Cohort studies, designed to search for links between risk factors and health outcomes, are increasingly collecting genomic data. Thus, it is important to consider designing those studies in ways that maximize the usefulness of the data. Speakers in the first session were asked to explore current gaps and opportunities regarding data collected in large cohort studies and what elements could enable a robust drug discovery toolbox. Speakers noted that genetics studies at the national level in countries such as Iceland and Finland can reveal valuable lessons about the potential of national registry data for research, along with constraints associated with gathering and analyzing large quantities of genotypic and phenotypic data. Many cohort studies are designed as longitudinal studies that collect data from the same group of participants over an extended period of time. Another approach to cohort studies is used by companies that offer direct-to-consumer genetic testing, meaning genetic tests that are marketed directly to the public. Each of these types of cohort studies offers important lessons in cohort study design and implementation. Issues that arise when designing large cohort studies include challenges concerning sharing data, protecting individual privacy, and overcoming analytical and computational barriers.
BIOBANK STUDIES IN FINLAND
Several large cohort studies in northern European countries have demonstrated the potential of genetic bioresources to enhance drug discovery and development. In Finland, a combination of linguistic and geographic isolation has resulted in a population that can trace much of its DNA back to bottleneck events in the 16th century, during which time internal migrations gave rise to isolated regional subpopulations. Over the years, these small subpopulations expanded, but without the genetic influence of outsiders. As Mark Daly, the chief of the Analytic and
Translational Genetics Unit at Massachusetts General Hospital and the co-director of the Program in Medical and Population Genetics at the Broad Institute, pointed out, members of the Finnish population have approximately one-third as many rare variants in their genomes as do the members of most other European populations. In addition, some rare variants in the Finnish population have increased to unusually high frequencies. For example, this population is enriched in loss-of-function variants that lower lipoprotein(a) levels and are strongly cardioprotective, providing, as Daly said, “precise clues to interventions that could be safe and effective.”
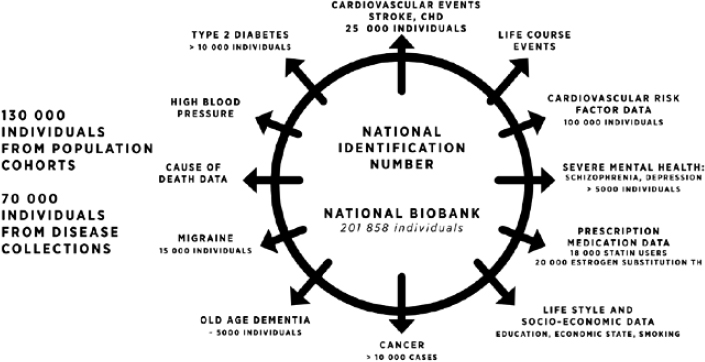
Health data are most useful when they have been collected consistently in a large population over many years so as to track outcomes over long periods of time, Daly said. Finland, like other Nordic countries, has an enormous amount of national registry data, all coordinated with individual identification. This makes it possible to track all hospitalizations, outpatient visits to specialty clinics, and prescription medications that an individual has taken, Daly said. For example, the THL Biobank in Finland1 now has samples from about 130,000 individuals from population cohorts and 70,000 individuals from disease collections (see Figure 2-1). The data in this biobank enable studies on a wide range of human diseases. “If you are interested in a specific phenotype, or a response to a specific medication, that information can not only be fleshed out over the life history of two individuals but over hundreds of thousands,” Daly said. This enables new study designs that are difficult to do in a traditional academic environment, such as searching for adverse outcomes using prescription registry data, including adverse outcomes in people with specific genetic backgrounds, he said.
Another key element to effective cohort studies is extensive population participation and genotype-based recall, Daly said. Once a gene has been determined to play a role in a disease, understanding that gene’s function can require additional studies in genotype-based or phenotype-based populations. Finland’s Biobank Act of 20132 has made genotype-based recall very easy, Daly said, because it enables recall access to all individuals in the biobank, even those whose samples were collected years earlier. For example, Daly and his colleagues have identified loss-of-function
__________________
1 For more information on Finland’s THL Biobank, visit https://www.thl.fi/fi/web/thlfien/topics/information-packages/thl-biobank (accessed May 23, 2016).
2 The Biobank Act of Finland can be found at http://www.findex.fi/fi/laki/.kaannokset/laki/kaannokset/2012/en20120688.pdf (accessed May 23, 2016).
NOTE: CHD = coronary heart disease.
SOURCE: Mark Daly, National Academies of Sciences, Engineering, and Medicine workshop presentation, March 22, 2016.
mutations in the SLC30A8 gene as being strongly protective against type II diabetes (Flannick et al., 2014). These mutations in SLC30A8 are extremely rare, but their existence in Finland and Iceland permitted focused and highly targeted genotype-based recall studies.
Even though the studies required subjects to have a meal and then undergo numerous blood draws over 3 hours, more than 75 percent of the eligible individuals in the biobanks made themselves available for the recall study. The Finnish population tends to participate in recall studies because they have high levels of education, access to their own medical data, and a great deal of trust in their doctors and researchers, Daly said.
Biobank studies should not be viewed in isolation or as competitors with other studies, Daly said. On the contrary, genetic cohort studies point to the value of widespread national and international collaborations because no one cohort or country can generate all the needed insights on a disease. Also, patient communities focused on specific diseases can often provide the sequenced genomes or exomes of more individuals than will be diagnosed with those diseases in many biobank studies. Most primary disease discoveries will continue to emerge from collaborations among investigators, and biobanks will then allow deeper exploration of these findings. “Insights into human genetics generally have
come and will continue to come from the extremes of the population—individuals with severe disease. We need to embrace data from the extremes of the population, and we need to then use biobanks . . . to allow much deeper exploration of those findings,” Daly said.
GENETIC BIORESOURCES AND DISCOVERY EFFORTS IN ICELAND
As in Finland, a great deal is known about the genetic diversity of the population in Iceland. deCODE genetics,3 a company based in Iceland, has been working to collect and curate genotypic and medical data on the Icelandic population, reported Kári Stefánsson, the chief executive officer and founder of deCODE genetics. Approximately half of the Icelandic population, 160,000 individuals, volunteered to have their DNA genotyped using an Illumina chip, he said. The whole genome has been sequenced in 20,000 Icelanders to a median depth of 30× coverage, and imputed variants are known down to a frequency of 0.01 percent in a cohort composed of 390,000 living or deceased Icelanders, Stefánsson said. A list is available of 16,000 Icelanders who are homozygous for a loss of function mutation in at least 1 of 1,800 genes, he added.
In addition, much is known about the phenotypes of the Icelandic cohort participants, including information on diseases, surgical procedures, prescriptions, mental disorders, and weight and height, along with educational attainment, socioeconomic status, and even memberships in associations—all of which can be used in studies in combination with data on genetic variants.
Researchers have been using the data that are available in Iceland in a few different ways. First, they are looking at common genetic variants that can either increase or decrease the risk of specific diseases. Stefánsson and his colleagues at deCODE have associated a large number of complex illnesses, including diabetes, osteoporosis, dementia, and cancer, with common variants in sequences (Stacey et al., 2015; Steinberg et al., 2015; Steinthorsdottir et al., 2014; Styrkarsdottir et al., 2016). These common variants usually have a subtle effect on the risk of disease, but an individual can carry a collection of them whose confluence creates a relatively high genetic risk.
Researchers at deCODE have also been looking at rare variants to
__________________
3 For more information on deCODE genetics, see http://www.decode.com/research (accessed July 8, 2016).
identify genes that can be used as drug targets in a wide variety of conditions and diseases. This research can readily link diversity in sequence to diversity in phenotype, Stefánsson said. In addition, the genetic data from the Icelandic population have facilitated research on the potential side effects of therapeutic manipulation of the protein encoded by a particular genetic target. For example, antibodies that block the activity of PCSK9 cause an increase in the recycling of the low-density lipoprotein receptor and thus a reduction of low-density lipoprotein cholesterol levels in the blood (Blom et al., 2016). However, questions have arisen about whether reducing the activity of PCSK9 through antibody treatment might increase the risk of dementia or diabetes. By examining a loss of function PCSK9 variant (p.R46L), which is present in the Icelandic population, researchers found no evidence of such risks and a slightly longer lifespan, Stefánsson said. Additional work using the Icelandic biobank samples has uncovered another variant that has a greater protective effect for coronary artery disease, he added (Nioi et al., 2016).
Using Biobank Data for Alzheimer’s Disease Research
Another example of how genetic biobank data can aid in the drug discovery process involves research on Alzheimer’s disease, Stefánsson said. In August 2012, deCODE researchers and their colleagues announced the discovery of a coding mutation (p.A673T) in the amyloid protein precursor (APP) gene that confers protection against Alzheimer’s disease and cognitive decline (Jonsson et al., 2012). Furthermore, when the APP mutation was examined in a cohort of nursing home residents in Iceland who had not been diagnosed with Alzheimer’s disease, the variant was associated with a slowing of normal cognitive decline. Using a similar process, the researchers discovered a variant in another gene, TREM2, that results in a predisposition to Alzheimer’s disease. TREM2 could be a particularly useful target in Alzheimer’s disease (Jonsson et al., 2013).
Data from genetic bioresources also can help answer the question of when to begin treating a chronic disease such as Alzheimer’s, Stefánsson said. One approach is to group individuals based on carrier status in a particular gene, such as apolipoprotein E-4 (APOE-4) and look for signs of early cognitive decline as an indicator for when to begin treating. The use of genetic markers for patient stratification in clinical trials could increase the likelihood of successful drug development, Stefánsson said.
IMPROVING GENOMICS AND COHORT STUDIES
As part of their presentations and during the panel discussion following Session I, Baras and Stefánsson identified several potential improvements that could strengthen genomics as a research tool for drug discovery:
- Better annotation of intergenic sequences could provide a greater understanding of the mechanisms of disease caused by common variants. (Stefánsson)
- Improvements in technology could allow for longer reads on whole genomes. Curating longer-read data from large populations would help detect rare variants and replicate novel gene discoveries. (Baras, Stefánsson)
- Understand the basic underlying biology of human diseases. (Stefánsson)
- Better capabilities for biological and clinical target validation. (Baras)
- Novel computational methods for handling challenges with large datasets and rare variants. (Baras)
LONGITUDINAL STUDIES AT THE COMMUNITY HEALTH LEVEL
Longitudinal health data from patient populations, in combination with genetic data, are another source of valuable information. For example, the Inova Translational Medicine Institute (ITMI), in Falls Church, Virginia, has generated roughly 8,000 whole-genome sequences with matched expression, methylation, microRNA, clinical, and survey data from families that come to the Inova health center for care, said Joe Vockley, the chief operating officer, chief scientific officer, and senior vice president at the institute. Many of ITMI’s studies focus on issues in neonatal and pediatric care, and anyone who delivers a child within the Inova health system has the opportunity to enroll in a longitudinal study, he said.
ITMI’s study designs integrate physicians, genomics researchers, and bioinformatics scientists, and ITMI has standardized collection practices to maximize the usefulness of the data. The data gathered include clinical, family history, and survey information (including nutritional, behav-
ioral, and environmental data). Every sample at ITMI that is collected for the biobank is done so using standard operating procedures to ensure quality and consistency. For example, warm and cold ischemic time is minimized, meaning that biospecimens are cooled quickly after collection for long-term storage. Once samples are collected and cooled, they undergo barcoding and are stored at a constant temperature. The institute also has built a robust infrastructure for data analysis, using both an on-site supercomputer and cloud-based storage, with computing and other resources provided to researchers for accurate and integrated analysis of datasets.
In the past, a challenge for longitudinal studies has been the failure to adhere to high standards for data generation, Vockley said. For example, to save time, laboratories often generate data from collected samples using commercially available kits, but companies frequently update or modify their kits, and researchers do not always properly document or disclose these modifications. These modifications to research tools can significantly change the data, making them difficult to integrate and interpret, Vockley said. Another related challenge is the high number of potential biomarkers of human disease discovered in longitudinal studies and cited in peer-reviewed literature; Vockley noted only a small proportion of those are validated biomarkers. Therefore, researchers need to be aware that findings related to biomarkers in published studies can sometimes be incorrect, he said.
The community health care system in the United States is an excellent place to launch longitudinal cohorts, Vockley said, because it sees a great deal of patients. In 2014, approximately 23 million people were seen at community health centers supported by the Health Resources and Services Administration (HRSA, 2014). Tissues from large numbers of patients can be easily collected, biobanked, and made available for drug discovery research, he observed.
GENETICS-GUIDED DRUG DEVELOPMENT AT REGENERON
Studying human genetic data collected from longitudinal studies is one approach that Regeneron Pharmaceuticals is taking to accelerate the discovery and development of new drugs, said Aris Baras, the vice president and co-head of the Regeneron Genetics Center. Regeneron is incorporating human genetics into its research and development paradigm using a three-pronged approach:
- Target discovery: Identify new drug targets and pathways
- Indication discovery: Identify new indications for drug targets and programs
- Biomarkers: Use pharmacogenetics to predict an individual’s response to drugs
A major bottleneck that is slowing the development of new drugs is identifying well-validated drug targets, Baras said. The pharmaceutical industry as a whole is looking at approximately 500 to 1,000 genes (accounting for less than 5 percent of all genes) as targets for medicines, he said, and Regeneron is currently pursuing 200 different targets, with support from a variety of sectors, including human genetics, mouse genetics, molecular biology, biochemistry, and clinical pharmacology. The company also uses principles from human genetics to identify links that can help expand drug indications, and it uses pharmacogenetics to identify patients who may be more responsive or who may have a predictable adverse effect to a particular medicine.
Regeneron’s Collaboration with Geisinger
To support its research goals, Regeneron has taken an integrated approach to drug discovery, relying on general population studies, family-based studies, research on founder populations, and studies of phenotype-specific cohorts. One example of this approach is the partnership that Regeneron has developed with the Geisinger Health System in Pennsylvania. The goal of the partnership with Geisinger is to link genetic mutations with real-world outcomes in order to make actionable discoveries. Geisinger is a community-based health system that serves more than 2.5 million patients and that has used the same electronic health record (EHR) system for more than 20 years. Therefore, it has the world’s largest longitudinal population in which iterative callback phenotyping and sample collection have been operationalized, Baras said.
The DiscovEHR Study, a collaboration between Regeneron and Geisinger, is a large population-based project whose goal is to sequence the DNA of more than 250,000 participants. To date, Regeneron and Geisinger have developed a large number of high-quality, EHR-derived phenotypes for genetic analyses, Baras said, as well as a growing library of more than 2,000 quantitative and qualitative traits that are available
for high-throughput and in-depth analyses. Examples of traits for which in-depth datasets are available include coronary artery disease and lipid metabolism, chronic obstructive pulmonary disease and asthma, and bariatric traits and liver histology. Within the subjects sequenced to date, carriers of heterozygous loss-of-function variants have been found in more than 92 percent of genes, and the study has already found more than 1,300 genes with homozygous loss-of-function indications, including many Regeneron drug targets, Baras said. Increasing the number of sequenced individuals and expanding to studies in founder populations will boost the ability to understand the consequences of these mutations across many diseases, he added.
Using Genetics to Guide Drug Discovery
Through general population cohort studies, including the DiscovEHR Study, it was found that loss-of-function mutations in the ANGPTL3 and ANGPTL4 genes result in lower levels of triglycerides and a lower risk of coronary artery disease than is found in noncarriers (Dewey et al., 2016; Helgadottir et al., 2016). Both ANGPTL3 and ANGPTL4 are endogenous inhibitors of lipoprotein lipase, which mediates triglyceride metabolism and triglyceride-rich lipoproteins (Ono et al., 2003; Sukonina et al., 2006). Based on these findings, Regeneron developed an antibody to ANGPTL3 (evinacumab) which is now in late-stage development for dyslipidemias. The ANGPTL3 and ANGPTL4 variants also appear to have a protective effect for type II diabetes, which Regeneron researchers are now pursuing.
Researchers at Regeneron who are interested in pulmonary arterial hypertension, a cardiometabolic disorder, rely on family-based, Mendelian approaches that involve genetic data gathered from large pedigrees. Pulmonary arterial hypertension is characterized by high blood pressure in the pulmonary artery, which carries blood from the heart to the lungs; to date only 10 genes are known to be associated with pulmonary arterial hypertension (NLM, 2016). In looking at about 70 families and 190 singletons with familial pulmonary arterial hypertension enriched for pediatric onset, researchers from Columbia University Medical Center discovered rare deleterious variants in the TBX4 gene, Baras said. The hope is that Regeneron researchers can take genetic insights like those and convert them to drug discovery opportunities and therapeutics for patients, he said.
As a final example, Baras shared information about Regeneron’s efforts to study fibrodysplasia ossificans progressiva (FOP), a rare disorder that results in heterotopic ossification leading to severe disability and
early mortality. Most FOP patients have a mutation in a bone morphogenic protein type I receptor gene, ACVR1R206H, which promotes heterotopic ossification. While examining all of the ligands for this receptor, researchers at Regeneron discovered that Activin A induces ossification in mice that carry the ACVR1R206H mutation. Based on this finding, a neutralizing antibody to Activin A is now in clinical development.
In summarizing Regeneron’s work, Baras laid out several guiding principles for genetics-guided drug discovery:
- Leverage multiple approaches to maximize opportunities for gene discovery, including research in large general populations, Mendelian genetics and families, founder populations, and disease cohorts with deep phenotyping.
- Gather deep phenotypic data, including longitudinal data and quantitative traits.
- Emphasize large-effect coding variations.
- Take advantage of multi-ethnic cohorts.
- Use analytical strategies to interrogate wide-ranging biological questions.
Human genetics has proven itself to be a valuable way of optimizing and improving drug development, Baras concluded. “You cannot pursue modern drug discovery and development without incorporating human genetics into your R&D approaches,” he said.
DRUG DISCOVERY THROUGH DIRECT-TO-CONSUMER GENETICS
Another source of genomic and phenotypic data is personal genetics companies that offer direct-to-consumer genetic tests. For example, when consumers send a sample of their saliva to the genetic testing company 23andMe, they have the option of giving the company permission to use their data for research purposes. More than 80 percent of people do agree to have their data used for these purposes, said Richard Scheller, the chief scientific officer and head of therapeutics at 23andMe. “We are receiving over 2 million data points per week, and people have answered over 345 million survey questions,” he said.
23andMe currently genotypes about 600,000 SNPs (single-nucleotide polymorphisms) per sample and from that they are able to use Minimac, an
imputation software program, to expand the data to 15 million SNPs. The company has genotyped more than 1.2 million people, 77 percent of whom have been Euro-American, 10 percent Latino, 5 percent African-American, 4 percent East Asian, 2 percent South Asian, and 2 percent other. About 52 percent of customers are female, with a fairly even distribution of ages. The company uses a series of question to gather a wide variety of phenotypic data (see Box 2-1).
The company then provides a report to its customers with 35 carrier status reports. (This number is down from more than 200, Scheller said, as the company responded to concerns expressed by the Food and Drug Administration about the accuracy and comprehension of the health information being provided to consumers.4) Other reports cover ancestry, wellness, and other traits.
__________________
4 The warning letter from the U.S. Food and Drug Administration to 23andMe, dated November 22, 2013, is available at http://www.fda.gov/ICECI/EnforcementActions/WarningLetters/2013/ucm376296.htm (accessed October 14, 2016), and the close-out letter, dated March 25, 2014, is available at http://www.fda.gov/ICECI/EnforcementActions/WarningLetters/ucm391016.htm (accessed October 14, 2016).
23andMe is trying to encourage people to become very engaged in their own genetics, Scheller said, and the goal is for non-experts to learn about their traits and how their genetics affects their health.
Self-Reported Health Information as a Discovery Tool
23andMe also uses data from consenting customers to do genome-wide and phenome-wide association studies, with the former focused on diseases of interest and the latter focused on SNPs or genes of interest. Because customers report their own health information through a series of questions, Scheller said, 23andMe wanted to carefully examine and validate the data and ensure that this approach was scientifically sound. One way that the company did this was by looking at a variant in the CYP1A2 gene, whose product is responsible for more than 95 percent of the primary metabolism of caffeine (Kalow and Tang, 1993). When 23andMe researchers examined the participants who indicated that drinking a caffeinated beverage in the evening makes sleeping difficult, they found an enrichment of CYP1A2 variants, indicating that in that instance the self-reported phenotype information corresponded very well to genetic data. They went on to find other genetic variants that were protective against getting jittery from drinking caffeine. “By asking [our participants] simple questions, we are able to see pathways that control how caffeine is metabolized,” Scheller said.
Recently, researchers at 23andMe published their analysis of genetic associations of self-reported “morningness,” or the preference to rise and to rest early (Hu et al., 2016). In analyzing data from a cohort of nearly 90,000 23andMe participants, the authors identified 15 loci that were associated with morningness, seven of which were located near genes involved in circadian rhythms. As further confirmation that self-reported information is useful, a survey question about psoriasis validated most of the well-known genetic associations with the condition and found several new associations that were not significant in other genome-wide association studies, Scheller said.
Self-reported medical data have enabled the efficient replication of more than 180 genetic associations (Tung et al., 2011), which, Scheller said, “is reassuring because I came to 23andMe to try to use this data to find drug targets.” The database now contains large cohorts of people with specific conditions, including Parkinson’s disease, cancer, depression, asthma, cardiovascular disease, and colorectal cancer, which are now the subject of genome-wide and phenome-wide association studies.
23andMe also has the ability to contact subpopulations to ask if they will participate in a study. “We have a target that we believe might be interesting in non-allergic asthma, but we needed to understand the natural history of the disease better before we could design a clinical trial,” Scheller said. “So we sent out an email to 80,000 of our customers [with] asthma, and in 2 weeks we set up a survey where 8,000 people will answer questions once a month for 6 months.”
PARTICIPATION, DATA SHARING, AND PRIVACY
A major consideration in collecting information from large cohorts is assuring the participants that their data will remain private. In single studies and collaborative studies where data are shared, patients need to know that their information will not be mishandled, Stefánsson said.
Once data privacy is assured, an argument can be made that data sharing among cohort members and among researchers is important. The sharing of information has led to many discoveries and new treatments, and patients should be willing to continue to share data so that the next generation can have access to improved health care, Stefánsson explained. Reflecting this perspective, Daly speculated about shifting to an opt-out rather than an opt-in model for patient participation in biobanks and research, where the default would be sharing. He also wondered if the academic model of publishing is slowing the dissemination and use of results.
A question was raised about participant compensation for participation in genetics-based cohort studies. “It is considered to be a violation of elementary fundamental principles of bioethics to pay people large sums of money for participation in research, and I think it is really inadvisable to propose to do that,” Stefánsson said. Baras observed that some people are very willing to participate in follow-on research, though not everyone is.
Researchers, too, can be seen as having an obligation to share data among themselves, in part because it increases the rate at which new drugs can be delivered to patients, said Lon Cardon, the senior vice president of alternative discovery and development and head of target sciences at GlaxoSmithKline. “Those groups that learned to share earliest got their findings first,” he said. There has been extensive data sharing in psychiatric genetics, though the topic has turned out to be extraordinarily difficult, Stefánsson said.
Several workshop participants commented that sharing is common in
many areas of genomic research. Regeneron, like all companies, must work within the confines of local laws and regulations, but sharing data with collaborators happens all the time, Baras said. Stefánsson pointed out that companies like Regeneron and Amgen are collaborating on genetics research, while successfully competing in drug development. “It’s not just a willingness to collaborate,” he said. “People are doing it.”
Nordic countries do have some restrictions on the use of genetic data, Stefánsson emphasized. For example, laws in Iceland prohibit exporting genetic data or giving people free access to the data that have been gathered. But researchers nevertheless collaborate extensively and productively using the Icelandic data. “Anyone who comes to Iceland can have access to this data and work with [investigators] there,” he said. Performing genomics research studies in the United States can be easier than doing so elsewhere, Baras said, because privacy laws and informed consent procedures are well established and constantly being refined. In certain circumstances, health systems in the United States share their data and health records, including with private companies, Baras said. By contrast, other countries may have laws about data leaving the country that require special procedures.
However, data sharing among investigators can heighten concerns among cohort participants about privacy, Stefánsson said. “It is a formidable challenge to convince people to participate at a high rate at the same time as you are making the primary data available to a very large number of people,” he said. “You have to find some sort of a middle ground, [such as] sharing summary-level data.” To some extent, people have a tendency to overestimate the dangers posed by sharing genetic data. Sharing stories widely about the success of genomics-driven drug discovery may take some of the fear out of it, Baras said.
Technological solutions may reduce concerns about the risk of data sharing. People are not going to accept putting all of their genomic and medical data on the Internet, Daly said, but novel informatics and computational solutions could generate results while addressing privacy concerns. It would be challenging to enact a law like Finland’s Biobank Law5 here in the United States, Daly said. “It would take quite a shift in understanding,” he said. Educating the public on the potential benefits of genomics-driven drug discovery is critical for combating the distrust, Scheller said. The 23andMe experience has demonstrated that many people are willing to have their data used for research and to become en-
__________________
5 The Biobank Act of Finland can be found at http://www.finlex.fi/laki/kaannokset/2012/en20120688.pdf (accessed July 12, 2016).
gaged with genetic information. “If they feel as though they are part of the process, they will be engaged and will let you use their data,” Scheller said.
Another important issue with sharing of genotypic and phenotypic data is the quality of the data. Several international disease-focused consortia have addressed data quality challenges by agreeing on effective diagnostic and general criteria, Daly said, and those criteria have led to harmonious research tools that can be used across populations in the same disease state. Data quality issues are not insurmountable, Stefánsson said; even self-reported data from 23andMe is of high quality and has led to a lot of discoveries.
















