardt, T. Chim, I. Berry, I. Bogoch, and M. Cetron. 2013. Potential for the international spread of Middle East respiratory syndrome in association with mass gatherings in Saudi Arabia. PLoS Currents 5.
Kondgen, S., H. Kuhl, P. K. N’Goran, P. D. Walsh, S. Schenk, N. Ernst, R. Biek, P. Formenty, K. Matz-Rensing, B. Schweiger, S. Junglen, H. Ellerbrok, A. Nitsche, T. Briese, W. I. Lipkin, G. Pauli, C. Boesch, and F. H. Leendertz. 2008. Pandemic human viruses cause decline of endangered great apes. Current Biology 18(4):260-264.
Koo, D., and S. B. Thacker. 2010. In Snow’s footsteps: Commentary on shoe-leather and applied epidemiology. American Journal of Epidemiology 172(6):737-739.
Koplow, D. A. 2003. Smallpox: The fight to eradicate a global scourge. Berkeley, CA: University of California Press.
Lau, S. K., P. C. Woo, K. S. Li, Y. Huang, H.-W. Tsoi, B. H. Wong, S. S. Wong, S.-Y. Leung, K.-H. Chan, and K.-Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proceedings of the National Academy of Sciences of the United States of America 102(39):14040-14045.
Lebarbenchon, C., J. D. Brown, and D. E. Stallknecht. 2013. Evolution of influenza A virus H7 and N9 subtypes, eastern Asia. Emerging Infectious Diseases 19(10):1635-1638.
Lederberg, J. 2000. Infectious history. Science 288(5464):287-293.
Le Poder, S. 2011. Feline and canine coronaviruses: Common genetic and pathobiological features. Advances in Virology. doi:10.1155/2011/609465.
Li, F. 2013. Receptor recognition and cross-species infections of SARS coronavirus. Antiviral Research 100(1):246-254.
Li, G., Q. Chen, K. M. Harmon, K. J. Yoon, K. J. Schwartz, M. J. Hoogland, P. C. Gauger, R. G. Main, and J. Zhang. 2014a. Full-length genome sequence of porcine deltacoronavirus strain USA/ia/2014/8734. Genome Announcements 2(2):e00278-14.
Li, Q., L. Zhou, M. Zhou, Z. Chen, F. Li, H. Wu, N. Xiang, E. Chen, F. Tang, D. Wang, L. Meng, Z. Hong, W. Tu, Y. Cao, L. Li, F. Ding, B. Liu, M. Wang, R. Xie, R. Gao, X. Li, T. Bai, S. Zou, J. He, J. Hu, Y. Xu, C. Chai, S. Wang, Y. Gao, L. Jin, Y. Zhang, H. Luo, H. Yu, X. Wang, L. Gao, X. Pang, G. Liu, Y. Yan, H. Yuan, Y. Shu, W. Yang, Y. Wang, F. Wu, T. M. Uyeki, and Z. Feng. 2014b. Epidemiology of human infections with avian influenza A (H7N9) virus in China. New England Journal of Medicine 370(6):520-532.
Liu, D., W. Shi, Y. Shi, D. Wang, H. Xiao, W. Li, Y. Bi, Y. Wu, X. Li, and J. Yan. 2013. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: Phylogenetic, structural, and coalescent analyses. Lancet 381(9881):1926-1932.
Lozano, R., M. Naghavi, K. Foreman, S. Lim, K. Shibuya, V. Aboyans, J. Abraham, T. Adair, R. Aggarwal, S. Y. Ahn, M. Alvarado, H. R. Anderson, L. M. Anderson, K. G. Andrews, C. Atkinson, L. M. Baddour, S. Barker-Collo, D. H. Bartels, M. L. Bell, E. J. Benjamin, D. Bennett, K. Bhalla, B. Bikbov, A. Bin Abdulhak, G. Birbeck, F. Blyth, I. Bolliger, S. Boufous, C. Bucello, M. Burch, P. Burney, J. Carapetis, H. Chen, D. Chou, S. S. Chugh, L. E. Coffeng, S. D. Colan, S. Colquhoun, K. E. Colson, J. Condon, M. D. Connor, L. T. Cooper, M. Corriere, M. Cortinovis, K. C. de Vaccaro, W. Couser, B. C. Cowie, M. H. Criqui, M. Cross, K. C. Dabhadkar, N. Dahodwala, D. De Leo, L. Degenhardt, A. Delossantos, J. Denenberg, D. C. Des Jarlais, S. D. Dharmaratne, E. R. Dorsey, T. Driscoll, H. Duber, B. Ebel, P. J. Erwin, P. Espindola, M. Ezzati, V. Feigin, A. D. Flaxman, M. H. Forouzanfar, F. G. Fowkes, R. Franklin, M. Fransen,
M. K. Freeman, S. E. Gabriel, E. Gakidou, F. Gaspari, R. F. Gillum, D. Gonzalez-Medina, Y. A. Halasa, D. Haring, J. E. Harrison, R. Havmoeller, R. J. Hay, B. Hoen, P. J. Hotez, D. Hoy, K. H. Jacobsen, S. L. James, R. Jasrasaria, S. Jayaraman, N. Johns, G. Karthikeyan, N. Kassebaum, A. Keren, J. P. Khoo, L. M. Knowlton, O. Kobusingye, A. Koranteng, R. Krishnamurthi, M. Lipnick, S. E. Lipshultz, S. L. Ohno, J. Mabweijano, M. F. MacIntyre, L. Mallinger, L. March, G. B. Marks, R. Marks, A. Matsumori, R. Matzopoulos, B. M. Mayosi, J. H. McAnulty, M. M. McDermott, J. McGrath, G. A. Mensah, T. R. Merriman, C. Michaud, M. Miller, T. R. Miller, C. Mock, A. O. Mocumbi, A. A. Mokdad, A. Moran, K. Mulholland, M. N. Nair, L. Naldi, K. M. Narayan, K. Nasseri, P. Norman, M. O’Donnell, S. B. Omer, K. Ortblad, R. Osborne, D. Ozgediz, B. Pahari, J. D. Pandian, A. P. Rivero, R. P. Padilla, F. Perez-Ruiz, N. Perico, D. Phillips, K. Pierce, C. A. Pope, 3rd, E. Porrini, F. Pourmalek, M. Raju, D. Ranganathan, J. T. Rehm, D. B. Rein, G. Remuzzi, F. P. Rivara, T. Roberts, F. R. De Leon, L. C. Rosenfeld, L. Rushton, R. L. Sacco, J. A. Salomon, U. Sampson, E. Sanman, D. C. Schwebel, M. Segui-Gomez, D. S. Shepard, D. Singh, J. Singleton, K. Sliwa, E. Smith, A. Steer, J. A. Taylor, B. Thomas, I. M. Tleyjeh, J. A. Towbin, T. Truelsen, E. A. Undurraga, N. Venketasubramanian, L. Vijayakumar, T. Vos, G. R. Wagner, M. Wang, W. Wang, K. Watt, M. A. Weinstock, R. Weintraub, J. D. Wilkinson, A. D. Woolf, S. Wulf, P. H. Yeh, P. Yip, A. Zabetian, Z. J. Zheng, A. D. Lopez, C. J. Murray, M. A. AlMazroa, and Z. A. Memish. 2012. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 380(9859):2095-2128.
Luke, T. C., E. M. Kilbane, J. L. Jackson, and S. L. Hoffman. 2006. Meta-analysis: Convalescent blood products for Spanish influenza pneumonia: A future H5N1 treatment? Annals of Internal Medicine 145(8):599-609.
Martin, V., X. Zhou, E. Marshall, B. Jia, G. Fusheng, M. A. FrancoDixon, N. DeHaan, D. U. Pfeiffer, R. J. Soares Magalhaes, and M. Gilbert. 2011. Risk-based surveillance for avian influenza control along poultry market chains in South China: The value of social network analysis. Preventive Veterinary Medicine 102(3):196-205.
Memish, Z. A., N. Mishra, K. J. Olival, S. F. Fagbo, V. Kapoor, J. H. Epstein, R. Alhakeem, A. Durosinloun, M. Al Asmari, A. Islam, A. Kapoor, T. Briese, P. Daszak, A. A. Al Rabeeah, and W. I. Lipkin. 2013. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerging Infectious Diseases 19(11):1819-1823.
Meteyer, C. U., D. Barber, and J. N. Mandl. 2012. Pathology in euthermic bats with white nose syndrome suggests a natural manifestation of immune reconstitution inflammatory syndrome. Virulence 3(7):583-588.
Meyer, B., M. A. Müller, V. M. Corman, C. Reusken, D. Ritz, G. Godeke, E. Lattwein, S. Kallies, A. Siemens, and J. van Beek. 2014. Antibodies against MERS coronavirus in dromedary camels, United Arab Emirates, 2003 and 2013. Emerging Infectious Diseases 20(4):552-559.
Mickleburgh, S. P., A. M. Hutson, and P. A. Racey. 2002. A review of the global conservation status of bats. Oryx 36:18-34.
Monadjem, A., P. J. Taylor, W. Cotterill, and M. C. Schoeman. 2010. Bats of southern and central Africa: A biogeographic and taxonomic synthesis. Johannesburg, South Africa: Wits University Press.
Morens, D. M., and A. S. Fauci. 2013. Emerging infectious diseases: Threats to human health and global stability. PLoS Pathogens 9(7):e1003467.
Morens, D. M., and A. S. Fauci. 2014. Chikungunya at the door—déjà vu all over again? New England Journal of Medicine 371(10):885-887.
Morens, D. M., G. K. Folkers, and A. S. Fauci. 2004. The challenge of emerging and re-emerging infectious diseases. Nature 430(6996):242-249.
Morens, D. M., J. K. Taubenberger, and A. S. Fauci. 2013. Pandemic influenza viruses—Hoping for the road not taken. New England Journal of Medicine 368(25):2345-2348.
Murray, C. J., T. Vos, R. Lozano, M. Naghavi, A. D. Flaxman, C. Michaud, M. Ezzati, K. Shibuya, J. A. Salomon, S. Abdalla, V. Aboyans, J. Abraham, I. Ackerman, R. Aggarwal, S. Y. Ahn,
M. K. Ali, M. Alvarado, H. R. Anderson, L. M. Anderson, K. G. Andrews, C. Atkinson, L. M. Baddour, A. N. Bahalim, S. Barker-Collo, L. H. Barrero, D. H. Bartels, M. G. Basanez, A. Baxter, M. L. Bell, E. J. Benjamin, D. Bennett, E. Bernabe, K. Bhalla, B. Bhandari, B. Bikbov, A. Bin Abdulhak, G. Birbeck, J. A. Black, H. Blencowe, J. D. Blore, F. Blyth, I. Bolliger, A. Bonaventure, S. Boufous, R. Bourne, M. Boussinesq, T. Braithwaite, C. Brayne, L. Bridgett, S. Brooker, P. Brooks, T. S. Brugha, C. Bryan-Hancock, C. Bucello, R. Buchbinder, G. Buckle, C. M. Budke, M. Burch, P. Burney, R. Burstein, B. Calabria, B. Campbell, C. E. Canter, H. Carabin, J. Carapetis, L. Carmona, C. Cella, F. Charlson, H. Chen, A. T. Cheng, D. Chou, S. S. Chugh, L. E. Coffeng, S. D. Colan, S. Colquhoun, K. E. Colson, J. Condon, M. D. Connor, L. T. Cooper, M. Corriere, M. Cortinovis, K. C. de Vaccaro, W. Couser, B. C. Cowie, M. H. Criqui, M. Cross, K. C. Dabhadkar, M. Dahiya, N. Dahodwala, J. Damsere-Derry, G. Danaei, A. Davis, D. De Leo, L. Degenhardt, R. Dellavalle, A. Delossantos, J. Denenberg, S. Derrett, D. C. Des Jarlais, S. D. Dharmaratne, M. Dherani, C. Diaz-Torne, H. Dolk, E. R. Dorsey, T. Driscoll, H. Duber, B. Ebel, K. Edmond, A. Elbaz, S. E. Ali, H. Erskine, P. J. Erwin, P. Espindola, S. E. Ewoigbokhan, F. Farzadfar, V. Feigin, D. T. Felson, A. Ferrari, C. P. Ferri, E. M. Fevre, M. M. Finucane, S. Flaxman, L. Flood, K. Foreman, M. H. Forouzanfar, F. G. Fowkes, M. Fransen, M. K. Freeman, B. J. Gabbe, S. E. Gabriel, E. Gakidou, H. A. Ganatra, B. Garcia, F. Gaspari, R. F. Gillum, G. Gmel, D. Gonzalez-Medina, R. Gosselin, R. Grainger, B. Grant, J. Groeger, F. Guillemin, D. Gunnell, R. Gupta, J. Haagsma, H. Hagan, Y. A. Halasa, W. Hall, D. Haring, J. M. Haro, J. E. Harrison, R. Havmoeller, R. J. Hay, H. Higashi, C. Hill, B. Hoen, H. Hoffman, P. J. Hotez, D. Hoy, J. J. Huang, S. E. Ibeanusi, K. H. Jacobsen, S. L. James, D. Jarvis, R. Jasrasaria, S. Jayaraman, N. Johns, J. B. Jonas, G. Karthikeyan, N. Kassebaum, N. Kawakami, A. Keren, J. P. Khoo, C. H. King, L. M. Knowlton, O. Kobusingye, A. Koranteng, R. Krishnamurthi, F. Laden, R. Lalloo, L. L. Laslett, T. Lathlean, J. L. Leasher, Y. Y. Lee, J. Leigh, D. Levinson, S. S. Lim, E. Limb, J. K. Lin, M. Lipnick, S. E. Lipshultz, W. Liu, M. Loane, S. L. Ohno, R. Lyons, J. Mabweijano, M. F. MacIntyre, R. Malekzadeh, L. Mallinger, S. Manivannan, W. Marcenes, L. March, D. J. Margolis, G. B. Marks, R. Marks, A. Matsumori, R. Matzopoulos, B. M. Mayosi, J. H. McAnulty, M. M. McDermott, N. McGill, J. McGrath, M. E. Medina-Mora, M. Meltzer, G. A. Mensah, T. R. Merriman, A. C. Meyer, V. Miglioli, M. Miller, T. R. Miller, P. B. Mitchell, C. Mock, A. O. Mocumbi, T. E. Moffitt, A. A. Mokdad, L. Monasta, M. Montico, M. Moradi-Lakeh, A. Moran, L. Morawska, R. Mori, M. E. Murdoch, M. K. Mwaniki, K. Naidoo, M. N. Nair, L. Naldi, K. M. Narayan, P. K. Nelson, R. G. Nelson, M. C. Nevitt, C. R. Newton, S. Nolte, P. Norman, R. Norman, M. O’Donnell, S. O’Hanlon, C. Olives, S. B. Omer, K. Ortblad, R. Osborne, D. Ozgediz, A. Page, B. Pahari, J. D. Pandian, A. P. Rivero, S. B. Patten, N. Pearce, R. P. Padilla, F. Perez-Ruiz, N. Perico, K. Pesudovs, D. Phillips, M. R. Phillips, K. Pierce, S. Pion, G. V. Polanczyk, S. Polinder, C. A. Pope, 3rd, S. Popova, E. Porrini, F. Pourmalek, M. Prince, R. L. Pullan, K. D. Ramaiah, D. Ranganathan, H. Razavi, M. Regan, J. T. Rehm, D. B. Rein, G. Remuzzi, K. Richardson, F. P. Rivara, T. Roberts, C. Robinson, F. R. De Leon, L. Ronfani, R. Room, L. C. Rosenfeld, L. Rushton, R. L. Sacco, S. Saha, U. Sampson, L. Sanchez-Riera, E. Sanman, D. C. Schwebel, J. G. Scott, M. Segui-Gomez, S. Shahraz, D. S. Shepard, H. Shin, R. Shivakoti, D. Singh, G. M. Singh, J. A. Singh, J. Singleton, D. A. Sleet, K. Sliwa, E. Smith, J. L. Smith, N. J. Stapelberg, A. Steer, T. Steiner, W. A. Stolk, L. J. Stovner, C. Sudfeld, S. Syed, G. Tamburlini, M. Tavakkoli, H. R. Taylor, J. A. Taylor, W. J. Taylor, B. Thomas, W. M. Thomson, G. D. Thurston, I. M. Tleyjeh, M. Tonelli, J. A. Towbin, T. Truelsen, M. K. Tsilimbaris, C. Ubeda, E. A. Undurraga, M. J. van der Werf, J. van Os, M. S. Vavilala, N. Venketasubramanian, M. Wang, W. Wang, K. Watt, D. J. Weatherall, M. A. Weinstock, R. Weintraub, M. G. Weisskopf, M. M. Weissman, R. A. White, H. Whiteford, N. Wiebe, S. T. Wiersma, J. D. Wilkinson, H. C. Williams, S. R. Williams, E. Witt, F. Wolfe, A. D. Woolf, S. Wulf, P. H. Yeh, A. K. Zaidi, Z. J. Zheng, D. Zonies, A. D. Lopez, M. A. AlMazroa, and Z. A. Memish. 2012. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990-2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 380(9859):2197-2223.
NIAID (National Institute of Allergy and Infectious Diseases). 2013. Middle East respiratory syndrome coronavirus (MERS-CoV) research: Current status and future priorities. Bethesda, MD: NIAID.
NRC (National Research Council). 2004. Biotechnology research in an age of terrorism. Washington, DC: The National Academies Press.
OIE (World Organisation for Animal Health). 2014a. Bluetongue. http://www.oie.int/fileadmin/Home/eng/Media_Center/docs/pdf/Disease_cards/BLUET-EN.pdf (accessed September 4, 2014).
OIE. 2014b. The OIE PVS pathway. http://www.oie.int/support-to-oie-members/pvs-pathway (accessed September 8, 2014).
OIE. 2014c. The OIE tool for the evaluation of performance of veterinary services (OIE PVS tool). http://www.oie.int/support-to-oie-members/pvs-evaluations/oie-pvs-tool (accessed June 10, 2014).
Olson, S. H., J. Parmley, C. Soos, M. Gilbert, N. Latorre-Margalef, J. S. Hall, P. M. Hansbro, F. Leighton, V. Munster, and D. Joly. 2014. Sampling strategies and biodiversity of influenza A subtypes in wild birds. PLoS ONE 9(3):e90826.
Pantin-Jackwood, M. J., P. J. Miller, E. Spackman, D. E. Swayne, L. Susta, M. Costa-Hurtado, and D. L. Suarez. 2014. Role of poultry in the spread of novel H7N9 influenza virus in China. Journal of Virology 88(10):5381-5390.
Pavade, G., L. Awada, K. Hamilton, and D. E. Swayne. 2011. The influence of economic indicators, poultry density and the performance of veterinary services on the control of high-pathogenicity avian influenza in poultry. Revue Scientifique et Technique 30(3):661-671.
Perera, R. A., P. Wang, M. R. Gomaa, R. El-Shesheny, A. Kandeil, O. Bagato, L. Y. Siu, M. M. Shehata, A. S. Kayed, Y. Moatasim, M. Li, L. L. Poon, Y. Guan, R. J. Webby, M. A. Ali, J. S. Peiris, and G. Kayali. 2013. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveillance 18(36):pii=20574.
Pfeiffer, D. U., M. J. Otte, D. Roland-Holst, K. Inui, N. Tung, and D. Zilberman. 2011. Implications of global and regional patterns of highly pathogenic avian influenza virus H5N1 clades for risk management. Veterinary Journal 190:309-316.
Pfeiffer, D. U., M. J. Otte, D. Roland-Holst and D. Zilberman. 2013. A one health perspective on HPAI H5N1 in the Greater Mekong sub-region. Comparative Immunology Microbiology and Infectious Disease 36(3):309-319.
Preidt, R. 2010. Dengue re-emerges in Florida. USA Today. http://usatoday30.usatoday.com/news/health/2010-07-15-dengue-fever_N.htm (accessed September 4, 2014).
Qi, X., D. Jiang, H. Wang, D. Zhuang, J. Ma, J. Fu, J. Qu, Y. Sun, S. Yu, and Y. Meng. 2014. Calculating the burden of disease of avian-origin H7N9 infections in China. BMJ Open 4(1):e004189.
Raboud, J., A. Shigayeva, A. McGeer, E. Bontovics, M. Chapman, D. Gravel, B. Henry, S. Lapinsky, M. Loeb, and L. C. McDonald. 2010. Risk factors for SARS transmission from patients requiring intubation: A multicentre investigation in Toronto, Canada. PLoS ONE 5(5):e10717.
Renn, O. 2005. Risk governance: Towards an integrative approach. Geneva, Switzerland: International Risk Governance Council.
Reperant, L. A., T. Kuiken, and A. D. Osterhaus. 2012. Influenza viruses: From birds to humans. Human Vaccines & Immunotherapeutics 8(1):7-16.
Reusken, C., C. van den Wijngaard, P. van Beek, M. Beer, R. Bouwstra, G. J. Godeke, L. Isken, H. van den Kerkhof, W. van Pelt, W. van der Poel, J. Reimerink, P. Schielen, J. Schmidt-Chanasit, P. Vellema, A. de Vries, I. Wouters, and M. Koopmans. 2012. Lack of evidence for zoonotic transmission of Schmallenberg virus. Emerging Infectious Diseases 18(11):1746-1754.
Reusken, C. B., B. L. Haagmans, M. A. Muller, C. Gutierrez, G. J. Godeke, B. Meyer, D. Muth, V. S. Raj, L. Smits-De Vries, V. M. Corman, J. F. Drexler, S. L. Smits, Y. E. El Tahir, R. De Sousa, J. van Beek, N. Nowotny, K. van Maanen, E. Hidalgo-Hermoso, B. J. Bosch, P. Rottier, A. Osterhaus, C. Gortazar-Schmidt, C. Drosten, and M. P. Koopmans. 2013. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: A comparative serological study. Lancet Infectious Diseases 13(10):859-866.
Rubin, C. 2014. Making One Health a reality: Crossing bureaucratic boundaries. Microbiology Spectrum 2(1):OH-0016-2012.
Rupprecht, C. E., A. Turmelle, and I. V. Kuzmin. 2011. A perspective on lyssavirus emergence and perpetuation. Current Opinion in Virology 1(6):662-670.
Russell, C. A., J. M. Fonville, A. E. Brown, D. F. Burke, D. L. Smith, S. L. James, S. Herfst, S. van Boheemen, M. Linster, E. J. Schrauwen, L. Katzelnick, A. Mosterin, T. Kuiken, E. Maher, G. Neumann, A. D. Osterhaus, Y. Kawaoka, R. A. Fouchier, and D. J. Smith. 2012. The potential for respiratory droplet-transmissible A/H5N1 influenza virus to evolve in a mammalian host. Science 336(6088):1541-1547.
Salaam-Blyther, T. 2014. The 2014 Ebola outbreak: International and U.S. responses. Congressional Research Service. http://fas.org/sgp/crs/row/R43697.pdf (accessed November 11, 2014).
Samji, H., A. Cescon, R. S. Hogg, S. P. Modur, K. N. Althoff, K. Buchacz, A. N. Burchell, M. Cohen, K. A. Gebo, M. J. Gill, A. Justice, G. Kirk, M. B. Klein, P. T. Korthuis, J. Martin, S. Napravnik, S. B. Rourke, T. R. Sterling, M. J. Silverberg, S. Deeks, L. P. Jacobson, R. J. Bosch, M. M. Kitahata, J. J. Goedert, R. Moore, and S. J. Gange. 2013. Closing the gap: Increases in life expectancy among treated HIV-positive individuals in the United States and Canada. PLoS ONE 8(12):e81355.
Scobey, T., B. L. Yount, A. C. Sims, E. F. Donaldson, S. S. Agnihothram, V. D. Menachery, R. L. Graham, J. Swanstrom, P. F. Bove, J. D. Kim, S. Grego, S. H. Randell, and R. S. Baric. 2013. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proceedings of the National Academy of Sciences of the United States of America 110(40):16157-16162.
Seimon, T. A., D. G. Miquelle, T. Y. Chang, A. L. Newton, I. Korotkova, G. Ivanchuk, E. Lyubchenko, A. Tupikov, E. Slabe, and D. McAloose. 2013. Canine distemper virus: An emerging disease in wild endangered Amur tigers (Panthera tigris altaica). mBio 4(4):e00410-13.
Shi, Z., and Z. Hu. 2008. A review of studies on animal reservoirs of the SARS coronavirus. Virus Research 133(1):74-87.
Simonsen, L., P. Spreeuwenberg, R. Lustig, R. J. Taylor, D. M. Fleming, M. Kroneman, M. D. Van Kerkhove, A. W. Mounts, and W. J. Paget. 2013. Global mortality estimates for the 2009 influenza pandemic from the glamor project: A modeling study. PLoS Medicine 10(11):e1001558.
Skinner, J. D., and C. T. Chimimba. 2005. The mammals of the southern African sub-region. Boston, MA: Cambridge University Press.
Smith, K. M., S. J. Anthony, W. M. Switzer, J. H. Epstein, T. Seimon, H. Jia, M. D. Sanchez, T. T. Huynh, G. G. Galland, and S. E. Shapiro. 2012. Zoonotic viruses associated with illegally imported wildlife products. PloS ONE 7(1):e29505.
Spellberg, B. 2008. Dr. William H. Stewart: Mistaken or maligned? Clinical Infectious Diseases 47(2):294.
Swenson, S. L., L. G. Koster, M. Jenkins-Moore, M. L. Killian, E. E. DeBess, R. J. Baker, D. Mulrooney, R. Weiss, J. Galeota, and A. Bredthauer. 2010. Natural cases of 2009 pandemic H1N1 influenza A virus in pet ferrets. Journal of Veterinary Diagnostic Investigation 22(5):784-788.
Taylor, L. H., S. M. Latham, and M. E. J. Woolhouse. 2001. Risk factors for human disease emergence. Philosophical Transactions of the Royal Society of London, Series B: Biological Sciences 356(1411):983-989.
Tong, S., P. Dale, N. Nicholls, J. S. Mackenzie, R. Wolff, and A. J. McMichael. 2008. Climate variability, social and environmental factors, and Ross River virus transmission: Research development and future research needs. Environmental Health Perspectives 116(12):1591-1597.
Tong, S., Y. Li, P. Rivailler, C. Conrardy, D. A. Castillo, L. M. Chen, S. Recuenco, J. A. Ellison, C. T. Davis, I. A. York, A. S. Turmelle, D. Moran, S. Rogers, M. Shi, Y. Tao, M. R. Weil, K. Tang, L. A. Rowe, S. Sammons, X. Xu, M. Frace, K. A. Lindblade, N. J. Cox, L. J. Anderson, C. E. Rupprecht, and R. O. Donis. 2012. A distinct lineage of influenza A virus from bats. Proceedings of the National Academy of Sciences of the United States of America 109(11):4269-4274.
Trock, S. C., S. A. Burke, and N. J. Cox. 2012. Development of an influenza virologic risk assessment tool. Avian Diseases 56(Suppl 4):1058-1061.
Tsunemitsu, H., and L. J. Saif. 1995. Antigenic and biological comparisons of bovine coronaviruses derived from neonatal calf diarrhea and winter dysentery of adult cattle. Archives in Virology 140(7):1303-1311.
Tsunemitsu, H., Z. R. el-Kanawati, D. R. Smith, H. H. Reed, and L. J. Saif. 1995. Isolation of coronaviruses antigenically indistinguishable from bovine coronavirus from wild ruminants with diarrhea. Journal of Clinical Microbiology 33(12):3264-3269.
USDA/APHIS (U.S. Department of Agriculture/Animal and Plant Health Inspection Service). 2010. Assistance to Dairy Farms and Facilities. http://www.aphis.usda.gov/publications/wildlife_damage/content/printable_version/faq_dairy_farms.pdf (accessed November 7, 2014).
USDA/ARS (U.S. Department of Agriculture/Agriculture Research Service). 2014. Honey bees and colony collapse disorder. http://www.ars.usda.gov/News/docs.htm?docid=15572 (accessed September 2, 2014).
USGS (U.S. Geological Survey). 2013. Ranavirus. http://www.nwhc.usgs.gov/disease_information/other_diseases/ranavirus.jsp (accessed September 2, 2014).
USGS. 2014. White-nose syndrome. http://www.nwhc.usgs.gov/disease_information/white-nose_syndrome (accessed September 2, 2014).
Van Doremalen, N., T. Bushmaker, and V. Munster. 2013. Stability of Middle East respiratory syndrome coronavirus (MERS-CoV) under different environmental conditions. Euro Surveillance 18(38):20590.
Vijgen, L., E. Keyaerts, P. Lemey, E. Moës, S. Li, A. M. Vandamme, and M. Van Rast. 2005. Circulation of genetically distinct contemporary human coronavirus OC43 strains. Virology 337(1):85-92.
Vlasova, A., and L. J. Saif. 2013. Biological aspects of the interspecies transmission of selected coronaviruses. In Viral infections and global change, edited by S. K. Singh. Hoboken, NJ: Wiley Blackwell Press. Pp. 393-418.
Vlasova, A. N., D. Marthaler, Q. Wang, M. R. Culhane, K. D. Rossow, A. Rovira, J. Collins, and L. J. Saif. 2014. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013-February 2014. Emerging Infectious Diseases 20(10):1620-1628.
Wang, Q., J. Qi, Y. Yuan, Y. Xuan, P. Han, Y. Wan, W. Ji, Y. Li, Y. Wu, J. Wang, A. Iwamoto, P. C. Woo, K. Y. Yuen, J. Yan, G. Lu, and G. F. Gao. 2014. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe 16(3):328-337.
Wesley, R., R. Woods, and A. Cheung. 1991. Genetic analysis of porcine respiratory coronavirus, an attenuated variant of transmissible gastroenteritis virus. Journal of Virology 65(6):3369-3373.
WHO (World Health Organization). 2008. International Health Regulations (2005). Geneva, Switzerland: WHO.
WHO. 2009. Frequently asked questions about the International Health Regulations (2005). http://www.who.int/ihr/about/FAQ2009.pdf (accessed September 2, 2014).
WHO. 2011a. Pandemic influenza preparedness framework. Geneva, Switzerland: WHO.
WHO. 2011b. Strengthening response to pandemics and other public-health emergencies: Report of the Review Committee on the Functioning of the International Health Regulations (2005) and on Pandemic Influenza (H1N1) 2009. Geneva, Switzerland: WHO.
WHO. 2013. Human infection with influenza A (H7N9) virus in China. http://www.who.int/csr/don/2013_04_01/en (accessed June 5, 2014).
WHO. 2014a. Avian influenza. http://www.who.int/mediacentre/factsheets/avian_influenza/en (accessed June 3, 2014).
WHO. 2014b. Frequently asked questions on Middle East respiratory syndrome coronavirus (MERS CoV). http://www.who.int/csr/disease/coronavirus_infections/faq/en (accessed June 11, 2014).
WHO. 2014c. Human infection with avian influenza A (H7N9) virus—Update. http://www.who.int/csr/don/2014_06_10_h7n9/en (accessed June 10, 2014).
WHO. 2014d. Middle East respiratory syndrome coronavirus (MERS-CoV) summary and literature update–as of 23 July 2014. http://www.who.int/csr/don/2014_07_23_mers/en (accessed September 9, 2014).
WHO. 2014e. WHO: Ebola response roadmap update. http://apps.who.int/iris/bitstream/10665/132834/1/roadmapupdate8sept14_eng.pdf?ua=1&ua=1 (accessed September 10, 2014).
WHO. 2014f. Middle East respiratory syndrome coronavirus (MERS-CoV) – Saudi Arabia. http://www.who.int/csr/don/23-february-2015-mers-saudi-arabia/en/ (accessed February 25, 2015).
WHO MERS-CoV Research Group. 2013. State of knowledge and data gaps of Middle East respiratory syndrome coronavirus (MERS-CoV) in humans. PLoS Currents Outbreaks 1:1-30.
World Economic Forum. 2014. Global risks 2014. http://www3.weforum.org/docs/WEF_GlobalRisks_Report_2014.pdf (accessed September 5, 2014).
Wu, Y., and G. F. Gao. 2013. Lessons learnt from the human infections of avian-origin influenza A H7N9 virus: Live free markets and human health. Science China Life Sciences 56(6):493-494.
Xiang, N., F. Havers, T. Chen, Y. Song, W. Tu, L. Li, Y. Cao, B. Liu, L. Zhou, L. Meng, Z. Hong, R. Wang, Y. Niu, J. Yao, K. Liao, L. Jin, Y. Zhang, Q. Li, M. A. Widdowson, and Z. Feng. 2013. Use of national pneumonia surveillance to describe influenza A (H7N9) virus epidemiology, China, 2004-2013. Emerging Infectious Diseases 19(11):1784-1790.
Xu, C., F. Havers, L. Wang, T. Chen, J. Shi, D. Wang, J. Yang, L. Yang, M. A. Widdowson, and Y. Shu. 2013. Monitoring avian influenza A (H7N9) virus through national influenza-like illness surveillance, China. Emerging Infectious Diseases 19(8):1289-1292.
Yang, S., Y. Chen, D. Cui, H. Yao, J. Lou, Z. Huo, G. Xie, F. Yu, S. Zheng, Y. Yang, Y. Zhu, X. Lu, X. Liu, S. Y. Lau, J. F. Chan, K. K. To, K. Y. Yuen, H. Chen, and L. Li. 2013. Avian-origin influenza A (H7N9) infection in influenza A (H7N9)-affected areas of China: A serological study. Journal of Infectious Diseases 209(2):265-269.
Yang, Y., L. Du, C. Liu, L. Wang, C. Ma, J. Tang, R. S. Baric, S. Jiang, and F. Li. 2014. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proceedings of the National Academy of Sciences of the United States of America 111(34):12516-12521.
Yu, H., J. T. Wu, B. J. Cowling, Q. Liao, V. J. Fang, S. Zhou, P. Wu, H. Zhou, E. H. Lau, D. Guo, M. Y. Ni, Z. Peng, L. Feng, H. Jiang, H. Luo, Q. Li, Z. Feng, Y. Wang, W. Yang, and G. M. Leung. 2014. Effect of closure of live poultry markets on poultry-to-person transmission of avian influenza A H7N9 virus: An ecological study. Lancet 383(9916):541-548.
Zaki, A. M., S. van Boheemen, T. M. Bestebroer, A. D. Osterhaus, and R. A. Fouchier. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New England Journal of Medicine 367(19):1814-1820.
Zhao, J., K. Li, C. Wohlford-Lenane, S. S. Agnihothram, C. Fett, J. Zhao, M. J. Gale, R. S. Baric, L. Enjuanes, and T. Gallagher. 2014. Rapid generation of a mouse model for Middle East respiratory syndrome. Proceedings of the National Academy of Sciences of the United States of America 111(13):4970-4975.






















































































































