The United States has a strategic interest in identifying and responding effectively to potential disease outbreaks, whether they result from the spread of naturally occurring pathogens or are due to accidental or intentional release of infectious microorganisms. The web of systems needed to achieve this goal range from environmental monitoring to clinical and public health epidemiology to intelligence community assessments of bioterror threats. Implementing methods to detect the presence of pathogens of concern in the environment forms one component of the strategy. As defined by Homeland Security Presidential Directive 21, biosurveillance is
the process of active data-gathering with appropriate analysis and interpretation of biosphere data that might relate to disease activity and threats to human or animal health—whether infectious, toxic, metabolic, or otherwise, and regardless of intentional or natural origin—in order to achieve early warning of health threats, early detection of health events, and overall situational awareness of disease activity. (White House 2007)
The biosurveillance umbrella captures multiple activities of threat awareness, detection, characterization, and information sharing in clinical and environmental settings. The National Biosurveillance Science and Technology Roadmap (NSTC 2013), for example, describes over 20 programs, networks, and activities from across the government that contribute to the National Strategy’s goals. Table 1-1 lists selected documents that address strategic priorities for biosurveillance activities. BioWatch, the focus of this report, is one component of the overall system.
TABLE 1-1 Selected Federal Strategy Documents Related to Environmental Biosurveillance
| Year | Document | Description |
| 2004 | Homeland Security Presidential Directive 10: Biodefense for the 21st Century | Describes pillars of national biodefense preparedness including threat awareness, prevention and protection, surveillance and detection, and response and recovery (White House 2004) |
| 2007 | Homeland Security Presidential Directive 21 | Establishes a strategy for public health and medical preparedness (White House 2007) |
| 2009 | National Strategy for Countering Biological Threats | Provides a planning framework and describes seven objectives to reduce threats posed by biological risks (NSC 2009) |
| 2011 | A National Strategy for CBRNE Standards | Articulates goals to enhance coordination on standards and to establish conformity assessment capabilities (NSTC 2011) |
| 2012 | National Strategy for Biosurveillance | Promotes strengthened biosurveillance capabilities (White House 2012) |
| 2013 | National Biosurveillance Science and Technology Roadmap | Identifies research and development needs to enhance the effectiveness of biosurveillance activities (NSTC 2013) |
BIOWATCH DETECTS AIRBORNE RELEASES OF SPECIFIC BIOLOGICAL AGENTS
The BioWatch program is part of a layered approach to rapidly detect and respond to exposure to airborne biological agents of concern. A brief overview of the program is presented below and additional information is provided throughout the report. Certain details that the committee used to inform its analysis have been determined by the government to be exempt from public release under 5 U.S.C. § 552(b)(7); further information on these aspects is provided in Appendix A.
BioWatch is an environmental monitoring system developed to detect and characterize genomic material from a selected set of aerosolizable pathogens.1 The program is operated by the Department of Homeland
___________________
1 The decision on which pathogens are included for detection in the program is informed by risk assessments such as the Biological Terrorism Risk Assessment (BTRA) and material threat assessments for specific agents. These risk assessments are developed by the DHS Science and Technology Directorate as part of DHS’s mandate under several Homeland Security Presidential Directives. The BTRA has generally been undertaken at 2-year intervals, and DHS reportedly reviews the alignment of BioWatch with the BTRA when a new assessment is released (GAO 2012b).
Security (DHS) and managed through its Office of Health Affairs. The DHS Science and Technology Directorate also supports program goals by undertaking activities in research and development that contribute to its operations. BioWatch was first deployed in 2003 in outdoor urban settings, amid concerns about potential damage that would be caused by a bioterrorism attack that could affect significant numbers of people.2 The current system (sometimes referred to as the “Generation 2” or “Gen-2” system) includes collectors in over 30 jurisdictions and makes use of both outdoor and indoor collectors; systems also are deployed during selected special events. By 2013, reportedly more than 7 million polymerase chain reaction (PCR) assays had been conducted during routine daily operations to monitor for genetic material from the set of microbial agents for which BioWatch tests (IOM and NRC 2014).3
BioWatch comprises a network of components, processes, and participants that interact to capture genetic information from aerosol samples and determine whether and what actions should be taken based on the results. The context of BioWatch operations and the complexities and limitations of the role it plays should be appreciated. To detect an intentional release of biological material from air samples involves a host of factors affecting overall system performance. Environmental conditions, such as wind speed and direction, and particle parameters, such as size and density, will alter the transport and dissemination of biological aerosols. The location of collection devices and the spacing between collectors relative to the location of a released plume will affect signal strength at the collection device and whether more than one device will register a detection. The method of collection, particle size cutoff, and air sampling volume will affect the amount of biological material that is present in a sample. Finally, characteristics of the pathogen itself, such as the infectious dose and whether or not the agent is viable, will influence whether exposure to a particular quantity of an airborne microorganism is likely to produce disease.
From a practical standpoint, decision makers will want to know whether the BioWatch system can reliably detect airborne biological material at a level that would indicate the occurrence of a release of the size and scope for which BioWatch was designed. This question can be roughly divided into two parts: (1) what amount of material from a tested
___________________
2 BioWatch’s mission supports goals articulated in the 2003 State of the Union Address of an “early warning network of sensors to detect biological attack” (Bush 2003). BioWatch’s mission is to “Provide, maintain and support a continuous aerosol bio-terrorism monitoring capability in selected metropolitan areas” (Walter 2014a).
3 The locations of BioWatch air collectors and certain operational details of the program are not considered public information. See Appendix A for additional program and operational details.
agent needs to be deposited at the collection device to indicate with high probability that a release has occurred (or alternatively, what probability of a release is indicated by the presence of x amount of biological material at the collection device), and (2) what is the probability that the assays used in testing the sample will be able to reliably detect this level (or a lower one) of material.
The committee was not asked to examine and does not have information on the first of these components—the minimum size of a release event that BioWatch should be able to detect and how this might translate to a predicted minimum amount of material at a collector. Rather, the scope of this report is limited to the second part: How can the assays that test for the presence of a biological agent be validated so as to give the BioWatch program and its stakeholders confidence in their performance? This piece is in many ways the most straightforward. As a result, it is also worth noting that high confidence in the PCR assay is only one dimension of the system, and should not give users an overly optimistic sense of the performance of the system as a whole without further system testing.
The main processes in BioWatch are shown in Figure 1-1, which depicts steps from filter collection to assay interpretation and decision making.
BioWatch collectors continuously draw air through a dry filter where particulates are deposited. These filters are collected manually at regular intervals, usually once a day, and taken to participating diagnostic laboratories for analysis. The laboratories that analyze the filters collected from BioWatch air samplers are generally co-located with state and local public health laboratories, which maintain a dedicated BioWatch capacity. Most of these public health laboratories are members of the Centers for Disease Control and Prevention’s (CDC’s) Laboratory Response Network (LRN), which forms part of the national infrastructure to respond to public health emergencies and to potential bioterrorism incidents.
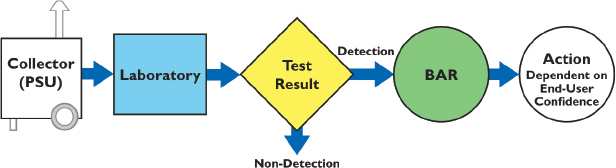
FIGURE 1-1 Schematic of the steps in the BioWatch process from air collection to assay interpretation and decision making. PSU = portable sampling unit; BAR = BioWatch Actionable Result. SOURCE: Walter (2014a).
At the laboratories, DNA is extracted from the air filters and targeted regions from the genomes of the pathogens of interest are amplified and detected using the method of real-time PCR (see Box 1-1). For these analyses, a set of PCR primers specific to DNA from the microorganisms of BioWatch concern is used for the PCR assay. For each tested pathogen, the laboratories use an initial screen targeting a single nucleic acid target sequence (a signature) for preliminary identification of candidate microorganisms. If this initial screen yields a positive result, a secondary panel of assays directed against three or more additional nucleic acid signatures is used to verify the presence of the agent.4 The BioWatch screening and secondary PCR assays currently are used in an integrated fashion by laboratories in the program (e.g., a BioWatch Actionable Result [BAR] generally is not called only on the basis of the results of the screening assay).
Possible Outcomes Arising from the PCR Assay
Based on the readouts from the PCR assays and the associated assay decision algorithms, there are four possible outcomes:
- True positive: the assay indicates the presence of the tested agent’s DNA, and the agent’s DNA is indeed present in the sample.
- True negative: the assay does not detect the agent, and the tested agent is indeed not present in the sample.
- False positive: the assay indicates that the tested agent is present in the sample, but in fact, the agent is not in the sample.
- False negative: the assay indicates that the tested agent is not present in the sample, but in fact, the agent is in the sample.
No assay will ever be free of false positive and false negative results, and these results are dependent on how one defines these terms in practice. The potential for fundamental limitations of a test’s design (tradeoffs between sensitivity and specificity, for example) and instrument and operator failures means that some likelihood of error will always be present. Understanding the nature, origins, and probability that a potential error may occur assist in the interpretation of results and provide a measure of confidence for the user.
___________________
4 The report uses the term “secondary assay” throughout to refer to this step, recognizing that it actually entails the performance of several individual PCR assays to detect several distinctive genetic signatures for the pathogen. BioWatch program materials frequently refer to this as the “verification assay” or verification assay panel; the committee prefers the term “secondary” to distinguish the assay from the processes of assay validation and verification.
BOX 1-1
Real-Time PCR as a Pathogen Detection Method
PCR is a widely used nucleic acid amplification technique, and its inventor, Kary Mullis of Cetus Corporation (Emeryville, CA), was awarded the Nobel Prize in Chemistry in 1993. PCR involves the polymerization of nucleotides in a reaction performed and mediated by enzymes and thermal conditions, resulting in a theoretical doubling of the amount of nucleic acid in each cycle of PCR. Realtime PCR adds a detection strategy to the amplification process—a common method is through the use of fluorescence, as shown in the figures below. A series of temperature changes is used to separate strands of DNA, allow primers and a probe to hybridize to complementary sequences in the target region for amplification, and enable polymerase to synthesize new nucleic acid in this region. As the polymerase synthesizes the new nucleic acid strand, it degrades the probe, releasing a fluorescent reporter whose emission can be detected. The greater the fluorescent signal, the greater is the amount of target in a sample. Other technologies can use a DNAbinding dye instead of a fluorogenic probe to measure DNA amplification. The fluorescent signal can be monitored at each cycle to create curves such as those below, with associated threshold cycle (Ct) values for when the signal crosses the background threshold. Alternatively, the strategy of end point detection, measuring only the final signal rather than the intermediate amplification curves, may be used. In this case, results are frequently reported as median fluorescence intensity values.
PCR methods can be used to detect nucleic acid from agents of interest such as those tested by BioWatch. Using a pathogen’s genetic sequence and genetic information from related microorganisms (such as closely related but nonpathogenic species to reduce crossreactivity), specific sets of primers and probes are designed. The goal is to amplify and detect a DNA target (an amplicon) that is distinctive for the tested organism. The curve of the detected fluorescence provides information on whether the DNA signature of the pathogen is present in the sample as well as information on the starting amount of target DNA. For BioWatch, there are multiple steps from a pathogen in the air to a sample of extracted DNA used in a PCR, and so the relative amount of DNA in the PCR sample can be informative but does not directly provide the original pathogen concentration.
During a PCR assay, a false positive detection could result from a number of factors, such as cross-reactivity of assay primers and probes (both types of short oligonucleotides that hybridize to target DNA) or the presence of a closely related but as-yet unsequenced organism occurring naturally in the environment. As was emphasized to the committee dur-
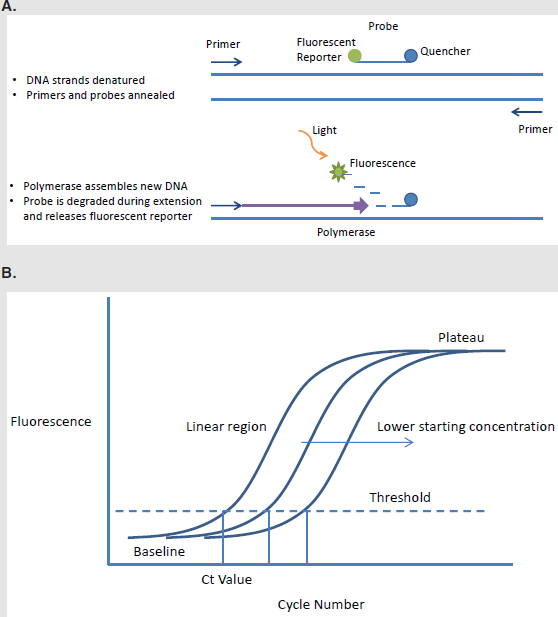
BOX FIGURE. Depiction of a method of realtime PCR that makes use of a fluorescent reporter molecule. A. The fluorescence of the probe is normally quenched. During amplification, the probe is degraded, and emissions from the fluorescent reporter can be detected. B. If the sample contains the signature DNA for amplification, the fluorescent signal will increase with each cycle. A threshold when signal crosses background is set, and the cycle number at which this threshold is reached is reported as the Ct value.
ing its data-gathering and has been highlighted in other reports, the public health system is called upon to do many things with limited resources. Responding to a detection that turns out to be a false positive places considerable drain on jurisdictional resources and could undermine confidence in the system when a true positive occurs. Over the history of the
program, PCR assay positive results that appear not to have been true biothreat agent detections have been reported (Garza 2012; U.S. House of Representatives 2012a), resulting in changes to certain assays and decision algorithms. In the current BioWatch system, a sample yielding a positive result in the screening assay undergoes a secondary assay for the presence of several additional target sequences. A BAR is not normally called until after this secondary step, reducing the likelihood that jurisdictions and federal partners will mount a full-scale response that turns out to be unnecessary.
On the other hand, a false negative result could be catastrophic if a pathogen release is missed. At the collector, factors such as air temperature, relative humidity, and wind may influence how airborne particles are deposited on the filter, all of which can affect the type and amount of material available as a starting point for analysis. Because the filters capture all particulates of a given size and not only genetic material, the mixture of material can be complex and may contain molecules that present challenges for nucleic acid extraction, may cause loss and degradation of the sample, or are capable of inhibiting PCR amplification. Methods used for extracting DNA from the filter and preparing the sample for downstream analysis likewise affect the input material for the PCR assay. In the PCR assay itself, the presence of molecules that inhibit PCR, operational errors, instrument failures, or other errors in PCR amplification and detection could all produce a negative result despite the actual presence of pathogen DNA. Additionally, the concentration of the pathogen on the filter may be below the threshold of detection of the given assay. To help identify errors resulting from instrument failures or contamination, samples are run with sets of positive and negative controls. However, since specific sequences are targeted by the primers and probe, a failure to detect also could be the result of mutations in the pathogen’s targeted areas, loss of targeted sequences (e.g., in assays that target a bacterial plasmid), or the result of a previously unknown or manipulated strain of the pathogen. Addressing these concerns poses a greater challenge and is discussed as part of PCR assay performance validation in Chapter 2.
A point is sometimes raised that the PCR assays used by BioWatch do not provide information on agent viability, and thus positive detection results do not necessarily indicate the presence of an active risk to public safety and health. Viability information is extremely useful in making further decisions on what public health actions may be necessary. On the other hand, the committee noted that detecting the presence of DNA from even nonviable agents on a BioWatch filter is a situation that law enforcement, public health, and others need to be aware of and that should be further investigated. It could, for example, indicate the presence of a failed release. Thus, the committee sees an important role
for the BioWatch assays separate from the issue of additional testing to characterize whether the agents are viable. The committee understands that the BioWatch program may use RNA assays in viability testing as part of additional investigations following a BAR. RNA testing also can be used to detect the presence of DNA-based organisms, including bacteria or parasites (Murphy et al. 2012). The sample collection method would need to be one that preserves the presence of RNA, which is more labile than DNA. The report does not delve further into this strategy.
It is also important to emphasize that BioWatch is designed to detect very rare events, but ones that would be of high consequence should they occur. The fact that any assay will have some associated false positive and false negative rate, combined with the very low probability of a biological agent release, implies that most BARs will result from detections that are not actual releases or attacks. This type of event has been observed over BioWatch’s history and is inherent in the nature of this type of system. As a result, it is critical to be able to analyze a BAR in the context of other sources of information in order to inform decision making.
BioWatch needs to serve public safety and health as a tool for high-visibility decision making. Because there are serious consequences from both false positive and false negative results being passed forward, the program has defined acceptable limits for these rates as part of testing within its quality assurance program (which includes the set of steps from DNA extraction from a filter sample through to the interpretation of a result following PCR amplification and detection; see Appendix A for these limits and a discussion of what they encompass). However, the overall system performance will not be better than the performance of any of the individual components. Because real-time PCR is the core detection method currently used by BioWatch, assay performance characteristics, including false positive and false negative rates, provide valuable information to the program’s managers and users.
BioWatch Actionable Result
The responsibility for declaring a BAR following the positive results of the PCR assays rests with the director of the local laboratory that conducted the sample analysis, or an appropriate designee, and technical consultation with partners in DHS and CDC can occur before making the determination. As described to the committee, a BAR indicates that nucleic acid considered distinctive for a tested organism is present on a collected filter and can provide qualitative indications of the concentration of material along with the location and time of filter pickup. The BioWatch program considers a BAR to be a detection that activates notification protocols and warrants some form of response from the affected
jurisdiction and the federal government. The response usually involves a local conference call to discuss the BAR and begin to obtain information on the local response strategy, and subsequent national conference call between the jurisdiction and federal partners to share information and put the BAR into context. As was presented to the committee, a BAR “does not necessarily mean that a terrorist attack has occurred; a viable biological agent was released; the agent is infectious; [or] there is a risk to the public’s health” (Walter 2014a). Post-detection situational assessment activities (e.g., further environmental sampling, analysis of additional threat-related information, or pathogen characterization) are frequently undertaken to provide data to better understand the circumstances and potential risk to public health to inform decision makers. Figure 1-2 shows the scope and approximate time line of BioWatch operations.
The BioWatch program relies on the expertise and collaboration of the public health community and other stakeholders for analytical laboratory capacity and, more importantly, in determining appropriate responses to a positive assay result. The network of BioWatch partnerships required
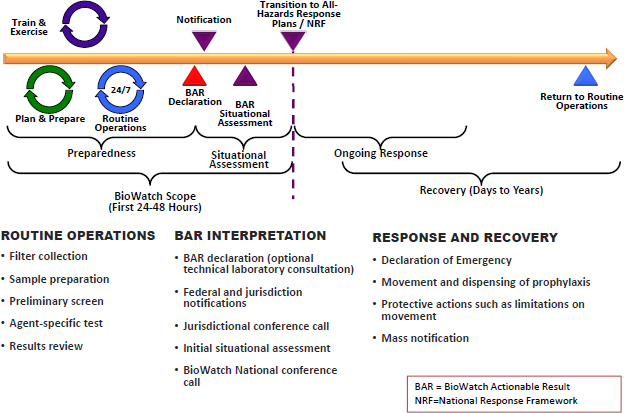
FIGURE 1-2 The BioWatch program maintains routine operations in support of preparedness goals and operates in the early notification and assessment stages of a potential event involving release of an aerosolized pathogen. Another important component of response and recovery is cleanup.
SOURCE: Walter (2014a).
for successful operation includes DHS, state and local officials from public health, law enforcement, emergency management in the BioWatch jurisdictions, and federal partners such as national laboratories, CDC, the Environmental Protection Agency (EPA), the Department of Defense (DOD), and the Federal Bureau of Investigation (FBI). The EPA is involved in additional environmental sampling activities after a BAR and any necessary decontamination, while the Department of Justice and FBI would play significant roles in any investigations of alleged pathogen release and attribution.
PCR ASSAY STANDARDS CONTRIBUTE TO PROGRAM SUCCESS
BioWatch’s mission is to provide timely warning of the presence of a set of targeted pathogens of concern. Success in this mission will be a useful tool for decision makers. It is therefore critical that the BioWatch participants and the user community have trust in the results. An important element of this trust is that the PCR assays have performance characteristics that have been validated and that provide sufficient confidence to program stakeholders. This point is the central topic of this report.
An assay performance standard describes the minimal requirements that must be met for the assay to be considered acceptable and describes how testing to validate this performance is to be carried out. PCR performance standards include items such as the bacterial or viral targets, limits of detection (LOD) for a target, reproducibility of detection, dynamic range of detection, the strains that the assay will detect, including strains or closely related strains it should not detect, and associated confidence limits of the results. A useful BioWatch standard is required to be “executable in a reasonable time, affordable, [and to] maximize confidence in results” (Walter 2014a). A standard or set of standards also needs to be acceptable and transparent to the program’s many stakeholders, including DHS; the BioWatch program; the CDC and public health laboratories; state, local, and federal decision makers; and private companies involved in developing BioWatch technology.
The BioWatch program can leverage existing data and guidance to inform the development, testing, and validation of the PCR assays it uses. Relevant recommendations on the development and validation of PCR assays for biothreat agent detection have been developed by the Stakeholder Panel on Agent Detection Assays (SPADA) to support public safety actions and through the Public Health Actionable Assay (PHAA) approach to support public health decision making, efforts that have both been led by DHS’s Science and Technology Directorate. Approaches used by other environmental monitoring programs also may be relevant to BioWatch. For example, the U.S. Postal Service operates an automated
Biohazard Detection System aerosol monitoring system to detect Bacillus anthracis in mail processing facilities, and the DOD deploys environmental monitoring at selected military locations (e.g., Portal Shield).
None of these assay approaches, however, was developed specifically to support BioWatch’s mission. As a result, BioWatch has used a modification of the assay standards recommended by SPADA to test the performance of BioWatch screening and secondary PCR assays. When the program conducted testing of vendor systems as part of efforts to develop autonomous technology,5 it similarly modified approaches recommended by SPADA to design its testing strategy.
Despite federal multiagency efforts on assay performance and assay performance equivalency, there currently does not appear to be a clear and well-described set of PCR standards, associated validation protocols, and reference materials that are agreed upon and accepted by DHS, the BioWatch program, and the relevant stakeholder partners including the CDC, public health laboratories in the LRN, and those entities involved in developing BioWatch technology.
Given the fundamental nature of the BioWatch program’s reliance on PCR performance characteristics, the significant investments that the United States makes in BioWatch as a defense against large-scale releases of biological pathogens, and the need for the system’s users to interpret and have confidence in its results, DHS asked the National Research Council and the Institute of Medicine to examine the types of performance data, performance criteria, and data analyses that the community requires. A committee appointed by the National Academies was asked to examine PCR performance criteria and validation measures and to evaluate how existing or proposed PCR assay standards meet the program’s requirements in terms of performance, cost, and capacity to provide actionable information to leaders in public health. The committee was asked to provide guidance on standards for validation and verification of PCR reactions such that these standards can provide a reasonable measure of confidence to federal, state, and local public health officials and key stakeholders in the BioWatch program. To accomplish this goal, the committee examined current BioWatch protocols and the process used
___________________
5 A proposed “Generation 3” acquisition of autonomous PCR-based detection systems for BioWatch was cancelled by DHS in 2014. See Appendix A for information on the assay and system testing conducted by BioWatch.
to assess the performance of these protocols, and was asked to provide guidance on whether improvements could be made. DHS also requested that the committee consider, to the extent feasible, the implications for PCR assay performance of combining multiple nucleic acid targets into a single PCR (referred to as multiplex PCR). Box 1-2 provides the committee’s full statement of task.
There are important limitations in the focus of the report that should be recognized by program stakeholders. A comprehensive PCR assay is a process with multiple steps, only one of which is the PCR amplification. Most of the performance characterization and validation strategies
An ad hoc committee will be convened to conduct a study and prepare a report that will evaluate and provide guidance on appropriate standards for the validation and verification of polymerase chain reaction (PCR) tests and assays in order to ensure that adequate performance data are available to public health and other key decision makers with a sufficient confidence level to facilitate the public health response to a BioWatch Actionable Response (BAR).
Specifically the ad hoc committee will:
- Determine PCR assay test and evaluation criteria that will provide a reasonable measure of confidence to federal, state, and local public health officials and key stakeholders.
- Identify and evaluate the Stakeholder Panel on Agent Detection Assays (SPADA), the Public Health Actionable Assays (PHAA), and any other existing and proposed standards applicable for use in defining the performance (validation and verification) of PCR assays for the BioWatch and other programs to ensure confidence as identified in Subtask 1, above. Standards are to be evaluated in terms of performance, cost, and public health applicability.
- Examine current PCR protocols used by the BioWatch program and other relevant biosurveillance programs and determine if the processes used to assess the performance of these protocols and assays are adequate to meet the standards identified in Subtask 2, above.
- Determine whether improvements could be made by adopting changes based on the evaluation in Subtasks 2 and 3, above.
- Determine if any existing standards approach is conducive, taking into consideration cost, schedule, and data requirements, to measuring performance of a PCR assay in multiplexed format.
- In the event that no approach currently in existence is judged to be appropriate in Subtask 5, above, provide recommendations for aspects that a standard must include to measure performance of multiplex PCR technology.
examined by the committee involve dimensions of the PCR (e.g., the limits of detection of the reaction or the performance of the reaction against inclusivity and exclusivity panels). A detection platform is always associated with the reaction in order to detect the results, and therefore assay performance testing specifies which detection platform is used. However, the report does not compare or recommend one type of detection platform versus another. Additional information on the focus and limitations of the report is detailed in Box 1-3.
BOX 1-3
Limitations of This Report
For a BioWatch sample to make it from an airborne particle to an identified and interpreted signal on a computer leading to an action, it must go through numerous steps, only one of which is the PCR reaction. The input to the PCR reaction is critical to the success and ultimately to the interpretation of the PCR result, as is the way that the PCR amplification process is detected. The thermal cycling conditions and reagents used are also important factors in obtaining a successful PCR product. All of these steps and issues can cause false positive and false negative results in a PCRbased test, even if the PCR reaction is optimized.
The report focuses on performance characterization and validation of the PCR. It comments on aspects of the PCR assay that precede the reaction, including the complications posed by environmental background and the effect of sample preparation. However, the report does not make recommendations on
- The utility or effectiveness of the method of sample collection, such as air filters or air collectors;
- Particular sample processing methodology, including specific methods to extract and purify the nucleic acids from the sample, other than to suggest rigorous validation of all processes;
- How to address the effects of materials that can affect a PCR, such as road or brake dust, pollen, dirt, heavy metals, or other substances, or how to address the effects of background biological organisms and nucleic acids that may or may not be amplified in the PCR process, other than to recommend a rigorous examination of environmental background;
- Particular thermal cycling conditions or instrumentation;
- Specific sources of PCR reagents, or specific methods of storing or dispensing reagents;
- Method of detection of the PCR products, or the size or other particularities of the PCR products; and
- The algorithm for interpreting the results from the PCR.
Since its inception, BioWatch has been the subject of a number of reports and congressional hearings that have explored its costs, effectiveness, and roles (e.g., GAO 2009, 2010, 2011, 2012a; IOM and NRC 2011). This report does not address broad programmatic questions on the function of BioWatch and how it fits into the country’s public health and security enterprises. Rather, the report discusses performance characteristics of PCR and explores the implications for the BioWatch program and its stakeholders of different types of assay standards approaches. The report seeks to provide guidance to the program to inform decisions made on standards for the PCR assays it uses.
Chapter 1 introduces BioWatch, its mission, and the motivation behind the current study. Chapter 2 discusses the development and use of assay performance standards and provides background information on federal environmental biosurveillance efforts. Chapter 3 explores several of the core components of real-time PCR performance standards and the committee’s analysis of tradeoffs arising from different types of standards approaches. Chapter 4 returns to the operational context of the program and the experience of stakeholders in making use of assay results, as well as the important role of a quality assurance program as part of validation. Chapter 5 considers potential impacts on BioWatch of the changing technology landscape, including additional assay performance considerations with multiplexed PCR assays and the implications of continued advances in next-generation sequencing technology. Finally, Chapter 6 summarizes the committee’s conclusions and recommendations.
This page intentionally left blank.
















