In order to realize a future of biological, chemical, and combined approaches being viewed as equally viable options for chemical manufacturing, a number of technical challenges must be overcome. As discussed previously, the use of biological systems for chemical manufacturing has already attained fairly widespread use in some specific sectors, but by comparison to traditional chemical manufacturing it is still a relatively small market. The promise shown by these previous successes, however, is significant.
One key technical consideration that has been less well integrated into planning and processing for bio-based methods than for traditional chemistry is the ability to model and fully understand the entire manufacturing process when considering the use of biological systems for chemical manufacturing. The characteristics of biological behavior make this a daunting task, but relatively recent advances in life sciences and chemical engineering make it attainable if the many factors that could cause progress to stagnate are avoided. In order to facilitate biomanufacturing for chemical production, a series of conclusions and roadmap goals are presented and discussed in this chapter. The discussion is organized into three broad categories: feedstocks, enabling transformations, and integrated design toolchain.
The feedstocks section discusses the promising array of feedstocks currently used in manufacturing as well as the array of opportunities that are possible with key technological advances. Starch and other simple
sugars obtained from biomass are the most widely used feedstocks today, and the use of cellulosic biomass is expanding. There are still many challenges associated with using recalcitrant cellulosic material for manufacturing, but there are potential solutions to this issue as discussed herein. Although the discussion is focused on different forms of biomass, the discussion is not limited to biomass. There is active work in facilitating the use of syngas, methane, and carbon dioxide in manufacturing as well.
The enabling transformations section discusses the science, technology, and engineering knowledge and tools required to transform the feedstock material into a useful product or intermediate. One of the major engineering considerations is related to fermentation and processing that are required for biological production of chemicals. Fermentation can be facilitated in many ways, but it typically represents a large capital expense that must be overcome in order to begin production. To mitigate this capital expense, the ability to reliably and efficiently scale up processes is very important.
This section continues to discuss the research and development needed to facilitate chemical transformations. The majority of this section discusses synthetic biology and the use of chassis and pathways to develop microorganisms for use in chemical manufacturing. Although this work is ongoing in a number of sectors, the use of microorganisms for chemical manufacturing could be more widespread. This portion describes the priority research needs to enable chemical transformations using biological systems.
The final section discusses the overall needs in measurements science and technology in relation to the research and development needs discussed in this chapter.
Carbon in the form of fermentable sugars is the primary raw material, and often the largest single input cost, for the biological production of chemicals. In the case of large-volume chemicals, sugar costs can represent the majority of the total product costs. In the extreme case of biofuels, sugar costs represent as much as 65 percent of the total product costs.96 By contrast, for industrial enzyme and specialty chemical production, the overall cost of the carbon source is a small fraction of the total costs. The feedstock cost is so small for these products that changes to feedstock price are negligible.
For today’s fermentation, the source of carbon is overwhelmingly dextrose derived from the starch in grain. In Brazil, abundant sucrose
from sugarcane is used instead of dextrose. For the biological production of chemicals to reach its full potential, more abundant, more diverse, and less costly sources of carbon are needed.
Cellulosic material derived from agricultural residues, forestry by-products, and even dedicated energy crops are both abundant and diverse. Conversion of cellulose to fermentable sugars is the subject of active research and development. A number of challenges must be overcome if cellulose-based sugars are to become fully substitutable for starch-derived sugars, and with a cost advantage.
Today’s agricultural economy has well-functioning markets for grain trading, and a well-established infrastructure for the production, transportation, and storage of the commodities. None of this exists for fermentable sugars derived from cellulosic feedstocks. As the first cellulose-based ethanol plants are coming on line, individual plants develop their own technology and (local) markets for originating cellulosic material. The cost of the cellulosic feedstock must be kept low—much less than $100/ton—in order for it to be a viable alternative to grain.
Cellulosic sugars are not the only alternative to starch-based carbon. Methane and methane derivatives are also potentially attractive feedstocks for bioprocessing. Abundant shale gas has dramatically increased the supply and reduced the price of methane.
Multiple Generations of Feedstocks
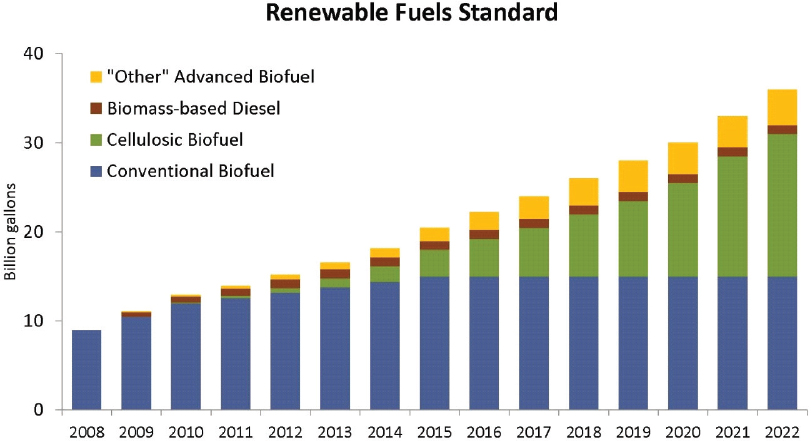
As mentioned above, the first-generation source of carbon for fermentation has been starch derived from grain. The U.S. ethanol industry has been built on grain feedstocks, and all current biological production of chemicals relies on grain. In the United States today, nearly 40 percent of the corn crop is consumed in nonfood or feed uses, primarily for the production of fuel ethanol.97 Although this feedstock has served the industry well, there are limitations to the supply of grain and concerns about competition with the food and feed uses for grain. These concerns were anticipated in the Renewable Fuels Standard created by the 2007 Energy Independence and Security Act, which mandated dramatic expansion of the use of cellulose as a feedstock for fuel ethanol. The Renewable Fuels Standard mandated that, beyond 2010, most of the increases in fuel ethanol would derive from cellulosic sugar sources (Figure 4-1).
Ultimately, it is the land on which the grain is grown that is scarce. The supply of high-quality, arable land is finite, in the United States and globally. Yields per unit area will continue to increase, through improvements in agronomic practices, breeding of higher-yielding cultivars, and
the application of agricultural biotechnology. Yields can be expected to improve by 1 to 2 percent per year in the developed world, and by somewhat higher rates in the developing world, where the yield baseline is lower. The projected rates of yield improvement will ensure an adequate supply of our food and feed needs. Alternative sources of carbon are needed to realize our full ambitions for the biological production of chemicals.
Agricultural residues will be the first source of cellulose used in the biological production of chemicals. A current generation of cellulosic ethanol plants will rely on corn stover (stalks, leaves, and cobs) as the exclusive source of carbon. Other sources of cellulose are available from agriculture; wheat straw, rice straw, and sugarcane bagasse are all examples.
The use of cellulosic biomass as a source of sugars for fermentation requires a multistep process to digest the cellulose. The first step of the processing is size reduction to facilitate the flow of materials and to increase surface area for subsequent chemical steps. The second step is exposure to acid or base at elevated temperature, to break down the cellulosic structure and to unlock it from the lignin. The third and final step is saccharification, usually achieved using a cocktail of cellulases and hemicellulases, to hydrolyze the polysaccharides, yielding a mixture of simple sugars for fermentation.
The resulting sugar stream is a mixture of five-carbon and six-carbon sugars. Concentrations of sugars are much more dilute than that used in today’s dextrose-based fermentation. The stream also contains recalcitrant polysaccharides, lignin, and other solids. For production of fuels or large-volume chemicals, economics require that the sugar stream be used without refining, separations, or concentration. Furthermore, economics dictates that the host fermentation organism be engineered to consume both five- and six-carbon sugars. Finally, the cost of cellulases and hemicellulases used for saccharification is a significant element. Enzyme efficiency must increase, and overall cost contribution from enzymes must decrease, as essential elements of cost reduction for sugars from cellulosic biomass.
Enzymes are eliminated entirely in an alternative process, based on supercritical CO2 that is being developed yielding a cleaner, more concentrated sugar stream, with a somewhat higher associated cost.98
Lignin constitutes about 20 percent of corn stover mass. It is currently recovered and valued as a fuel. As the use of cellulosic feedstocks expands, strategies are needed to derive additional value from lignin so that it can be used as a co-product rather than a waste stream from fermentation.
Beyond residues from agriculture, the forestry industry also produces residues that are a potential source of sugars for fermentation. The “hard” cellulose that constitutes wood is characterized by its higher hemicellulose and lignin contents (lignin approaching 40 percent by mass for some wood).
Forestry residues exist in large quantities, and they are often readily available at the saw mill for further processing. The disadvantages of woody biomass derive from its high lignin content. The lignin requires, and complicates, the intensive mechanical size-reduction operation. Because of these difficulties, woody biomass is considered recalcitrant in its release of sugars suitable for fermentation. New technology that would improve the release of sugars from wood could have significant economic value.
Dedicated energy crops will also play a significant future role as a source of carbon. Cropping patterns change slowly. It is unlikely that land used for today’s crops will be converted to production of an energy crop. This said, annual crops such as sorghum have great potential to be a future source of cellulosic feedstock. Sorghum is well adapted to the more arid conditions of the U.S. western Great Plains. It is a versatile crop, for which the agronomic systems are well established. Sorghum has been bred to develop varietals with high yield of grain, or for cane sugar content, or to maximize biomass yield.
While the timeline for deregulation of biotech agricultural traits extends well over a decade, on that longer time horizon, additional technology can be brought to bear to produce sugars from cellulose. Use of advanced breeding techniques and transgenic traits can lead to cultivars designed for biomass disassembly into its constituent sugars. Modifications to the level of lignin and the nature of the hemicellulose content will lead to less recalcitrant biomass, yielding more fermentable sugars per ton, and further reducing the cost of usable cellulosic sugars.
Perennial grasses may also be adapted to cultivation on marginal land not currently used for row crops. As such, they have the potential to augment biomass supply without competing for today’s agricultural land. Switchgrass, for example, is native to the United States and can yield large quantities of biomass per unit area. Grasses also offer greater flexibility in the timing of harvest, when compared to row crop residues. The principal disadvantage of perennial grasses is the 2 to 3 years needed to establish the crop. This constitutes a significant economic penalty at startup.
Fast-growing trees also hold potential as sources of fermentable sugars, in much the same way that they provide feedstocks for pulping processes. This source faces the twin challenges of the long time needed to establish the crop, and the challenge associated with the high lignin content of “hard” cellulose sources.
Cheap, abundant natural gas (whose composition is essentially methane with trace amounts of other hydrocarbons) from unconventional sources is revolutionizing the U.S. energy and feedstocks landscape. Natural gas is replacing the products of naphtha crackers as the preferred feedstock for many chemical products. In addition to unconventional gas, there are also biological sources of methane from landfill gas or the biological digestion of biomass. Methane and its derivatives such as methanol, syngas, or formate all have potential as carbon sources for fermentation.
Despite the potentially attractive costs of C1 feedstocks, considerable technical challenges exist. Two-phase gas-liquid fermentation reactors are complex and costly. Both methane and hydrogen are sparingly soluble in aqueous media. Gas-liquid mass transfer is a significant impediment to high volumetric productivity in the fermenter. However, at least three demonstration-scale syngas-to-ethanol facilities are operating today. Additional process engineering and host organism research are needed to expand the economic viability of C1 feedstocks for the biological production of chemicals. Additional advances in C1s are coming from ICI, INEOS Bio, LanzaTech, and Newlight Technologies.
Key Conclusions
Conclusion: Improvements in availability of economically feasible and environmentally sustainable feedstocks are necessary to accelerate the production of fuels and high-volume chemicals via bioprocessing.
Conclusion: Improvements in the availability, reliability, and sustainability of biofeedstocks including
- cellulosic feedstocks from plants, including plants engineered for disassembly with special attention to low-cost saccharification;
- full use of lignin co-product from feedstocks;
- utilization of dilute sugar streams;
- ability to convert complex feedstocks into clean, fungible, usable intermediates via biological pathways;
- dramatic lowering of environmental impact;
- utilization of methane, methane derivatives, carbon dioxide, and formate as feedstocks; and
- use of noncarbon feedstocks (e.g., metals, silicon)
would increase the range of economically viable products, provide more predictive levels and quality of feedstock, and lower barriers to entry into the biological production of chemicals.
Conclusion: Improving the basic understanding of C1-based fermentation, including both host organism and fermentation processes, is enabling in light of the increased availability of natural gas in the United States.
Roadmap Goals
- Within 4 years, for biological processing, achieve widespread use of novel sources of carbon, such as fermentable sugars derived from soft cellulose at a full cost less than $0.50 per kilogram of substrate.
- Within 7 years, for biological processing, achieve widespread use of novel sources of carbon, such as fermentable sugars derived from soft and hard cellulose at a full cost less than $0.40 per kilogram.
- Within 10 years, for biological processing, achieve widespread use of diverse sources of carbon, such as lignin, syngas, methane, methanol, formate, and CO2, in addition to fermentable sugars derived from soft and hard cellulose at a full cost less than $0.30 per kilogram.
Economic challenges have slowed the industrialization of biology. To accelerate the use of industrial biology for the production of chemicals, overall economics must be improved.
The product targets for industrial biotechnology must be selected using economics as a primary factor. It is difficult for a bioprocess to compete directly with large-volume chemicals produced from common petrochemical feedstocks in fully depreciated assets. High-valued specialties that take advantage of the high specificity of biology are advantaged. In the case of molecules that cannot be practically produced using conventional chemistry, an economically feasible bioprocess has no competition. For chemicals with a value of less than $20/kg, the market size must justify production of more than 1 kt/year.
Based on committee background and interactions with industry experts, for commodity chemicals, having a value of $2-5/kg, the potential market must be as large as 50 kt/year. For such products, both feedstock costs and capital costs are critical considerations. Hence, both product costs and capital costs must be reduced for industrial biology to compete effectively with conventional petrochemical processing. Moreover,
it is recognized that bioprocesses should be viewed as complementary to thermochemical processes, rather than competing with them. In the future, many chemicals will be produced by a combination of biological and conventional chemical synthetic steps.
The host organism is generally viewed as the most important determinant of the economics of a biological production process. The biocatalyst determines three important economic parameters: the production rate, product titer, and yield from feedstocks. These factors greatly influence both the product costs and the plant capital expenses required. High-productivity, high-efficiency bioprocessing is needed to accelerate the industrialization of biology to produce chemical products. A step-change improvement in space-time yields for bioprocessing is essential to achieving needed reductions in product and capital costs. A typical fermentation reactor will produce 3-5 g/L-hr of product. This is at least one order of magnitude lower than that achieved in a typical chemical reactor. Such improvement can only come from more productive host organisms, combined with improvements in process engineering.
A bioprocessing facility for chemical production consists of a series of operations. Fermentation assets represent the largest capital expense in bioprocessing, but there are several essential operations. Feedstock pretreatment may be needed if the feedstock is anything other than a clean sucrose or glucose stream. Pretreatment is discussed in the preceding section on feedstock. Feedstock pretreatment operations may be integrated with fermentation or performed remotely. Pretreatment is followed by fermentation. Fermentation normally includes the use of seed fermenters to grow the biocatalyst cell population before its introduction into the production-scale fermenters. Following fermentation, separation is needed to remove the product from the cells and fermentation broth. This is accomplished through a variety of filtration or centrifugation steps. Finally, concentration and purification of the product is achieved using ultrafiltration, extraction, evaporation, distillation, ion exchange, and other processes. It is important to note that separation steps can be some of the most expensive steps in the manufacturing process and should be considered carefully.
Fermentation assets represent the largest capital expense in bioprocessing. Chemical production is normally done in an aerobic fermenter, equipped with cooling coils to maintain temperature and with agitation for both mixing and to facilitate gas-liquid mass transfer of oxygen and heat transfer for cooling. A typical aerobic fermentation plant for production of a specialty chemical typically costs $200,000/m3 and produces
0.1-1 g/L-hr. A large-volume chemical produced via aerobic fermentation costs typically $50,000-100,000/m3 and produces 1-5 g/L-hr. In contrast, an anaerobic corn ethanol plant, operating at a vastly larger scale, costs typically $7,500/m3 (including dry-mill saccharification) and produces 3-5 g/L-hr.
Fermentation has been conducted in batch mode for a long time. Batch fermentation gave way to “fed-batch” reactors in which the carbon source and co-factors needed to grow the biocatalyst, maintain its metabolism, and deliver the product, which was done on a continuous basis.
Improvements have been made in fermenter performance through better agitation, heat transfer surfaces, and better gas-liquid contacting. Better heat and mass transfer have led to larger fermenters that can operate efficiently. Space-time yield remains low because of constraints of the microorganism and temperature and shear limitations.
Historically, host organisms have been selected and engineered to optimize productivity in terms of production rate, fermentation titer, and product yield (per unit feedstock). Additional characteristics of host organisms are needed that are developed in tandem with the overall process development. For example, the need to maintain a sterile fermenter environment contributes significantly to energy costs in the form of steam needed for sterilization. Organisms capable of operating in a less sterile environment, or having tolerance to allow for pH-based versus steam sterilization, would reduce product costs. Host organisms that exhibit greater temperature or shear resistance, or that require less oxygen, would contribute to improved space-time yield. Hosts exhibiting better strain stability can be adapted to continuous fermentations and longer, more productive batch fermentation.
Little attention has been paid to the continuous removal of the product. In typical batch fermentations, the end point is determined by the loss of productivity of the production host, which, in turn, is caused by the deleterious effects of accumulated metabolites, including the targeted product. Continuous removal of metabolites can reduce the costs associated with growing the host cells—both the costs of the carbon substrate, and the less productive fermenter hours, during the cell growth phase of the batch.
The chemical process industry evolved from batch reactors to continuous processes. The reasons for this were improved uniformity (elimination of batch-to-batch variation) and enhanced process control. The two are related, but the ultimate driver has been economics. It is hard to imagine the petrochemical process industry, operating at its enormous scale, without highly efficient continuous processes. Industrial biotechnology, true to its origins in brewing, has clung to batch and fed-batch fermentation processes. The development of continuous fermentation is important
to improving the economics of industrial biology. This must be done in tandem with the development of host organisms built to this purpose.
The ability to build predictive models at the level of individual metabolic pathways, at the level of whole-cell metabolism, and at the level of the overall fermenter operation is a significant need. Available modeling tools for fermenters are helpful in constructing mass and energy balances, and flowsheeting of fermentation processes. Dynamic modeling tools that predict the effects of perturbation at the cell or fermenter level are a gap. These tools would be useful for development of batch, fed-batch, and especially continuous fermentation.
Improvements in the host organism are essential to high-productivity, high-efficiency fermentation processing. While the host may be the most important determinant of the economics of a bioprocess, improvements in the engineering of the bioprocess are also a significant factor, with clear impact on both capital costs and operating costs. It must be recognized that the development of the host organism and of the bioprocess must be done in concert.
Process scale-up represents a key challenge and a potential hurdle to production of chemicals and fuels. The challenge of translating the host organism performance across scales, starting from microtiter, to small-scale fermenters, and eventually to production-scale fermentation is a significant one. Getting this right can assist rapid progress of the field. As the promise of synthetic biology starts to deliver, and the design-build-test-learn cycle (described below) begins to churn, high-throughput screens are needed to select the variants to be used in higher-scale testing. These decisions can be helped by assay protocols that can mimic at the microscale the performance of the strain during large-scale fermentation. For a specialty product, fermentation may occur at the 1,000-L scale, whereas a large-volume chemical could be produced in a fermenter of >100,000 L. Bioethanol is typically produced in fermenters of 1 million liters, or larger. The goal is to scale from the microtiter to the production scale, as quickly as possible, with the fewest number of intermediate scales of testing and rework. This challenge requires an interdisciplinary effort that includes chemical engineering, cell physiology, automation, statistics, and modeling.
The use of enzymes in the production of biochemicals, or organic fine chemicals, has been practiced commercially for many decades. Early
embodiments made use of naturally occurring enzymes, isolated from living organisms. As recombinant technologies developed, from the 1970s more efficient enzymes were developed that improved the process economics of enzyme-mediated reactions and broadened the base of applications. Enzyme catalysts are produced via fermentation, via the process described above. Enzymatic catalysis is typically used to effect reactions such as hydrolysis, aminolysis, amidation, or resolution of racemic mixtures. Typical commercial uses include a broad range of alcohols, amines, amino acids, and organic acids.
Enzyme-mediated reactions can be carried out at high yield. The stereo- and regioselectivity of enzymes heightens their utility. Increasingly, these reactions can be conducted in organic solvents, further broadening the use of enzymes. While enzyme-mediated reactions are often performed via homogenous catalysis, the development of stable, engineered enzymes has increased the ability to immobilize enzymes on a variety of substrates.99
The potential to conduct complex, multistep biocatalysis outside the cell offers tremendous promise. Cell-free processing is just this: the activation of complex biological processes without the use of living cells.100 In practice, cell extracts have been used for many years to conduct simple reactions, along the lines of the enzyme-mediated reactions described in the section above. Cell-free processing utilizes the biochemicals of the cell, without the disadvantages of the cell’s metabolism. The biocatalyst organism is grown via fermentation. The cells are then lysed, destroying the cells but allowing the biochemistry of the enzymes and co-factors to persist. The advantages of cell-free processing include the ability to add, or to remove, catalysts and/or reagents, and reduced effects of toxicity, because cell viability is not a concern. Energy and mass transfer may be enhanced by the absence of cell walls. The reaction medium is homogeneous, facilitating measurements of concentrations without concern about gradients across the cell wall. Cell-free bioprocessing is not without its challenges. Metabolic networks that are essential to the desired synthesis must be maintained. Co-factors must be recycled, to make the processes economical. To date, production rates remain modest. Complex, multistep syntheses have not been achieved. Operations at a scale suitable for large-volume chemicals are still to be demonstrated, but, given the great potential and numerous advantages of this technology, its development is likely to continue.101
Additional Bioprocessing Operations
A number of unit operations are required downstream of the fermenter, enzyme-catalyzed reactor, or cell-free bioreactor. As in any chemical production, product separation and purification are necessary steps. These steps add operating and yield costs and represent a significant capital cost for the facility. Thermal separation processes are both energy and water intensive. Greater efficiency is needed to reduce the capital and operating costs of thermal separations. Use of alternative separations technologies such as extraction and membranes should be expanded. Lower-cost, cleanable membranes can reduce the costs of microfiltration and ultrafiltration. Separation processes adapted for continuous removal of product and other metabolites from batch fermenters is an additional need.
Fermentation processing requires the use of water. Water is used both as the fermentation process medium and as steam and cooling water in product recovery. The amount of water required per gallon of fuel ethanol has decreased from 5.8 gallons in 1998 to about 3 gallons today. Further improvements are needed in water reuse, with a goal of achieving near-zero net water usage.
While bioprocesses are often considered environmentally benign—“greener” than chemical plant operations—they do generate solid and liquid wastes. Dramatic increases in the use of bioprocessing will require disposal of larger quantities of these streams. Alternatives to current disposal methods will be required. Waste streams must be recognized for the additional value they can present. Co-product value will need to be derived from waste streams to improve the environmental footprint and to improve the economics of bioprocesses.
Key Conclusions
Conclusion: Aerobic, fed-batch, monoculture fermentation has been the dominant process for bioproduction of chemicals for many decades. Successful improvement efforts have focused on more productive host organisms. Little research has been conducted to improve the productivity of the fermentation process, by means of enhanced mass and heat transfer, continuous product removal, and more extensive use of co-cultures, co-products, and co-substrates.
Conclusion: The development of predictive computational tools based on small-scale experimental models that realistically predict performance at scale would accelerate the development of new products and processes for the production of chemicals via industrial biotechnology.
Conclusion: Unlike many traditional chemical processes, industrial biotechnology generates large aqueous process streams that require efficient mechanisms for product isolation and for efficient water reuse.
Roadmap Goals
- Within 3 years, achieve an operating process for an economically viable bioreactor that overcomes mass-transfer and separations limitations associated with gaseous feedstocks and/or gaseous products.
- Within 5 years, develop data-based modeling tools and scale-up technologies that enable reliable scale-up of any bioproduction process from 10 L to 10,000 L in less than 6 weeks.
- Within 7 years, consistently and reliably achieve fermenter productivity of 10 g/L-hr at steady state in a continuous fermenter, or following the growth phase in a batch fermentation.
- Within 5 years, for all bio-based aqueous processes, achieve 80 percent reuse of process water.
- Within 7 years, for all bio-based aqueous processes, achieve 90 percent reuse of process water.
- Within 10 years, for all bio-based aqueous processes, achieve 95 percent reuse of process water.
The core of an expanded industry emerging from the accelerated biological production of chemicals will consist of specialized organisms capable of producing a given compound at titers, productivities, and yields sufficient for economical production. These microbes will almost certainly be highly engineered, featuring many genetic modifications, including but not limited to insertion of genes encoding new enzymatic activities, deletion of genes encoding competing and undesired activities, and modification of genes to alter regulatory and feedstock, intermediate, and product tolerance processes. Hence, the core of this industry will consist not only of the microbes themselves but also of advanced methods for the facile production of these engineered organisms. The advances necessary to generate these next-generation production strains fall into several categories: first, the development of modeling and design tools capable of the predictive tailoring of pathways, genomes, and capabilities of industrial microorganisms, from discovery to large-scale fermentation; second, the underlying science and technology for genome manipula-
tion, including in organisms that are not part of the current pantheon of established production strains or that may yet be discovered in the wild; third, informative measurement techniques to assess the performance of engineered organisms and pathways; and finally, approaches to learn from previous efforts so as to repeat successes and avoid past failures.
The development of an engineered organism for the production of chemicals begins with a technical specification for the desired bioprocess, with particular emphasis on those aspects of the specification that influence the selection of host organism and metabolic pathway. The initial specification may include one or more of the following: (1) the chemical(s) to be produced, (2) the target price point of the finished chemical (e.g., dollars per kilogram), (3) the target volume of the chemical (e.g., metric tons per year), and (4) the target feedstock (e.g., glucose). These are most relevant to the host and pathway, as they establish the primary set of parameters and objectives around which strain engineering will commence. As proof of concept is established for biological production and the model needs expand to consider the full integration of process design and development, additional specifications may include (5) the quality specification of the finished product (e.g., purity); (6) the target titer, productivity, and yield (as determined from a technoeconomic model of the full bioprocess); (7) additional bioprocess considerations (e.g., batch versus fed batch versus continuous fermentation, use of co-solvent, aeration level) that may influence the design of the organism; and (8) designs that expedite scale-up and ongoing quality control measurements.
Engineering organisms for the production of chemicals thus requires modeling across many different levels of resolution, spanning from (re)design of host metabolism to support the carbon, energy, and co-factor needs of chemical production to design of the genetic sequences that encode the cellular machinery needed for manufacturing the chemical to the desired specification. Each of these levels presents its own set of technical challenges, needs, and opportunities. Further, biomanufacturing of any chemical compound will certainly require extensive strain engineering if the molecule is heterologous; however, even for hosts in which the molecule is a naturally occurring metabolite it is highly likely that additional modifications will be necessary to achieve a commercially viable process.
If the target molecule is not a known biological metabolite but its synthesis is believed to be accessible through biology, then a novel pathway must be designed to produce the product of interest from either an existing metabolic intermediate or a readily supplied carbon source. Once a pathway has been specified, the next step is to select the enzymes needed to catalyze each biosynthetic step. The mining, design, and evolution of discrete steps will lead to a functioning pathway, but typically
with low yield. After a functioning pathway has been established, the development process must continue in order to produce the ultimate, engineered microbe generating the desired product at specified rates, titers, and yields. As multilayered as the yield may be, it can ultimately be specified in terms of mass and energy balances, which will have concomitant impacts on the organism as a whole that must be taken into account.
Introduction: The Design-Build-Test-Learn Loop
An essential element of engineering biology is the application of the time-honored, iterative scheme of design-build-test-learn (DBTL) that is a hallmark of all engineering disciplines. Metabolic engineering first applied engineering principles toward strain construction for production of small molecules. Synthetic biology has endeavored to expand and greatly enhance the DBTL loop throughout all aspects of the engineering of biological systems. For a given desired bioprocess, this DBTL loop spans from the selection and tailoring of a suitable host and metabolic pathway, the enzymes that will constitute the pathway, the genetic systems that will express the enzymes, and the implementation plans for how to build and test what has been selected and tailored (design); to employing DNA synthesis, assembly, transformation, and genomic modification tools to generate the designed strain variants (build); to culturing these variants to assess the performance of the built strains, for example through approaches such as transcriptomics, proteomics, metabolomics, and one or another means of metabolic flux analysis102 (test); and finally to evaluating the resulting test data to determine whether the design was successfully realized and whether the initial design model(s) or build and test processes require further improvement (learn). Each aspect of this cycle will be considered in turn as it applies to the overall foundational science and driving conclusions that support the acceleration of biomanufacturing.
Fully Integrated Design Toolchain
Across each of the levels of resolution described at the outset, we note a common gap between the scientific design tools available today and the engineering design tools needed to achieve the envisioned future presented in this report. To date, most tools used in organism design are what are colloquially referred to as “pull” tools. Pull tools are tools that enable the user to ask and answer a specific question regarding a proposed design. For example, mFold allows a user to submit a nucleic acid sequence for secondary structure prediction. PROSITE allows a user to submit a protein sequence for known protein domain motifs.103 COBRA
allows users to use a genome-scale model to predict cellular metabolism under different conditions among other functionalities.104 Although each of these tools can be useful in the overall organism design process, they all require that the biological engineer formulate a specific question regarding a proposed design, identify and apply the tool that can answer that question, and then interpret the validity of the results. This approach limits the detection of flaws in a proposed design to those issues that the biological engineer opts to study—the engineer must “pull” information from each available tool. To realize the grand challenges presented in this report, it will increasingly be necessary to develop and deploy “push” tools that can preemptively provide useful information regarding potential flaws in proposed designs. For example, a comprehensive push tool for designed genetic sequences might scan the input nucleic acid sequence for gene expression regulatory motifs (promoters, transcription factor binding sites, ribosome binding sites/Kozak sequences, codon usage, translational pause sites, terminators, and RNase sites), structural motifs (DNA, RNA, and protein secondary and tertiary structure), as well as functional motifs at the protein level (known protein domains, signal sequences, and proteolytic cleavage sites) and “push” the summarized results of this analysis to the biological engineer. More sophisticated push tools may even be able to prioritize the results based on both estimated confidence in each prediction as well as the likelihood that each result might adversely impact organism performance. Push tools free the biological engineer from needing to query each design against a library of tools and instead rely on software to point out all potential issues in a proposed design. It should be clarified that push tools extend beyond merely more autonomous and integrated software systems. Push tools have the potential to notify the biological engineer, asynchronously with the engineer pulling information from an integrated system, of new concerns or opportunities as additional information or tool improvements emerge. For example, if a desired biosynthetic route is currently inaccessible because no known enzyme exists to perform a key step in the pathway, then a push tool could notify the engineer when such an enzyme is identified. Or, if new information indicates that a metabolic intermediate of a previously designed pathway poses a significant human health risk, a push tool notification could arrest the deployment of the designed biological system implementing that potentially hazardous pathway.
The realization of a fully integrated design toolchain will require the establishment of standardized software tool application programming interfaces (APIs) so that the tools can effectively send “push” notifications to each other, apply data-exchange standards that specify how the content of the notifications should be structured, and make use of standardized data repositories (relating in particular to bioprocesses, bioreactors,
organisms, pathways, enzymes, expression systems, and build and test methodologies) that design tools can pull information from. Standardization is a well-established concept and practice in synthetic biology, dating back to at least the development of the BioBrick DNA assembly.105 More recent efforts have sought to move standardization beyond physical DNA assembly to data-exchange and visual design representation standards, including the Synthetic Biology Open Language (SBOL) and its visual notation (SBOL Visual).42 Complementary efforts have sought to leverage and adapt other established standards, such as Digital Imaging and Communications in Medicine (DICOM),106 into the service of synthetic biology. Repositories of information concerning organisms, DNA sequences, and expression systems have begun to emerge, including the iGEM Registry of Standard Biological Parts,107 the ICE repository platform,108 the Virtual Parts Repository,109 the DNASU plasmid repository,110 and AddGene.111 The first three of these specific repositories have established APIs for design tools to access their contents, and efforts are under way to develop a standardized API across these repositories to enable a united “Web of Registries.” While these efforts demonstrate that some progress has been and is being made toward the establishment of the standardized APIs, data-exchange standards, and standardized data repositories that will be required to enable a fully integrated design toolchain, it is clear that much work remains (in particular around establishing repositories of experimental measurement and characterization data). It should also be noted that there is a delicate balance between the organic and prescriptive development of standards, namely that, although standards are essential to realizing the fully integrated design toolchain and new incentives (whether resource or social) are required for their development, there is a risk that prematurely institutionalizing a standard could create cumbersome legacy disincentives to make improvements that might adversely impact innovation and rates of progress. It is likely that if an integrated design toolchain is developed and becomes widely used, data standards for feeding this toolchain will follow naturally.
Key Conclusions
Conclusion: The development and use of a robust integrated design toolchain across all scales of the process—individual cells, cells inside reactor, and the fermentation reactor itself—is an important step in bringing biomanufacturing onto the same level as traditional chemical manufacturing.
Conclusion: The development of predictive modeling tools within and for integration across all scales of the process—individual cells,
cells inside reactor, and the fermentation reactor itself—would accelerate the development of new products and processes for the production of chemicals via industrial biotechnology.
Roadmap Goals
- Within 4 years, develop and demonstrate an integrated design toolchain for the design of a biomanufacturing process at and below the level of an individual organism (i.e., everything inside the cell).
- Within 7 years, develop and demonstrate an integrated design toolchain for designing a biomanufacturing process at and below the level of an individual biological reactor (i.e., everything inside the reactor).
- Within 8 years, develop and demonstrate an integrated design toolchain for designing an entire biomanufacturing process (i.e., everything from concept to product).
The first step in the design process is to select an appropriate metabolic pathway for biosynthesis. In this case, even the knowledge of an elucidated pathway for target synthesis does not necessarily render the choice of metabolic pathway obvious. For example, the isoprenoid/ terpenoid family of compounds can be produced using the mevalonate or nonmevalonate (DXP) pathway, or a hybrid incorporating elements of both.112 Similarly, succinic acid can be generated from either the oxidative or nonoxidative branch of the tricarboxylic acid cycle, or a hybrid of both.113
For more novel conversion steps, in which the enzymatic chemistry is validated but transformation of the specific substrate of interest has not been experimentally confirmed, new tools are needed to increase the predictability of proposed designs. Such tools would ideally provide a rank order for pathway designs based on predicted experimental feasibility. Factors to consider may include the chemical distance between known and target substrates,114 diversity of enzymatic sequences encoding the activity of interest, knowledge and understanding of reaction mechanism of target enzyme activities (to aid rational design of enzymes; see below), and extent of functional validation of substrate diversity and range.
For known enzymatic reactions, the design tool should include available experimental data to rapidly identify variants likely to possess the highest activity. It should be noted that in biology, context matters and thus typical experimental data, such as measurements of activity in idealized in vitro conditions, may not translate into high activity in the cellular host. Nonetheless, the integration of detailed biochemical information, where available, can aid the selection process. When enzymes are not identified that meet the target specifications, alternatives must be found. One option is the search for alternatives based on homology to known variants, for example, using BLAST alignments.115 This search method does not require isolated or functionally validated sequences but relies solely on similarity to suggest additional options. The advantage of this approach is that it facilitates access to the treasure trove of genomic and metagenomic data to access new variants; however, the disadvantage lies in the uncertainty associated with sequences that have not been functionally validated. Even as improvements in build throughput emerge, it is still desirable to avoid unnecessary synthesis of enzyme-encoding DNA sequences that fail to be functionally useful. Accelerated industrialization demands increased predictability to link protein sequence to enzymatic function. Design tools that improve the accuracy of functional prediction—and, ultimately, the ability to predict not just whether an enzyme will be active but how active it will be—can greatly accelerate the initial steps of establishing proof of concept for biosynthesis. The integration of pathway design and enzyme specification tools, resulting in exquisite computational tools that can reliably present a feasible de novo pathway toward a target compound, would herald a revolution in industrial biology as these tools would immediately and dramatically expand the scope of chemical compounds that would be candidates for biomanufacturing.
As indicated above, supporting and building on existing enzyme databases will accelerate efforts in enzyme design. As these databases are built out, data fields should be modified to include knowledge that is particularly relevant to pathway design, such as known side reactions, substrate specificities, allosteric controls, evolvability (based on phylogenetic or experimental knowledge), and potential functional analogues.
Implanting pathways and enzymes in a chassis for screening or production usually requires a (re)design of host metabolism and/or physiology to achieve the desired performance standard. Metabolic design objectives typically include reengineering of competing by-products and minimization of biomass formation. Both of these contribute to maximiz-
ing product yield. However, biosynthetic pathways often involve redox reactions such that electron flow has to be considered in combination with carbon flow. Additionally, specific transformations may require coupled reactions or the generation of activated substrates to provide the energy needed to catalyze thermodynamically unfavorable reactions. The fully integrated design toolchain should be able to satisfy these layered objectives, accounting for endogenous metabolism, heterologous product formation, and redox and energy balances to predict the optimal combination of genetic manipulations. To this end, it would be desirable to also have registries containing the characteristics of hundreds of host organisms and their phenotypes under a wide range of conditions, such as different temperatures, pressures, salinities, and carbon sources. Such a registry would be a public good in the same manner as PubMed and could be accessed to accelerate both corporate efforts and to provide fodder for the fuller development of systems biology tool sets for organismal and pathway design.
It will be especially important not to neglect systems-level effects on overall cellular physiology. A commonly viewed obstacle toward bio-based small-molecule production is toxicity, in which the product greatly or completely reduces cell viability and, in doing so, affects the production capacity of the host. These effects are often not easily classified in mass and energy balance equations and often manifest in both physical and biological ways. For example, a product may be inhibitory to enzymes in the pathway or to other endogenous reactions essential for cell performance. In this case, identification and incorporation of feedback-resistant enzymes may alleviate the most harmful effects. While certainly not trivial to implement, this form of toxicity has a clearly assigned biological cause and can be addressed as such. On the other hand, if the product associates physically with the cell membrane, disrupting membrane integrity and causing leakage of cytoplasmic contents, then this mode of toxicity must be understood on a more fundamental level to rationally propose a solution, perhaps through engineering the composition of the cell wall to withstand higher concentrations of the toxic production. In either case—or in combinations thereof—design tools are needed that can propose both a mechanism of toxicity and a means to address it, given knowledge of the system. It should be noted that adaptation and evolution could certainly be used to obtain strain variants with more tolerant phenotypes and, in this case, the design tool chain should be able to incorporate findings from these experiments to learn and thus implement that knowledge in future design scenarios.
While the design toolchain as described above is focused on the cellular organism, a fully integrated design process must operate across scales to incorporate bioprocess considerations. The strain that performs to specification will operate reliably as designed. These performance specifications can then be translated into well-established parameters, including, for example, observable product yield on substrate, product yield on biomass, and specific productivity, that have been successfully used for decades to model and design bioprocesses. As cellular behavior is more complex, for example, exhibiting dynamic behaviors through the incorporation of feedback control mechanisms, these behaviors can be modeled at the bioreactor scale to predict overall process performance, ultimately generating the predictions in titer, yield, and productivity that are necessary to evaluate the commercial viability of a process. Overall, models should predict cell behavior in culture over a wide variety of culture volumes and under a wide variety of bioprocess conditions. The systems biology–based registries imagined above would assist in building tools to eventually predict scale-up and scale-out.
The construction of new organisms for industrial biology applications can be further broken down into the identification, characterization, and modification of “chassis” for production, and the construction of appropriate pathways in these chassis for the production of a given compound.
The modification of chassis and the construction of new pathways will be greatly enabled by the ongoing revolution in DNA synthesis. To the extent that we remain on an exponential trajectory for the acquisition of longer and cheaper pieces of DNA, much larger and many more constructs can be generated and tested. Synthesis technologies will make the DBTL paradigm particularly powerful. That said, there is clearly a growing need for biofoundries that can scale the production of subgenomic assemblies or pathways. While public funding may lead to the establishment of more centers for synthesis, it is also possible that synthesis and assembly technologies can be developed to the point where DNA designs could be synthesized and assembled by bench-scale equipment (a “DNA printer”) in essentially every research lab.
Pathways are typically composed of a series of enzymatic transformations, integrated with central metabolism via sensors and regulatory interactions. In order to develop pathways capable of generating virtually
any small organic product of interest, it will be first and foremost necessary to enable the acquisition of enzymes that can carry out virtually any transformation. Such enzymes can likely come from three sources: first, mining phylogeny for novel enzymes; second, elaborating the catalytic activities and biophysical properties of known enzymes, by either design or selection; and finally, the generation of enzymes with wholly new properties not previously found in nature.
Bioinformatic approaches to mining new enzymes are already in vogue,116 and the integrated design toolchain described above is likely to continue to both fill informatics databases with alternatives and better target enzymes to new purposes and pathways. Although mining and characterization have yielded numerous parts that have proven to be useful for microbial engineering, in many cases the specific roles of parts or their performance in new contexts must be further optimized. Two methods have shown promise in the generation of parts for virtually any genetic circuit: computational design and directed evolution. Such methods are equally useful for proteins. The computational design of proteins has advanced to the point that it is now possible to generate novel protein folds and to frequently improve the functionalities of extant proteins, including their stabilities and interfaces with both small molecules and biopolymers. There have been several enabling improvements in protein design tools, most notably the widespread use of the Rosetta suite. In concert with the improvements in DNA synthesis that have been noted elsewhere, this has meant that it is frequently possible to redesign a given protein scaffold for novel structure, synthesize tens to hundreds of predicted variants, and quickly assay for those that have the required capabilities. Roadblocks that remain to future progress primarily have to do with improvements in physics-based approaches and algorithms that will better specify the energetics of interactions, especially with small molecules. As these barriers are overcome, it should be possible to redesign enzyme active sites to accommodate a wide range of substrates and co-factors, and thereby to more completely enable the development of virtually any transformative pathway. A reach goal would be the ability to design enzymes de novo for chemical reactions that currently have no biocatalytic equivalent.
Similarly, the directed evolution methods described for organisms also apply to enzymes, and there are a variety of techniques for altering enzyme properties. Directed evolution complements molecular design in that it can sieve through large numbers of molecules for those few with the required capabilities. However, directed evolution is frequently capable of sieving very large libraries of millions to billions of variants, thus partially obviating the need for design. On the other hand, the sequence spaces that are accessible by even small proteins are so large
that design tools have proven to be extremely valuable for delimiting the libraries that will be constructed for a given directed evolution experiment.
The key issue that restrains more widespread use of directed evolution as a means of optimizing parts is that novel selections or screens must be developed for each new molecular functionality. If an enzyme with new substrate specificities is desired, then either the enzyme’s functionality must be linked to cell growth or a high-throughput assay specific for that enzyme must be devised. In order to overcome these problems researchers have begun to develop more generalized schemes for directed evolution, such as phage-assisted continuous evolution117 and compartmentalized partnered replication, that attempt to generally connect the phenotype of a part with function in a system, thus enabling more modular selections. In this regard, improvements in rational design may enable smaller libraries of sequence space to produce desired activities with limited screening throughput.
Going beyond nature and beyond the capabilities of directed evolution is still mostly notional. It is possible that wholly new enzymes can be designed or selected that incorporate a variety of novel elements to carry out complex bioinorganic transformations. Similarly, the 20 amino acids available for enzyme chemistry can be greatly augmented by nonstandard amino acids that are better able to perform specific chemistries or that can “harden” proteins to the requirements of bioprocessing streams operating at high temperatures or under acidic conditions, intracellularly or in isolation.
This space is well populated (although not saturated) by industry. Between improvements in computational design and directed evolution, the prospect exists for taking a relatively small list of parts and endlessly morphing their function to suit the needs of industry. This in turn suggests that there will likely be productive niches within the corporate ecosystem devoted to parts improvement. Companies such as Codexis regularly develop novel enzymes for customers carrying out large-scale bioprocesses.118 It is not unreasonable to expect that if “conceptual barriers” between design and synthesis remain in place and are propagated, part of a future system will specify the characteristics of a part, rather than the part itself, and if those characteristics are not satisfied by something already in a database, then the specifications will be delivered to a parts foundry as a standing order.
Key Conclusion
Conclusion: Improvements in the ability to rapidly design enzymes with respect to catalytic activity and specific activity and engineer
their biophysical and catalytic properties would significantly reduce the costs associated with biomanufacturing and scale-up.
Roadmap Goals
- Within 7 years, have the ability to insert 1 megabase of wholly designed, synthetic DNA into the genome of an organism at an error rate of less than 1 in 100,000 base pairs, at a cost of $100, in 1 week.
- Within 7 years, have the ability to design de novo enzymes with new catalytic activities with a high turnover rate.
In the bioprocessing considered here, cells are the unit of engineering. Although enzymes or pathways can be embedded in cells, the cellular metabolism and physiology that supports chemical transformations are often critical aspects of bioprocess engineering and scale-up. While a great deal of basic metabolic engineering can take place in E. coli and other model organisms, these cellular “chassis” may not always be suitable for production.
The diversity of metabolic and physiological requirements for the production of different compounds necessitates a range of cellular chassis for metabolic engineering. For example, microorganisms with a naturally high tolerance for long-chain alcohols may be more suitable as hosts for new biofuel production, while strains with very low pH tolerance are advantageous for production of organic acids by minimizing downstream separation costs. The reason that E. coli, S. cerevisiae, and other model organisms are so highly used is the extensive repertoire of genetic tools available for these hosts. As a result, the correlation between genomic, proteomic, metabolic, and other information is relatively complete and is already laid down into systems biology models that are increasingly being quantified (as apparent from the Design Toolchain described above). Therefore, it is critical that additional foundational research be carried out on the systems biology and physiology of organisms that are better suited to bioprocess engineering and production.119 Beyond capturing the genome sequences of laboratory strains, sequencing greater numbers of microorganisms that are actually involved in production should prove useful. Ancillary proteomic and metabolic analyses, and follow-on quantitative and predictive models for these systems as a whole, will provide fodder for grafting new enzymes and pathways to these chassis and therefore for producing a new cornucopia of compounds at the industrial scale.
As we garner better understanding of industrially relevant chassis, new tools for the manipulation of organismal genomes will become increasingly important. This is especially true because of the limitations on transformation and because the breadth of different chassis under consideration will require more generic mechanisms for undertaking site-specific genome modifications. In this regard, the ongoing innovations with CRISPR-derived systems promise to revolutionize the modification of many organisms, including those relevant for chemical production, either via targeted genomic editing or via regulation of individual pathways by catalytically inactive, programmable ribonucleoproteins such as dCas9.120 There are other systems for programmable site-specific modification, including Targetrons,121 TALENS,122 and zinc-finger endonucleases;123 modifications of all of these systems often allow the site-specific insertion or mutation of genes, as well as their deletion. Overall, continued advances in these areas promise to widen the reach of methods like MAGE,124 in which there is iterative optimization of function across the entire organismal genome.
In contrast to these methods, many synthetic biologists have focused on developing orthogonal systems that can operate beside or on top of extant genomes. Such orthogonal systems may come to represent very large, programmable subsystems with their own replication, transcription, and translation capabilities, as well as internally programmed regulatory and metabolic pathways. In essence, episomes carrying these features would be subgenomes that would both direct their own function and redirect their host’s genome toward a desired functionality, such as the production of a particular metabolite or compound. To promote the development of this new generation of programmable, self-sufficient episomes may require a renaissance in plasmid and epsiome biology. Indeed, this may be an area where synthetic biology can provide modules that go well beyond regulation or metabolism. Into the future, it should be possible to take a toolbox of standardized and orthogonal origins, polymerases, promoters, ribosomes, and encoded amino acid biosynthetic and charging capacities and create made-to-order episomes for any of a variety of industrially relevant bacteria. The addition of CRISPR or other elements would allow these subgenomic control systems to finely control host expression.
Following site-specific genome engineering or the introduction of subgenomic control systems on episomes, the stabilization of an engineered chassis would be paramount. Most organisms have evolved not to produce a metabolite or compound in great yields, but instead to grow and survive. Redirecting metabolic flux for human purposes is usually an evolutionary dead end. Thus, either the rate of mutation and genetic change must be greatly reduced, or the engineered organisms or episomes
must be evolutionarily robust—able to retain function even in the presence of multiple mutations. For example, proteins may be engineered to tolerate multiple amino acid substitutions and would thereby exist on a large neutral fitness landscape that would greatly delay loss of function. When such proteins are expressed in a slow-evolving chassis that contains antimutator polymerases or enzymes that can remove nucleotide modifications even prior to incorporation, it may be possible to slow evolution to the point where it is no longer a consideration over the industrial lifetime of a biosynthesized product.
Paradoxically, before a chassis is fixed into an evolutionarily stable trajectory, directed evolution methods applicable to whole organisms will be of increasing importance. As systems biology approaches provide increasingly excellent “roadmaps” for metabolic and regulatory engineering in a wide variety of organisms, it should be possible to delimit what pathways, loci, or regulatory networks should be the focus of directed evolution. In the past, strain improvement via random chemical mutagenesis was one of the primary tools for generating a production strain. Now random or semirandom approaches to modifying organismal genomes, coupled with well-designed selections or the high-throughput screens described below, will allow organisms to be driven into more productive states. In particular, the sequence-directed approaches to manipulating organismal genomes described above will likely prove more useful not only for model-based manipulation but also for directed evolution. These include methods such as recombineering libraries (as embodied in MAGE) and Cas9/dCas9 libraries. Again, an issue with many of these approaches is that they are targeted largely to E. coli as a platform, and their use in nonstandard laboratory strains, especially those that may be of greatest importance for production, is limited. This will require the adaptation of these tools to new organisms, potentially via the development of broadly useful episomes for horizontal transfer, as described above. In this paradigm, the tools and libraries for site-specific or random modification might initially be created in a tractable chassis, such as E. coli, and then moved by horizontal transfer to a new host to execute.
Genetic designs are currently limited to approximately a dozen genes, whereas genomes consist of thousands and many of the potential products of biology will require large numbers of regulated genes. As such, as the desired products become more complex, so too will the need to push our design capacity to this scale. This will require pathway design involving dozens of genes that collectively build the desired product. This will have to be integrated into the broader cellular metabolism and cellular functions, for example, those involved in nutrient and feedstock acquisition (e.g., cellulases, nitrogen fixation), secretion and import of precursors, and stress response. These functions require precise timing as to the con-
ditions or order in which they are expressed as part of building a product or coordinating responses. This will require the ability to build synthetic regulation of a sophistication of the natural regulatory networks in cells. All of these genes are going to tax the host’s resources, which will require a better understanding of how to allocate cellular machinery. Collectively, these designs will require combining hundreds of DNA parts and being able to predict how they work in concert. All of these considerations will have to be integrated into future computer-aided design packages that facilitate the management of large genetic engineering projects. In essence, the domestication of an organism as a suitable chassis in industrial biotechnology, as E. coli is today.
Key Conclusions
Conclusion: Continued development of fundamental science and enabling technologies is required for the rapid and efficient development of organismal chassis and pathways.
Conclusion: Expanding the palette of domesticated microbial and cell-free platforms for biomanufacturing is critical to expanding the repertoire of feedstocks and chemicals accessible via bio-based manufacturing.
Conclusion: The design, creation, and cultivation of robust strains that remain genetically stable and retain performance stability over time in the presence of diverse feedstocks and products will reduce the costs involved in the use and scaling of biological production.
Roadmap Goals
- Within 2 years, achieve domestication (including >1 percent transformation competency, genetic and genomic modification tools) across five phenotypically diverse microbial types other than established models (such as E. coli and S. cerevisiae).
- Within 5 years, achieve domestication across an additional 10 or more industrially relevant recalcitrant microbial types and the ability to domesticate any other microbial type within 3 months.
- Within 7 years, develop the ability to achieve domestication in any new microbial type within 6 weeks.
- Within 7 years, have a suite of domesticated organisms (including cell-free systems) that can utilize diverse feedstocks and generate a range of products with high yield and productivity under various process conditions while maintaining process robustness.
Although the ability to design and evolve parts and circuits is of fundamental importance for the improved practice of synthetic biology, developing improved methods for measuring the results of experiments will perhaps have an even greater impact. Design and evolution can provide basal circuitry that frequently requires additional optimization. Improvements in design tools can reduce the number of circuits that need to be tested and can improve the overall quality of those circuits, while facile directed evolution methods allow an ever larger number of variants to be screened and selected for improved function. But in neither case will the tools developed cover all challenges; they will likely continue to run well behind the sheer size of the sequence spaces being explored.
By enabling the underlying data needed to create and continuously improve the design methods envisioned above, measurement technologies will play a strong role in the subsequent emergence, practice, and advancement of engineering biology. The comprehensive measurement of DNA, RNA, proteins, metabolites, their chemical and structural variants, and their interactions enabled the advances in molecular cell biology knowledge and methods that have brought us to our current state of capabilities and understanding. The new knowledge and technological advances in turn motivate new questions and unmet needs for measurement that must be addressed to enable the efficient and effective future pursuit of engineering biology. New innovations in measurement would help accelerate the DBTL cycle, improve predictive design, broaden the scope of directed evolution, support manufacturing development and process control, propagate standards, improve regulatory decisions, and ensure safe practices.
Many advances in measurement technology are driven by medical applications. These same advances, with modification and extension, can also be useful for engineering organisms. Creatively extending such measurement technologies to the needs of engineering biology in an application-specific way will be valuable.
A preeminent example of a revolution in measurement methods primarily motivated by biomedical research that simultaneously enables leaps in engineering biology is nucleic acid sequencing. Now well integrated into multiple parts of the DBTL cycle, current practice would be inconceivable without it. Advanced methods for manufacturing DNA constructs, characterization of the structure and stability of transformed genomes, quantification of the impact of genomic alterations on expressed transcripts, clarification of the behavior of regulatory elements, and identification of the genomic alterations accompanying phenotypes of interest on the basis of nucleic acid sequencing are prevalent. There remain, however, opportunities to usefully further extend high-throughput sequencing
by lowering its error rate to keep up with the very low and increasingly lower error rates of DNA synthesis; by improving its ability to delineate large-scale structural rearrangements as complex, large-scale, and precise genome engineering becomes more common; and by improving its sensitivity to single cells without compromising throughput to better discern the appearance and influence of heterogeneity among populations of cells. Improvements in error rate, read length, and sensitivity are also sought by biomedical researchers and clinicians concerned with complex diseases such as cancer. However, the requirements for throughput, data quality, data analysis methods, sample preparation, and integration of the results with complementary methods are quite distinct, leading to a divergence in the needed advances and their best implementation. These differences have already resulted in segmentation of platforms themselves across these very different fields. For example, some leading-edge single-molecule sequencing platforms have so far found more utility within studies of the microbial world than of mammalian systems.
The extraordinary utility of next-generation sequencing (NGS) makes it an ideal technology for many different types of measurements, beyond just sequencing genomes, constructs, and RNA expression levels. To the extent that protein and other analytes can be transduced into nucleic acids it may be possible to deconvolute extremely complex mixtures using NGS. For example, an antibody library tagged with unique DNA tags could be used to coordinately identify the presence and amounts of proteins on a cell surface or in a lysate. Transduction schemes for small molecules based on ligand-dependent nucleic acid conformational changes can also be envisioned. Protein modification states and epigenetic tags could be followed using similar implementations. The downside to such measurements is that they are not in real time and resolution may be lost through the transduction process.
Molecular sensing, molecular recognition, and cell signaling comprise a diverse set of fundamental biological processes. Commensurately, there is a diverse set of design options for engineering responses to environmental or internal cellular conditions. This flexibility is further increased by the success of taking a modular approach to the sensing process, making it easier to vary what is sensed and what happens as a consequence. Integration with designed cellular circuitry creates the potential for many options for control, memory, logical operations, and multiplexing. Building context-dependent sensors into microorganisms is one potential path to obtain subcellular measurements despite their small size. Overall, these phenomena can be used to help debug a living system under development, to provide feedback to living cells, or as subsystems within an ex vivo measurement solution for research, production, diagnostics, or environmental monitoring. In some cases, biosensor systems can be run
in vitro and thus be exported to cell-free systems in solution or on solid supports. Conversely, advanced cell-free systems can be used to debug the biosensor before it is deployed in vivo. If the technical challenges can be overcome, then the number of situations in which biosensors can be expected to enable rapid, low-cost, high-throughput testing of individually engineered cells or, if desired, entire populations of engineered cells seems certain to increase.
Beyond sequencing, many additional measurements can be made to assess the performance of a given circuit. A field of measurement that is particularly essential to both engineering biology and the elucidation of human biology is metabolomics. The chemical industry’s interest in new biology-based routes for producing products is very much focused on metabolite production. Perhaps there is no better indicator than the nomenclature “metabolic engineering” for the traditional development of new organisms for better industrial bioprocessing. There is, as a consequence, a long history of developments that adapt metabolite measurements to the interests and needs of engineering biology. For example, there is already a rich diversity of laboratory and data analysis methods designed to identify and follow the pathways of metabolite production and modification. Because of the viability of progressing toward the creation of quantitative models for the associated chemical reactions in microorganisms, there is an especially close relationship between modeling and measurement in this field. They are advancing together and synergistically. Still, the most universal metabolite measurement solutions are too slow to meet the potential of the information gained while the highest-throughput methods require specialized optimization on a case-by-case basis. As is also true of proteomics measurements, faster, generalizable analysis of metabolites would greatly accelerate learning and the associated models that can encapsulate the results in ways that illuminate preferred steps throughout the engineering cycle. Here too, sensitivity to the level of single cells without compromising throughput will be of value. While there are important aspects of human biology that would also advance greatly with the advent of higher-throughput metabolomics, again there is a divergence of needs, especially because of the difference in scope, prior information, and sophistication of models between studies of industrially relevant microorganisms and human biology.
At the same time broadly useful measurement platforms are extended for use by biological engineers, synthetic biology is enabling and introducing new measurement paradigms particularly well suited for the needs of engineering biology. Circuits can be easily linked to readily observed reporters, such as green fluorescent protein, and high-throughput devices and methods that have already been developed, such as plate readers or fluorescence-activated cell sorting, can be used to parse performance.
Into the future, the development of additional reporters and analytical methods that can scale to even greater numbers may be important. In vivo measurements, genetic manipulations, intervention in regulatory processes, interference with molecular intermediates, such as expressed RNA transcripts, and chemical signal-induced alterations have been longstanding tools for generating and validating molecular biological hypotheses. Engineering biology brings a perspective of altering organisms for utilitarian purposes including the development of biosensor measurement devices at both the molecular and the organism levels for readout, feedback, and control.
In parallel, as circuits become increasingly complex there will be a need to increase the number of different parameters that can be measured in parallel, such as the expression of multiple genes or the production of multiple metabolites. This would argue for the development of higher-throughput methods that are capable of analyzing whole organisms or chemical mixtures, such as mass spectrometry or nuclear magnetic resonance. There are foundational technologies in miniaturization, micro- and nanofluidics, photonics, nucleic acid synthesis chemistry, and data analysis that could feed into and enable these desired advances in measurement solutions. Standards for metrics and materials will also facilitate convergence and deployment of the best methods along with ensuring reproducibility and transferability across laboratories, manufacturing sites, and institutions.
A second consideration in the development of analytical methodology for the assessment of synthetic circuitry is ensuring that the measurements being performed accurately reflect the performance of a given organism in an industrial setting. It does little good to optimize a circuit in the laboratory only to find that it does not work in a vat. In contrast, it is very difficult to carry out high-throughput experiments that scale to even small fermenters. Thus, it becomes important to rationally understand how the readouts of organismal metabolism scale from benchtop experiments to test bed to production. This in turn requires greater integration between systems modeling tools for gene expression and metabolism and analytical methods. The results of baseline experiments with a given chassis or circuit under a variety of conditions need to be compared with similar results in batch, or under different fermentation conditions, in order to develop feedback loops that will allow prediction of how augmentation of the chassis or circuit will perturb both the initial readings and the final performance.
Key Conclusions
Conclusion: The ability to rapidly, routinely, and reproducibly measure pathway function and cellular physiology will drive the development of novel enzymes and pathways, which are needed to increase the array of efficient and low-cost chemical transformations available for use in biomanufacturing.
Conclusion: The fall in cost and increase in throughput of measurement technologies should track that of strain engineering technologies and vice versa.
Roadmap Goals
- Within 4 years, develop the ability to routinely and reproducibly measure nucleic acids, proteins, and metabolites targeted to characterize 50 or more high-priority, selected model parameters for 2,000 strains and measure 1,000 or more parameters for 200 strains within 1 week at a cost no higher than the full cost of designing and building those strains.
- Within 10 years, have the ability to routinely and reproducibly measure 50 or more high-priority, selectable model parameters in vivo at the same cost and speed as above.