2
Advancing Patient Care and Research with Genomic Information
Important Points Highlighted by the Individual Speakers
- Integrating high-quality data into the health care system is a priority for ensuring that the best possible information is available for patient care and research. (Peterson, Risch)
- While a variety of genomic data sources exist, they are not readily accessible for use in the current electronic health record (EHR), and work to address the format of these data would create opportunities to use the information more effectively. (Risch)
- Medical information in the EHR coupled with gene sequencing information can be used as a discovery tool for identifying genetic variants associated with disease and for understanding individual response to therapeutics. (Peterson)
- Several ongoing efforts within government and the private sector are aimed at establishing data repositories for large-scale genomic information. These data could be used to demonstrate the power of rapid learning for improving patient care and informing health research. (Etheredge, Peterson)
- Data that are standardized, comparable, and consistent would facilitate the reuse of those data for discovery in multiple contexts beyond the original one. (Chute)
Ensuring the quality of the genomic data that are integrated into the health system is important to making certain that patient care is delivered and research is conducted with the best information available. Programs in the private sector, government, and at universities are currently in place or are being developed to generate genomic data that can be made
accessible to local and global communities so that they can use those data to enhance patient care and impact research progress.
THE TYPES AND QUALITY OF GENOMIC DATA
There are many sources and types of genetic and genomic data, said Neil Risch, the Lamond Family Foundation Distinguished Professor in Human Genetics, the director of the Institute for Human Genetics, and a professor and former chair of the Department of Epidemiology and Biostatistics at the University of California, San Francisco. The sources of data include tests for inborn errors of metabolism (such as newborn screening tests), chromosome studies (such as cytogenetic tests), array comparative genomic hybridization, DNA-based Mendelian disorder testing, and tumor sequencing. However, a major problem is that many of the results of these tests are not easily used in research because they are often represented as PDF files in the medical record. Furthermore, while most genetic tests are reliable, the quality of the results from some of the newer tests is not universally high. For example, a study of whole genome and exome sequencing found an error rate of 0.1 to 0.6 percent, depending on the platform and depth of coverage (Wall et al., 2014). “There still needs to be more work cleaning this up before next generation sequencing is what we would consider to be clinical grade,” Risch said.
If the information derived from genomic studies is to be reusable in multiple contexts, the data need to be standardized, comparable, and consistent, observed Christopher Chute, professor of medical informatics at the Mayo Clinic at the time of the workshop. For example, he said, the genomic data in the Database of Genotypes and Phenotypes (dbGaP) at the National Institutes of Health (NIH) are reasonably reusable, but the phenotypic data still lack comparability and consistency. “If we are going to generalize the research data, we need to do so in a way that we can pool [and] generate meta-analyses, reuse the data intelligently, and move on.” The Mayo Clinic has what Chute termed a “local dbGaP,” or the Mayo Genome Consortia (MayoGC), which pools genomic information from multiple studies across the Mayo Clinic and uses the information along with phenotype data that have been extracted from the electronic health record (EHR) for association studies.
EHRs as a Research Tool
Vanderbilt BioVU,1 a DNA databank and biospecimens repository linked to anonymized medical records, is being used to study the associations between genes and diseases and between genes and patient responses to medications. The resource has undergone considerable growth over the past decade, said Josh Peterson, an assistant professor of biomedical informatics and medicine at the Vanderbilt University School of Medicine. It now contains close to 200,000 samples, about 170,000 of which are adult and the remainder pediatric. About 90,000 have been genotyped with a high-density platform, usually a genome-wide association study or exome chip. Studying biobank data and corresponding phenotype data in EHRs can confirm known genetic associations and therefore be used as a discovery tool in genomics (Ritchie et al., 2010).
However, before BioVU is used as a discovery tool, the method needs to be validated, Peterson said. For example, in a study to predict cardiac events using genetic variants in patients receiving clopidogrel, 260 of 591 phenotyping cases were confirmed as “definite cases,” or patients who were prescribed clopidogrel following a myocardial infarction or percutaneous coronary and who then experienced one or more recurrent cardiac events (Delaney et al., 2012). Once the high-quality data were generated, an analysis of them demonstrated that adverse recurrent coronary events were correlated with CYP2C19 and ABCB1 but not with PON1 in that patients with specific variants of the first two genes were more likely to experience those events than control patients who did not have those variants.
The BioVU resource has also been used to link phenotype data with genomic data, which Peterson referred to as PheWAS data. For example, in an association study of single-nucleotide polymorphisms (SNPs) and EHR-derived phenotypes, IRF4, which was known to be linked with hair and eye color, was newly associated with actinic keratosis, a skin condition that may progress to cancer (Denny et al., 2013).
The Pharmacogenomic Resource for Enhanced Decisions in Care and Treatment program at Vanderbilt focuses on germline pharmacogenomic variants and has genotyped about 14,000 patients. Selected pharmacogenomic data are reported to the EHR so that providers receive clinical decision support that takes into account genomic variants. The quality of the genotyping data is very high, Peterson reported, including
_________________
1Vanderbilt Research, https://victr.vanderbilt.edu/pub/biovu/index.html?sid=194 (accessed March 4, 2015).
nearly 100 percent call rates for actionable variants and 100 percent concordance on repeat samples. This is important, he said, because low-quality data that can be obtained for trouble spots such as the highly polymorphic CYP2D6 loci can also be a problem for rapid learning health systems over time because of low replication accuracy.
The Electronic Medical Records and Genomics Network (Friedman et al., 2015) has also demonstrated that it is possible to use EHRs to do genomic research. Cohorts could be generated across multiple medical centers with shared algorithms in a reproducible and consistent way. The resulting studies have not been perfect, Chute said, “but they are clearly demonstrating that you can consistently and collaboratively leverage disparate and heterogeneous health records in a way that you can use that information for underlying research.” Bielinski et al. (2014) showed that MayoGC can be used successfully as a research tool to study genetic variants associated with bilirubin levels using data from individuals enrolled in three NIH-funded studies at the Mayo Clinic.
An area where BioVU has been particularly helpful for clinical implementation has been in creating warfarin dosing algorithms. Two commonly used algorithms are from the International Warfarin Pharmacogenomics Consortium and WarfarinDosing.org. The current difficulties in using genetic data to determine warfarin dosing may arise from the fact that errors in the algorithms are still too large and need to be reduced through further research, Peterson said. A new algorithm was developed at Vanderbilt in response to disparate results from studies in which warfarin dosing was guided by genetics, he said (Ray, 2013). Adverse events are tracked, but one limitation, Peterson acknowledged, is that re-contacting patients is not an option.
Genomic data are often de-identified prematurely, Chute said, but if the data are to maintain maximum usefulness, linkages need to be maintained between the clinical phenotypic data and the underlying genomic data. “I’m all for privacy and security,” he said, “but the importance of maintaining the consistency and linkage of the clinical information cannot be underestimated.”
The 100,000 Genomes Project
In the United Kingdom the 100,000 Genomes Project2 is intended to establish a genomic program that is transparent to patients, that will sup-
_________________
2The 100,000 Genomes Project, http://www.genomicsengland.co.uk/the-100000-genomes-project (accessed March 4, 2015).
port scientific and medical inquiry, and that will foster the development of an industry in genomics within the United Kingdom. By the end of 2017, the sequencing of 100,000 genomes will be complete, said Tom Fowler, the director of public health at Genomics England.
The genome sequences will be generated from National Health Service patients with rare inherited diseases, cancers, and pathogens. This specific focus was chosen, explained Fowler, because those were the three areas deemed most likely to result in expeditious translation from genomics research to practice. A key feature of the 100,000 Genomes Project is learning to use genomic technology and data in the health care system. For example, there is interest in “deep phenotyping” patients, or providing comprehensive detail about the components of patients’ phenotypes, because this may lead to improved diagnoses for diseases (Robinson, 2012).
The creation by the 100,000 Genomes Project of National Health Service centers for genomic medicine will result in samples and data being provided to the broader collaborative. Created at various institutions around the country, these centers are investing internal resources in this project, Fowler said. They also present an opportunity to move toward a hybrid approach to clinical care and research, in which both clinical care and research happen at once rather than being separate enterprises.
In addition to the genomic medicine centers, the 100,000 Genomes Project has created the Genomics England Clinical Interpretation Partnership (GeCIP), which is a mechanism for bringing the National Health Service and academic communities together to use the data that have been collected in order to analyze and assess how the genome dataset could be interpreted for clinical use. By opening up databases developed by individual researchers, the partnership will be able to take advantage of the capabilities of an entire community, including clinicians, and academic researchers, Fowler said. All generated data are contributed to the Genomics England Dataset and are available to all, with the intellectual property owned by Genomics England but freely licensed. The goal is to greatly accelerate the use of research-based results in health care (see Figure 2-1).
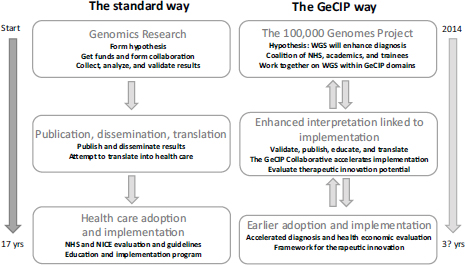
FIGURE 2-1 The Genomics England Clinical Interpretation Partnership (GeCIP) is intended to accelerate the adoption and implementation of research results into health care.
NOTE: GeCIP, Genomics England Clinical Interpretation Partnership; NHS, National Health Service; NICE, National Institute for Health and Care Excellence; WGS, whole genome sequencing.
SOURCE: Fowler, IOM workshop presentation on December 8, 2014.
ADVANCING RESEARCH AND PATIENT CARE
Several U.S. government initiatives are exploring ways to use genetic and genomic data to further research. Etheredge reported that the National Cancer Institute is developing and testing a new master protocol trial system3 in 200 collaborating centers which could become the basis for a much faster trial system for genetically informed research (Ledford, 2013). Genetic profiling is being used initially to determine which of five different treatment modalities will benefit a given patient the most so that the patient can be assigned to the most promising of five parallel treatment arms. The result of such assignments could be reduced cost and faster and smaller trials, since the cohorts can be organized genetically.
_________________
3Lung-MAP launches: First precision medicine trial from National Clinical Trials Network, http://www.cancer.gov/newscenter/newsfromnci/2014/LungMAPlaunch (accessed February 18, 2015).
Patient groups are enthusiastic, because the people in trials can get the best therapies available based on predictive models, and this type of approach could be greatly expanded, Etheredge said.
NIH is also working on a conceptual framework for what it calls The Commons,4 a cloud-based platform in which databases from publicly supported studies are shared among the biomedical research community—grantees, applicants, government agencies, the private sector, and others. Developing such a computing infrastructure would allow for the sharing of existing data in an accessible manner in order to foster the development of new ideas and knowledge by reusing data and avoiding duplication of studies. Grants include funds for curating and archiving databases and “vouchers” to allow researchers to access, analyze, and use the data, Etheredge said. The standards and data developed through the centers of excellence under the Big Data to Knowledge5 initiative would provide information that could be piloted as part of the emerging Commons. Some of the centers of excellence have a specific focus on genomics, and they are working to build an interoperable infrastructure. This would allow clinicians and researchers to share large-scale genomic data and to mine the information with computational engines that would inform research and, eventually, patient care.
There are other opportunities for large-scale data to be used in rapid learning systems. The Centers for Disease Control and Prevention (CDC) is expanding and enriching a genomics-enabled research system for epidemiology and public health science—for example, through its Human Genome Epidemiology Network, the HuGENet initiative. The goal of the program is to “translate genetic research findings into opportunities for preventive medicine and public health.”6 In collaboration with the Harvard Pilgrim Health Care Institute and Children’s Hospital, CDC has also developed a real-time national tracking and rapid learning network for public health emergencies, called EHR Support for Public Health, or ESPnet.7 Using data from initiatives such as these in learning systems could provide insights into how information could be used in preventive medicine and improving public health.
_________________
4The Commons, https://pebourne.wordpress.com/2014/10/07/the-commons/#_ftn2 (accessed March 12, 2015).
5Big Data to Knowledge (BD2K), http://bd2k.nih.gov/#sthash.hw61mFU4.dpbs (accessed March 13, 2015).
6Human Genome Epidemiology Network, http://www.cdc.gov/Genomics/hugenet/default.htm (accessed February 18, 2015).
7ESPnet, http://esphealth.org/ESPnet/images/overview.html (accessed February 18, 2015).
Other Genetic Research Resources
Kaiser Permanente started the Research Program on Genes, Environment, and Health (RPGEH), of which Risch is a lead co-investigator, to “examine the genetic and environmental factors that influence common diseases such as heart disease, cancer, diabetes, high blood pressure, Alzheimer’s disease, asthma, and many others.”8 The program uses Kaiser Permanente’s comprehensive EHR, supplemented with behavioral and demographic data from surveys, information on environmental exposures, and collected biospecimens, to study common diseases. To date, Kaiser Permanente has gathered about 200,000 saliva and blood specimens, along with survey data on demographics, health history, family history, smoking, alcohol use, diet, physical activity, and reproductive history. Although Kaiser Permanente is largely a clinical enterprise, it has invested in research, and the combination ultimately will translate into benefits for patients, Risch said.
RPGEH is intended to advance research by creating a large databank of genetic and other medical information along with lifestyle, demographic and environmental data that will be accessible to the Kaiser Permanente Division of Research and to collaborating scientists from other institutions. The long-term goal is to identify the genetic and environmental basis for common age-related diseases along with factors that influence healthy aging and longevity. The specific aims of the program, Risch said, are to conduct genome-wide genotyping of more than 675,000 markers on 100,000 participants in RPGEH; to assay telomere lengths for the same 100,000 samples; to develop customized genome-wide SNP arrays and use these arrays for genotyping; to merge, with patient consent, the genomic and telomere data with the EHR, survey, and environmental data in a research database; and to provide collaborative access to the data.
The group of subjects participating in RPGEH is 58 percent female and has an average age of about 65, said Risch. It is 78 percent white, 11 percent Latino, 8 percent Asian, and 3.5 percent African American, with more than half the participants having been members of Kaiser Permanente for 20 or more years. Comprehensive electronic records go back to 1995, with the physician notes being accessible from 2006. Information on cardiovascular disease, psychiatric disorders, cancer, diabetes, and other conditions is available for many thousands of people, along with
_________________
8The Research Program on Genes, Environment, and Health, http://www.dor.kaiser.org/external/DORExternal/rpgeh/index.aspx (accessed March 13, 2015).
data from electrocardiograms, magnetic resonance imaging, computerized tomography scans, mammographies, ophthalmologic exams, lipid panels, other serum chemistries, blood pressures, body mass indexes, and other health measures.
The genotyping was completed at the Institute for Human Genetics at the University of California, San Francisco, and it produced very high quality data, Risch said. Genome-wide association studies have led to the identification of more than 600 contributing genetic variants—approximately one-third of which were novel—which are associated with a variety of traits and diseases extracted from EHRs, ranging from blood pressure, cholesterol levels, and QT intervals to prostate cancer and diabetes. Data can be accessed in two ways: through a Web portal at Kaiser Permanente, where a committee reviews applications for the use of datasets by qualified researchers, and through dbGaP.9 In 2014, Kaiser Permanente made a large deposit of data into dbGaP—from 78,000 people who participated in the Genetic Epidemiology Research on Adult Health and Aging project, part of the RPGEH.10 The genetic data are housed in a separate database from the EHR data and are currently available only for research purposes.
Opportunities
Existing programs that generate data and foster research should be examined so that upgrades over the next couple of years could be planned to determine how they could facilitate rapid learning, Etheredge said. At FDA, the opportunities to leverage current programs into rapid learning systems that incorporate genomics could include national registries, standardized data, and coverage with evidence development initiatives, he said. The Sentinel11 system is accruing data on more than 50 million patients annually and has 380 million patient-years in its database. It could be extended into a tracking and registry system for effectiveness as well as safety. Clinical and scientific databases are being made publicly available, and oversight of predictive models could inform
_________________
9Resource for Genetic Epidemiology Research on Adult Health and Aging (GERA), http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000674.v1.p1 (accessed March 4, 2015).
10Kaiser, UCSF dump data from large genomic study into dbGaP, https://www.genomeweb.com/informatics/kaiser-ucsf-dump-data-large-genomic-study-dbgap (accessed March 13, 2015).
11FDA’s Sentinel Initiative, http://www.fda.gov/Safety/FDAsSentinelInitiative/ucm2007250.htm (accessed February 18, 2015).
the public about benefits and risks beyond patient package inserts.
PCORI is expanding its PCORnet capabilities in collaboration with NIH, FDA, and other agencies, Etheredge said. Potential upgrades could include identifying patient-centered research needs for genomics-enabled health care, with national work plans for who is accountable for answers to priority questions and by what time. PCORI also could engage patient groups, professional societies, health plans, hospital groups, accountable care organizations, and others for the collaborative funding of comparative effectiveness research using fast, affordable rapid learning systems. It could develop predictive models for patients and physicians to compare the benefits and risks of various options. The Department of Veterans Affairs also has plans to employ a learning health care approach with veterans who are diagnosed with non-small-cell lung cancer (Ray, 2015). The results from gene sequencing panels will be used to direct therapy, and the information will also be used for research purposes.
The Centers for Medicare & Medicaid Services (CMS) could support a genetics-enabled rapid learning center system for Medicare and Medicaid, Etheredge said. All cancer data in the systems could be collected and reported to a national privacy-protected cloud system, with coverage for genetic sequencing and analysis and predictive services. The CMS Innovation Center12 could test and advance best practices in genomics-enabled cancer care, using pay-for-performance to improve quality. Working with FDA, the center could use coverage with evidence development to support genomics-enabled medicine, such as with new cancer treatments, and it could collaborate with the American Society of Clinical Oncology on a rapid learning cancer system, Etheredge said.
_____________________
12The CMS Innovation Center, http://innovation.cms.gov (accessed February 18, 2015).










