5
Representing Genomic Information in the EHR Ecosystem
Important Points Highlighted by the Individual Speakers
- Transmission of genomic data within the health information ecosystem could be improved by defining data standards that would also foster interoperability and allow for scalability. (Aronson, Nolen)
- The DIGITizE Action Collaborative was established in 2014 to engage key stakeholders for developing a framework for data genomic standards so that this information is more easily integrated into EHR platforms for clinical use. (Aronson, Nolen)
- Setting data standards for genomic information can enable EHR systems—independent of where they are used—and users of these systems to easily understand the data across the medical community. (Nolen)
- Four use cases focused on pharmacogenomics will be the initial starting point for implementing the standards framework designed by the action collaborative. The goal of the effort is to pilot a project and demonstrate how genomic information can effectively flow through a health information technology system. (Aronson)
Connecting health and medical data from genomics to EHRs would be useful to furthering the understanding of disease, increasing the effectiveness of therapies and their safety, and improving health outcomes. However, as several workshop participants pointed out, health information technology systems are not currently represented in a structured, standards-based, and interoperable format. In the effort to use
genomic data successfully in the clinic and for research, defining standards for the data would improve interoperability and scalability by providing a common framework that could be understood across systems. Engaging key stakeholders in a collaborative effort to set these standards could improve the ability to integrate data into the EHR. Incorporating those data in a structured format could increase the knowledge gained about health from genomic information.
In 2014, the DIGITizE Action Collaborative1 was formed as an activity under the auspices of the IOM’s Roundtable on Translating Genomic-Based Research for Health. Its aim is to engage key stakeholders from the community in outlining and facilitating a framework for genomic data standards that could be uniformly implemented across health care systems. The action collaborative has brought together representatives of academic health centers, EHR vendors, government, laboratories, standards bodies, and patients to help facilitate the use of genomics in the clinic. The goal is to represent genetic information in a structured format that is interoperable between platforms. The collaborative’s members will evaluate their framework for genomic standards by implementing a pilot program to test the flow of information within the EHR ecosystem, which includes health centers, laboratories, and EHRs.
Today, individual hospitals are using genomics to improve the care of patients, but these individual efforts generally do not scale, said John David Larkin Nolen, a senior director and the general manager of the Laboratory Business Unit for Cerner Corporation. A learning health care system that works on a small scale may not be efficient or effective in a larger facility if the current state of the system is that it cannot handle the rapid growth of genomic knowledge, Nolen said. An effective system needs to work independently of the venue.
Clinicians typically obtain genomic results from a laboratory, perhaps with some guidance about how to interpret those results, Nolen said, but genomics produces too many data for clinicians to rely entirely on their expertise, which creates a need for decision support tools. Using these tools, physicians can navigate through a patient’s data, know what to do
_________________
1See http://www.iom.edu/Activities/Research/GenomicBasedResearch/InnovationCollaboratives/EHR.aspx (accessed March 19, 2015).
with that information, and decide how to take care of that patient—all while new information from research continually flows into the system.
The challenge, Nolen said, is to figure out what data to store, how to transmit and share the data, how to protect patient privacy, and how to be paid so that the benefits of genomics can be realized in any health care setting. One way to address the problem of transmitting large amounts of data among facilities and systems is to create a common vocabulary for the data. Once a standard vocabulary is established, the content of the information can be determined, and this will allow for improved portability of the data and connectivity among data users no matter where they are located. Many different stakeholder groups need to be involved in the establishment of standards, Nolen said, and they need to be guided by a plan on which the various stakeholders have agreed. The action collaborative provides a forum for bringing stakeholders together to decide on a common framework to move genomic data through the EHR ecosystem.
The goal of the action collaborative is to “accelerate the rate at which we develop clinical genomic and information technology support and deploy it,” said Sandy Aronson, executive director of information technology for Partners HealthCare Personalized Medicine. By continually evaluating the stakeholder groups represented (government agencies, health care providers, laboratories, information system vendors, standards bodies, patient representatives), the group makes certain that the stakeholders have the right expertise to achieve their goals.
The action collaborative members agreed that their starting point would be use cases that would focus their efforts on moving genomic data through the EHR ecosystem. After taking this small step, the group would still have to address significant challenges to “fundamentally improve patient care,” Aronson said, but the defined tasks would be achievable in a short time frame. The group will examine two pharmacogenomics examples (abacavir and Imuran), exploring four specific use cases for each:
- Incorporating genetic results into EHR user interfaces;
- Adding genetic tests in order sets;
- Using clinical decision support to identify when a test should be ordered (pre-test alert); and
- Using clinical decision support to identify when a drug order is inconsistent with a test result (post-test order alert).
The goal of the action collaborative, Aronson said, is not necessarily to publish papers but rather to establish the inter-institutional project management structures needed to deal with the interdependencies in this field (see Figure 5-1). Providers depend on laboratories for data. Laboratories and providers depend on information technology vendors to supply interoperable systems. To meet that requirement, vendors, which are usually competing organizations, depend on each other to establish interfaces among themselves. All stakeholders depend on standards bodies to provide the necessary standards and ontologies to enable interconnectivity to occur. The standards bodies depend on laboratory providers and, to some extent vendors to get the input and models that will make it possible for the clinical decision support to operate robustly.
The funding for generating the genetic data and delivering it through an interface that is interoperable generally flows through providers and laboratories. These entities in turn depend on government agencies to establish a reimbursement framework or to pay for generating and
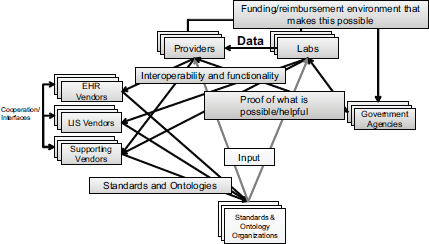
FIGURE 5-1 Interdependencies in the health care system complicate the establishment of inter-institutional project management structures.
NOTE: EHR, electronic health record; LIS, laboratory information system.
SOURCE: Aronson, IOM workshop presentation on December 8, 2014.
delivering the data through grants. Government agencies depend on laboratories and providers to demonstrate that funding will lead to progress. The action collaborative has individuals with expertise in each of these health information system interdependencies, Aronson said.
The group has sought to establish a framework that is detailed enough to enable the different players in this space to make progress efficiently. “Those interdependencies—and the connections between institutions—are what’s most holding us back,” Aronson said. The scope of genomics is huge, he pointed out, so the action collaborative is trying to focus its efforts on particular parts of the problem so as to make incremental progress. A general theme is to make sure that the data needed for clinical decision support transfer from the laboratory to the provider. “Until that transfer happens, nothing can start, so this is where we’re focusing our efforts,” he said.
Focusing on a clinical decision support rule would “add value and [allow us to] gain momentum,” Aronson said. As an example of how the rule would be used in practice, Aronson described the fourth case study, the post-test alert. In this case, a drug has been ordered for a patient on the basis of a genetic test, and the clinical decision support identifies whether that order is inconsistent with a pharmacogenomics result and alerts the clinician if a problem is detected. One rule that could add value, for example, would be to alert clinicians if they prescribe more than 50 mg of Imuran per day to adult patients who are low metabolizers of the drug. Some false positives will occur with chemotherapy situations, Aronson acknowledged, but enhancements could be added over time.
The action collaborative members decided that additional information needed to be added to the order messages that already are sent between laboratories and providers. For example, one piece of information that needs to be included in the order message is the two words at the top of the test report that summarize the information, such as “abacavir sensitivity,” Aronson said, and accompanying those two words would be descriptors such as “high metabolizer” or “low metabolizer.” An ontology needs to be established for which words are acceptable to use, he said. The Clinical Pharmacogenetics Implementation Consortium and ClinGen have taken on the task of establishing the ontology, and work with standards bodies will determine how the information will be coded and transported. Aronson said that those involved in the action collaborative, at “both at an organizational level and an individual level, really care about making sure
that we deliver the promise of genetic medicine to the patients and the families who could benefit from it.”
POTENTIAL NEXT STEPS AND CONSIDERATIONS
The four initial use cases are just a beginning, Nolen said. The action collaborative is starting with pharmacogenomics, but many other issues are waiting. For future projects, the action collaborative will continue to be a coordinating center that keeps projects organized and moving forward. The group will be “the common meeting spot for everyone to come together and help drive this,” Nolen said.
In particular, the vendor community can accelerate the process of integrating genomics into the EHR by delivering support to communities. Once the standards exist for vendors’ systems to talk with each other, Aronson said, many opportunities will open up to disseminate these systems. “The key is to think big but start small,” Nolen said. “While pharmacogenomics might not be that exciting to a lot of people in the room, it’s something that’s within easy grasp even without firm standards. It’s setting up connections, setting up the content, and pushing the data through.”
Nolen pointed out that similar problems exist in countries around the world, even where single-payer health systems make coordination easier. Even if the data are connected in a single country, he asked, “how do you [handle] the deluge of data that’s coming out of that sequencer? How do you move that into your system in a smart way that scales, that allows you to power up the decision process for your clinicians?” Lessons from other countries’ experiences could be valuable in the United States, Aronson said. “There’s no reason why knowledge and transaction shouldn’t be able to cross international boundaries and discoveries,” he said.
It is important to keep the payers engaged in order to get value out of the system, Nolen said. However, in some cases, action needs to occur before the costs of that action will be covered, he said. Reference laboratories, academic hospitals, community hospitals, and other institutions have all been eager to participate in the action collaborative regardless of reimbursement by payers because they realize that there is value in the form of cost savings and in providing better care for their patients. As Aronson said, “The goal of this work should be to make the incorporation of these genetic tests easier and more efficient.”






