3
Translation of Genomics for Patient Care and Research
Important Points Highlighted by the Individual Speakers
- Patients are very involved in their own health care and are producing their own health-related data, including genomic data. Understanding preferences for data use and communicating effectively in a fair and transparent way with the public about how information is used will be key to engaging the larger population in sharing their data for research. (Baker)
- Patient trust can be earned and maintained through good data practices, including establishing confidentiality policies, data encryption, and multifactor authentication. (Chute)
- Deriving clear standard consent language could reduce the burden on institutions, which today largely develop their own consenting mechanisms, and could provide transparent information for patients about the use of their data. (Baker, Chute, Fowler, Moss)
- A health care system in which an infrastructure supports complete learning cycles that encompass both the analysis of data to produce results and the use of those results to develop changes in clinical practices is a system that will allow for optimal learning. (Friedman)
- Using genomic data could improve population health and contribute to solving many care management problems. Starting with areas that can lead to a return-on-investment may encourage leaders of health systems to engage in these efforts. (Hill)
- Just-in-time information, guidelines for clinical action, and more information on the clinical utility of genetic testing would help physicians make effective use of genomic information and integrate it in their practices similarly to other medical test information. (Vassy)
Efforts are ongoing to facilitate the incorporation of genomics into health systems by understanding patient preferences, educating physicians, and addressing health care disparities. Designing a closed, platform-supported system could help enhance the flow of genomic information through a learning health system, increasing efficiency and bringing added value to patient care and health.
Patients are consumers of health products and services, said Dixie Baker, a senior partner at Martin, Blanck & Associates, and they are also the “primary source of the information and data that we need to create this learning health system.”
Today’s consumers are much more involved in their own care and in the health of their families than consumers in the past, Baker said. In January 2014, for example, nearly one-third of all U.S. smartphone owners (46 million unique people) used fitness and health apps.1 It has been predicted that by 2017, 30 percent of U.S. consumers will be wearing a device to track food consumption, exercise, heart rate, and other critical vital signs.2 More than 500,000 consumers have directly purchased DNA testing services, with no evidence of psychological harm and some evidence of positive behavior changes.3 Consumers willingly contribute their data and biological samples to medical research—when their permission is sought. But if the data are being used for research without permission, dramatic pushback can occur.
As computational power and the amounts of clinical and genomic data continue to grow, medical knowledge and the quality of health care will continue to increase. However, risks to personal privacy will also grow, creating the possibility that consumers will limit the amount and quality of the data that they make available to health care providers and to researchers. A high percentage of consumers are concerned about the
_________________
1Hacking health: How consumers use smartphones and wearable tech to track their health, http://www.nielsen.com/us/en/insights/news/2014/hacking-health-how-consumers-usesmartphones-and-wearable-tech-to-track-their-health.html (accessed March 4, 2015).
2Will an app a day keep the doctor away? The coming health revolution, http://www.forbes.com/sites/ciocentral/2013/09/08/will-an-app-a-day-keep-the-doctor-away-the-coming-healthrevolution (accessed March 4, 2015).
3Regulation: The FDA is overcautious on consumer genomics, http://www.nature.com/news/regulation-the-fda-is-overcautious-on-consumer-genomics-1.14527 (accessed March 4, 2015).
privacy and security of their medical information, Baker said. Many U.S. consumers with chronic conditions want to control their health information, but roughly half believe that they currently have very little control over that information.4 DNA is inherently unique to the individual, rendering it the ideal “biometric identifier”—1 of the 18 data elements of identifiability defined by the Health Insurance Portability and Accountability Act. Even without a name or phenotype linkage, DNA includes many clues for narrowing the identity possibilities—and it can be obtained from objects as ubiquitous as discarded coffee cups. Access to an individual’s DNA also poses a substantial privacy risk for blood relatives, who most likely did not consent to access.
Strong security protection, fair information practices, and “no surprises” will enable the learning health system to emerge, Baker suggested. Consumers generally do not want complex consent forms when they become involved in biomedical research. Rather, they want adherence to principles that have been adopted around the world to gain their trust, including transparency, being asked permission, having access to their own health information, and being provided with knowledge of how their information will be used.
The Platform for Engaging Everyone Responsibly
The nexus of issues associated with patient consent, patient privacy, and data access offers a variety of challenges to integrating genomics into the learning health care system. Patients need to have a way to indicate their preferences concerning their genomic information, said Scott Moss, a research informatics software developer with Epic. These issues are less technical. In the area of consent, for example, “there needs to be some consistent direction,” Moss said, possibly by creating a toolbox of best practices.
A new platform has been developed to address issues of sharing health information in a secure manner that takes into account individual preferences for data sharing and at the same time provides ease in accessibility of the data by the medical and research communities, Baker said. By using consent-management tools developed by Private Access, Genetic Alliance has developed a participant-centric research platform
_________________
4Accenture’s 2014 Patient Engagement Survey, http://www.accenture.com/us-en/Pages/insight-trends-life-sciences-consumer-perceptions-about-emr-benefits.aspx (accessed April 16, 2015).
known as the Platform for Engaging Everyone Responsibly5 (PEER) which enables individuals to make their health information available to researchers. It has three components: a data entry element, a privacy layer, and a data query module. The data entry component is accessed through a customizable consumer portal that can be embedded in any webpage. The entry allows access to de-identified health data and personal contact information. The data query component can be accessed through a researcher portal, which today takes place largely through a tool called RecruitSource (Terry et al., 2013). Searches, alerts, and access requests are handled in this section according to individually defined permissions. In between the data entry and the query components, a privacy layer serves as a filter to create and manage permissions for sharing patient data wherever the data reside and at any level of granularity. Participants establish their own sharing preferences based on a simple “stop light” metaphor: allow, deny, or ask me. PEER enables the expression of the full spectrum of personal views about privacy and sharing of health information, with the ability to adjust settings dynamically as one’s values and priorities change over time, Baker said. Individuals can define who can discover their information, who can download and use their information, and who can contact them directly if the situation makes it necessary to do so.
Genetic Alliance has created a virtual guide that people can use to establish their preferences. People can choose conservative, moderate, or liberal options, or they can go through and select each setting on their own. Among advocacy communities, about 85 percent of individuals release their data for all purposes, about 10 percent say “ask me,” and 5 percent decline to release their data, said Sharon Terry, the president and chief executive officer of Genetic Alliance.
Baker said that the PEER system is designed to increase the number of people involved in clinical trials. Customizable PEER entry points are easily embedded into any website, and a smartphone app provides mobile access. The system also makes it possible to engage people beyond the groups established for families with genetic diseases.
Alignment is a critical issue, not only between researchers and clinicians but among those groups and consumers, Baker said. Consumers are interested and eager to be involved in the overall ecosystem, but they face barriers to becoming involved. “We need to take that challenge on,” she said.
_________________
5Platform for Engaging Everyone Responsibly, http://www.geneticalliance.org/programs/biotrust/peer (accessed February 24, 2015).
Patient Trust, Consent, and Opting In
Establishing and maintaining patient trust is essential to demonstrating that patient information can be used securely and responsibly, Chute said, adding that the medical genomics community should “engage the public and our patients to trust that we are using genomic information respectfully and productively to enhance our understanding and discovery, and then establish an atmosphere of confidentiality,” he said. The way to maintain trust of patients and the community, he continued, is through consistent data practices, including establishing and enforcing confidentiality policies, maintaining encryption, separating identifiers after dataset linkage, prohibiting any clinical data on portable devices, and using multifactor authentication.
A specific barrier to genomic research is patient consent, said Chute and Gail Jarvik, who holds the Arno G. Motulsky Endowed Chair in Medicine and heads the Division of Medical Genetics and the Northwest Institute of Genetic Medicine at the University of Washington School of Medicine. The development of standard consent language is an obvious need, Chute said. Today, every academic research center and test development company generates its own consenting mechanisms. There is no reason why every academic medical center should have to reinvent clear language because this language could be widely shared, he said. In addition, communication among and between clinicians and families can be poor, yet these routes of communication are critical in conveying and understanding genomic information. Only a few percent of patients will have incidental findings from a genomic test, Jarvik said, yet they all need to be consented for incidental findings, and this takes time. Patients also need to be able to opt out of genomic data linkages, Chute added.
Many of the policy issues facing the 100,000 Genomes Project are similar to the policy issues related to the use of genomic information in the United States, Fowler said. Patients are granting consent to link to lifetime health records, with the information being pseudo-anonymized in a research dataset. However, the consent forms are overly complex, Fowler said, and they will need to be simplified as the project progresses. To further protect the privacy of patients, researchers have to use the data within a specific infrastructure.
So far, the greatest limitation on the conduct of research has been the opt-in system for patient involvement, said Risch. When the use of genetic data is considered standard care, and when genomic sequencing is performed as routinely as other clinical tests are (results from which are
broadly available for research), the need for opt-in may change, Risch observed. Aside from limited Mendelian carrier screening and cytogenetic studies, the use of genetic data in clinical research has required participant consent. Another current limitation for research, he said, is that data-analysis-only research proposals are not well received at the National Institutes of Health. However, ethical concerns are likely to still engender discussion, for example, regarding the return of results and their implications, the confidentiality and security of data, and the creation of genetic data for research purposes without current clinical relevance.
One way to address some of privacy concerns is to sort genetic information into categories that have different implications, Risch said. For instance, the information could be considered predictive when an individual already has symptoms and the purpose of genetic testing is to determine a diagnosis. When an individual has a family history of a disease but does not have the disease, testing may be indicated, and the findings could be considered incidental. Kaiser is exploring the use of this type of structure for genetic information and how it might be used in its system, Risch said. One thing that would help the clinical and research enterprises work together would be standards for representing clinical information in translational and discovery research. These standards, Chute said, should be aligned with, if not derived from, clinical standards, which would promote the secondary use of data for research.
Another issue involves not only patient access to results, which is governed by a number of regulations, but the responsibility of laboratories to maintain results over time. Will data be kept for a patient’s lifetime? If so, how will new interpretations of the data be conveyed to patients? “This is an area that needs a lot more exploration,” Peterson said.
PLATFORM-SUPPORTED, COMPLETE LEARNING CYCLES
In a health care system that can learn, every patient’s characteristics and experiences are available for study, best practice knowledge is immediately available to support decisions, and improvement is continuous and routine, said Charles Friedman, the Josiah Macy Jr. Professor at the University of Michigan Medical School. Such a system requires complete learning cycles, each of which consists of two halves, he said (see Figure 3-1). The first half includes the assembly and analysis of data and the interpretation of results once a decision is made to study a problem of
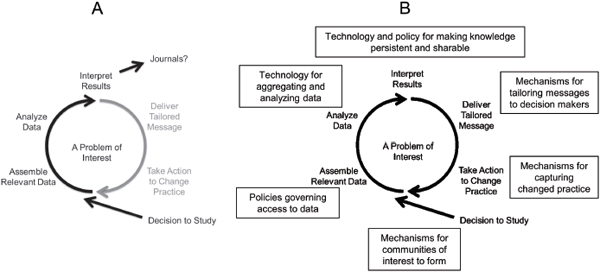
FIGURE 3-1 Components of the learning cycle and supportive platform: (A) an incomplete cycle (black arrows), and (B) a complete cycle with supportive infrastructure.
SOURCE: Friedman, IOM workshop presentation on December 8, 2014.
interest. The second half of the cycle includes the use of results to develop and deliver tailored messages, which in turn lead to action to change practice. This complete learning cycle then drives subsequent iterations of learning (Friedman et al., 2015). Currently, many projects that are establishing the basis for learning systems generally support only the first half of the learning cycle, not the second half.
Today, this learning cycle is often an open loop, Friedman said. Once the first half is completed, results are sent to journals to be published, after which it can take years to translate the results into practice (see Figure 3-1). “There seems to be a consistent belief that if we get the data and analytics side of this right, everything else is going to fall into place,” he said. “But everybody knows this isn’t true. In fact, maybe the harder part of the problem, as challenging as the data and analytics are, is the feedback side, where we’re engaged in needing to change human behavior.” The decision support systems now in place are a primitive and largely ineffectual version of what needs to be done to drive change and improvement through complete learning cycles, Friedman said.
Simultaneous Learning Cycles
To create health care systems that can learn, delivery systems and research networks must run many complete learning cycles simultaneously, Friedman said. This requires an infrastructure that makes learning effective, sustainable, and routine, with the accompanying economies of scale.
Friedman laid out the technology and policy components of a platform for such an infrastructure (see Figure 3-1). This sort of platform would have shareable and interchangeable components, thus distributing costs and making it possible for the platform to be built once and used repeatedly. Some learning cycles may cycle more slowly (e.g., clinical trials), while others would cycle faster (e.g., public health concerns such as disease outbreaks). “Without a platform, each learning cycle will develop its own—probably suboptimal—methods for learning, and there will be no economy of scale,” he said. “The cost of setting up the thirteenth platform will be equal to the cost of setting up the first.”
The development of the full platform could lead to a national or even a global learning system. As Friedman noted, the Internet grew out of a similar situation. A small kernel of standards was developed. Once those common standards were developed, everyone shared those standards while innovation continued to flourish around them. “We need some-
thing for the learning health system that is analogous to that small kernel of common standards to bind everything together, that will allow the kind of innovation at the edges that everyone wants to see,” Friedman said.
Only if knowledge is persistent can it be improved and shared effectively through this platform, Friedman said. It will also be necessary to have mechanisms in place to tailor messages to decision makers and to capture changing practices. “Everyone’s interests here are in alignment,” he said. “Everybody wants the same thing. The challenge is how to make these aligned interests into a set of activities that will bring the benefits that are commonly desired.”
Health Care Disparities
Groups adversely affected by disparities could be among those that benefit most from a learning health care system, observed both Baker and Jason Vassy, a primary care physician and clinician–investigator at Harvard Medical School, the Veterans Affairs Boston Healthcare System, and Brigham and Women’s Hospital. For example, patients who use emergency departments often tend to be from lower socioeconomic statuses, and more data might be in the system for them than for other people. “Depending on what clinical dataset you’re talking about, you might be able to improve some disparities,” Vassy said.
Alexander Ommaya, the senior director of implementation research and policy at the Association of American Medical Colleges (AAMC), brought up the importance of measuring and tracking inequalities. Health care systems can evaluate the impact of their interventions on disparities, he said. “To assume that, just because we provide care and [that] we’re providing it to everyone equally, we’re going to solve this problem is naïve.” Not everyone has access to the health care system. “We need to focus on it and develop specific interventions to address it.” Friedman observed that the problems that get attention are those around which communities of interest form and generate enthusiasm for solutions. If communities of interest form around reducing disparities, then learning cycles could take shape around those issues. “Let’s look at ourselves and decide what’s important,” Friedman said.
IMPROVING HEALTH WITH A KNOWLEDGE-BASED SYSTEM
More than half the drugs used in the United States do not work in the patients for whom they are prescribed, said Colin Hill, the chief executive officer and a co-founder of GNS Healthcare. “This, for me, points to why we need a learning health care system,” he said. “It’s not good enough to drive predictive models and diagnostics with data from electronic medical records and claims.” Genomics-related data will help clinicans understand a patient’s genetic variations and how they affect the response to therapy.
GNS Healthcare is a company that creates analytic solutions for improving population health in a cost-effective manner. These solutions are delivered as cloud-based software to health plans, health care providers, and pharmaceutical companies. The software handles data from EHRs, pharmacy and medical claims, patient registries, and other sources through machine learning platforms in order to determine what treatments will work for individual patients. As Hill put it, his company reverse-engineers causal networks so that patient characteristics can be combined with potential treatments to predict clinical and economic outcomes.
The company has applied this process to medication non-adherence, metabolic syndrome, preterm birth, and a variety of other health problems. For example, in a project with Aetna, GNS Healthcare developed a predictive model to identify which people are most likely to develop metabolic syndrome in the next year, along with the risk factors that contribute most to that development (Steinberg et al., 2014). Health care resources can then be targeted to those individuals where they can make the greatest impact. In addition, with heterogeneous data collected largely for other purposes, it was demonstrated that decreasing an individual’s waist circumference and improving an individual’s blood glucose levels produce the largest benefits for subsequent risk and medical costs for metabolic syndromes.
Genomic data could greatly extend the benefits of this approach, Hill said. For example, he described a project with Inova Translational Medicine Institute that combines sequence data with EHRs and other data types to predict which women are most likely to have a preterm birth. The goal is to identify the underlying causal mechanisms of preterm birth, predict personalized preterm birth risk, and accelerate the discovery of new diagnostic tools and treatments.
Gathering and analyzing more data from various populations could solve many care management problems, Hill predicted. What will ulti-
mately make this approach feasible will be the return on investment that it produces.
INNOVATION WITHIN HEALTH SYSTEMS
Building a learning health care system requires that research, educational, and health systems be coordinated, said Ommaya. But today the linkages among those need a solid infrastructure to support a learning health care system, he said.
As an example of trying to help build this infrastructure, Ommaya described the AAMC’s Research on Care Community (ROCC), which consists of researchers and clinical providers engaged in implementation research to improve the quality, safety, health equity, and outcomes of their patient populations. Since 2012, membership has more than doubled to roughly 250 people representing about 140 institutions.1 According to a survey performed shortly before the workshop, about 90 percent of ROCC members who conduct research are currently engaged in collaborative research projects with clinical colleagues, and about two-thirds are currently engaged in collaborative research projects with teaching faculty, Ommaya said.
ROCC also offers Learning Health System Champion and Pioneer Research Awards2 to encourage collaboration among researchers, educators, and health systems. Champion awards of $5,000 recognize best practices in research, health system, and education collaboration. Pioneer Awards of $10,000 support the implementation of systematic changes to enhance research. As an example, Ommaya discussed the Clinical Research Database at the Loyola University–Chicago Stritch School of Medicine, which is a large-scale, easy-to-use de-identified clinical data structure that provides population health information to everyone with EHR access. There is also an option to make a connection to the institutional review board submission system for access to identified data. Access to the database has alleviated the bottleneck that previously occurred when, prior to implementing the research database, requests were submitted to obtain data from the information technology department. The database features a Web application for casual users, such as faculty
_________________
1Research on Care Community, https://www.aamc.org/initiatives/rocc/about (accessed April 16, 2015).
2AAMC Learning Health System Champion and Pioneer Research Awards, https://www.aamc.org/initiatives/rocc/funding (accessed March 4, 2015).
members and in-house staff, and a number of tools for advanced users, such as analysts and the bioinformatics staff. The system contains data on 2 million patients and on more than 7 million patient encounters and has “really enhanced research activity at Loyola,” Ommaya said.
As another example, Ommaya mentioned the Community Engagement Studio, which provides an opportunity for researchers to recruit participants or “stakeholder experts” who represent the population of interest for a study. It conducts structured sessions that engage patients, consumers, and other non-academic stakeholders appropriate to the study needs including study design, recruitment, dissemination, implementation, and consent. The Community Engagement Studio also helps prepare participants to understand what research is and what their role in a given study will be. The program also offers training for researchers on how to engage with participants successfully.
The Learning Health System Champion and Pioneer Research Awards foster collaborations and the “type of capability that you need for the learning health system,” Ommaya said. This is a model that AAMC is evaluating for building capacity among their member institutions, particularly between the medical schools and the teaching hospitals.
Managing Priorities in a Health System
Health systems are being inundated with new forms of data—not just from genomics, but from gene expression profiles, proteomics, and other high-throughput technologies, said Fred Sanfilippo, the director of the Healthcare Innovation Program at Emory University and the Georgia Institute of Technology and a professor of pathology and laboratory medicine at the Emory University School of Medicine. Furthermore, many other factors besides a person’s genetics affect health outcomes, including behaviors, social circumstances, and environmental exposures.
A number of challenging issues surround the organization and function of academic health centers, such as how to establish priorities and allocate resources. For these institutions to become learning health systems, research, education, and clinical care need to be aligned, Sanfilippo said. Today, that is often not the case. The hospital system, faculty members, and universities typically have different missions, emphases, priorities, and values. This observation applies to other parts of these systems as well, including the financial, planning, communications, information technology, human resources, and community outreach components. As Friedman noted earlier in the session, researchers have generally been
given incentives to publish and not necessarily to establish connections with health systems. The leaders of health systems and the research enterprise are now thinking about ways to generate value from each other, which has created opportunities to bring these communities together.
One possible way to address the organizational challenges is to have a single leader oversee the enterprise, but that does not necessarily result in alignment, Sanfilippo said. A recent study of 84 university health systems found no correlation between the structural organization of the academic health center and its performance, either in the research, clinical, educational, or financial areas (Keroack et al., 2011). However, functional alignment, as measured by 12 different parameters, did correlate with those outcomes, with capital planning, strategic planning, and communication showing the highest correlation (the others were budgeting, financial reporting, program planning, chair hiring and firing, chair evaluation, medical directors, financial transfers, business development, and information systems).
More broadly, the culture of an institution can change outcomes, Sanfilippo said, echoing a point made earlier by Friedman. For example, when The Ohio State University Medical Center moved from a passive culture that lessened success to a constructive culture in which innovation and teamwork are encouraged, the academic, clinical, and financial performance improved dramatically (Sanfilippo et al., 2008). Each institution has a different culture, and changing that culture requires a different process, Sanfilippo said. “But the good news is it probably can be done.”
Promoting Health Services Research
In the Healthcare Innovation Program at Emory University and the Georgia Institute of Technology, it has been a priority to expand health services research and education, Sanfilippo said. Existing programs were scattered across units and institutions, and the resources needed to establish a health services center were limited. The goals of the initiative were to:
- Increase quality, scope, impact, and recognition
- Accelerate interactions across disciplines and units
- Engage other academic and health care organizations
- Develop new activities to accelerate collaboration
- Minimize expense and competition for resources and recognition
- Define measures for assessment and success
For an outlay of about $100,000, the program developed a number of bottom-up projects that were focused on bringing the key individuals together, Sanfilippo said. This included convening collaborations among those in the health system who were involved primarily in quality improvement with investigators in public health, the school of medicine, the school of nursing, the business school, and the school of law. Small seed grants, quarterly symposia, interest groups, research planning, and student–faculty networking nights were among the many steps taken to foster collaboration. Among the results were a significant increase in health services research funding and the engagement of more than 1,700 faculty members and students across more than 50 units, not just from Emory and Georgia Tech, but from other institutions as well.
Sanfilippo drew several lessons from the experience. One should focus on cross-unit and multidisciplinary teams and players, he said, and not duplicate or compete with existing activities. Small investments can yield significant returns, and cost–benefit accounting can justify the use of resources by demonstrating the return on investment and the benefits to patients, students, and staff members. Agreeing on measures of success at the outset is important, as is finding key internal and external advocates.
Alignment is particularly needed among strategic planning (in particular, the quality office), the informatics enterprise, and the research enterprise. “It’s a multibody problem,” Sanfilippo said.
USING GENOMIC DATA IN THE CLINIC
Many physicians report that they feel unprepared for genomic medicine, said Vassy. Among the many reasons that physicians cite, Vassy highlighted in particular a lack of genomic knowledge, low self-efficacy, EHRs that are not equipped to incorporate genomic information, and a lack of evidence for clinical utility.
The MedSeq Project was designed to explore this unpreparedness by engaging clinicians to use whole-genome sequencing results at Brigham and Women’s Hospital. The study involved two groups of 10 primary care providers caring for 100 generally healthy patients in their 40s through 60s and 10 cardiologists overseeing the care of 100 cardiomyopathy patients (see Figure 3-2). “We’re studying both the physicians and the patients in this process,” Vassy said. Patients in the two groups—the 100 generally healthy patients and the 100 cardiomyopathy patients—
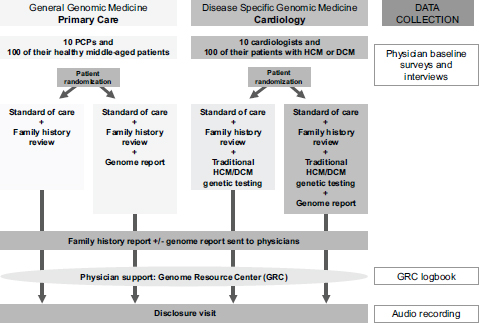
FIGURE 3-2 The MedSeq Project is studying the use of genomic data in the care of 100 healthy patients and 100 patients with hypertrophic cardiomyopathy and dilated cardiomyopathy.
NOTE: DCM, dilated cardiomyopathy; GRC, Genome Resource Center; HCM, hypertrophic cardiomyopathy; PCP, primary care physician.
SOURCE: Vassy, IOM workshop presentation on December 8, 2014.
were randomly assigned to either receive or not receive whole-genome sequencing; family history was reviewed with all patients. The genome reports were sent to the individuals’ physicians for review before they discussed the results with their patients. Physicians had access to a Genome Resource Center, where they could speak with medical geneticists, genetic counselors, and other specialists. The conversations between patients and their physicians were audio recorded.
The reports to the physicians of the genome sequencing (lamentably, still delivered in PDF form, Vassy noted) covered monogenic disease risk, carrier status, pharmacogenomics, and blood groups. Physicians also viewed 12 online educational modules that covered a variety of clinical genomic content, each about 15 minutes long (see Table 3-1). “We try to convey this in a brief setting that respects their time and their competing demands,” Vassy said.
TABLE 3-1 The 12 Online Genomics Educational Modules for Physicians Offered by the MedSeq Project
| Case | Clinical Content Area | Genomic Concepts |
| 1 | Familial hypercholesterolemia |
|
|
||
| 2 | Maturity onset diabetes of the young |
|
|
||
| 3 | Myotonic dystrophy |
|
|
||
| 4 | BRCA-related disease |
|
|
||
| 5 | Alzheimer’s disease |
|
|
||
| 6 | Cystic fibrosis |
|
| 7 | Hypertrophic cardiomyopathy |
|
| 8 | Clopidogrel pharmacogenomics |
|
| 9 | Vascular Ehlers–Danlos syndrome |
|
|
||
| 10 | Age-related macular degeneration |
|
| 11 | Atrial fibrillation |
|
| 12 | Thoracic aortic aneurysm |
|
In discussing the results of the study, Vassy focused on the primary care providers. Among the first 10 healthy patients, 3 had a monogenic
disease risk, with the three variants being classified as pathogenic, as likely pathogenic, and as of unknown significance with pathogenicity favored. Carrier variants were found in all 10 patients, with a mean of 2.2 such variants per person.
Examples of the questions physicians asked before they talked with patients were:
- Are there standard recommendations for counseling patients concerning the significance of their carrier status for their children?
- Would Ehlers–Danlos syndrome come up on the whole-genome sequence screen? There is a history of this in my patient’s family.
- Given that my patient’s directed screening for hypertrophic cardiomyopathy genes was negative, are there standard recommendations on the frequency and means for subsequent genetic reassessments?
According to Vassy, physicians did “pretty reasonable things” with the information they received. For example, one physician who was informed of a likely pathogenic variant for Romano–Ward syndrome, a condition that causes irregular heartbeat, ordered an electrocardiogram for the patient.
Physicians also were largely correct in their interpretations of the reports, Vassy said. They took the information from the report and added it to other information they had, such as the results of a physical or family history, to make decisions. They asked questions that the whole-genome sequence report prompted them to ask. They took information from the report and contextualized it for the individual patient. They understood some of the limitations of sequencing, both on the analytic side and regarding clinical decision making.
Physicians who were asked to be part of the study worried about the amount of time it would take, Vassy said, but the researchers tried to integrate the project into the physicians’ clinical care processes and not provide it as a separate research visit that was carved out of clinical time. Another issue was how to get genomic information back into the health care system. Providers would document it in their notes, but it might not be structured in a way that would be informative for the whole learning health care system.
In essence, the whole-genome sequencing results were “just like any other clinical test in medicine,” Vassy said. The physicians and patients engaged in shared decision making and clinical reasoning, just as they
would with other kinds of information. The concept of genetic exceptionalism did not hold in these exchanges.
What was learned from the study, Vassy said, was that in order to help translate genomic sequencing into improved patient outcomes, clinicians need just-in-time information, including test characteristics and limitations, guidelines or expert recommendations for decision making, and time limits on the validity of the information. “Genomic medicine is a rapidly evolving field, and what is true today may not be true 6 months from now, or 2 years, or certainly 5 years from now,” he said. The major barrier, he added, is the demonstration that genomics will improve clinical outcomes. Demonstrating the clinical utility of specific genomic tests would also help determine whether the benefits of new information will outweigh the risks for a particular patient and whether the information will change decisions. Clinicians are “going to want to know what difference does this make,” he said. “We need to provide that evidence for them before we can expect this to be implemented broadly.”


















