Multiple business models, including models in the precompetitive space, can support collaborations that use bioresources for discovery purposes. These models generally include multiple stakeholder groups across sectors, because each sector has strengths and resources that it can bring to a collaboration. Any given collaborative effort may be based in the private sector, the public sector, or the nonprofit sector, which includes patient advocacy groups. But in each case, as demonstrated by the examples discussed at the workshop, the goal of individuals in an effective collaborative model is to be part of and draw on broader systems of expertise and capabilities from a variety of stakeholder perspectives. Speakers in the third session were asked to examine potential precompetitive business models and investments that can support discovery efforts and opportunities across stakeholder groups.
ENGAGING PATIENTS FOR DRUG DISCOVERY
Pancreatic cancer is the only major type of cancer with a 5-year survival rate in the single digits and is a health threat in the United States and around the world, said Lynn Matrisian, the vice president of science and medical affairs at the Pancreatic Cancer Action Network (PanCan). In 2010, the network adopted a goal to double the survival rate for pancreatic cancer by 2020. PanCan has adopted a comprehensive approach to advancing research, which involves a central government affairs office in Washington, DC, affiliate networks in more than 60 U.S. cities, a research program to support translational and clinical research, and a patient services group. The patient services group, called Patient Central, operates a call center that receives more than 12,000 requests for information per year from patients and their families. Representatives at Patient Central can link callers to pancreatic cancer specialists in their area, give them information about educational events and webinars on pancreatic cancer, and help them find clinical trials in which they can enroll. Through this effort, the network aims to improve patient access to data and participation in clinical trial research.
Recognizing that genomic information can accelerate progress, PanCan has created a program called Know Your Tumor that over 2 years has enrolled more than 500 patients from across the United States. Patients are recruited not just from academic institutions but also from community settings, Matrisian said. Biopsies taken at the point of care are sent to a central location, where tissues are then allocated to different
molecular diagnostic companies. The results for the first group of patients have included information on a 343-gene panel designed for all solid tumors and an analysis of 23 separate proteins related to pancreatic cancer. This work has resulted in the identification of a diverse set of genes thought to be implicated in pancreatic cancer that can be used as a starting point for drug development programs (see Figure 4-1). The work can also be used to guide the selection of appropriate targeted treatments for an individual patient.
Different subtypes of cancer have distinct targets, treatment indications, biomarkers, therapies, and patient subpopulations, Matrisian said. However, she added, the approach taken by PanCan “leverages what we know in [other cancers] and takes advantages of the distinctions.” This approach would not be possible without large-scale genomic analysis and targeted therapies, she said.
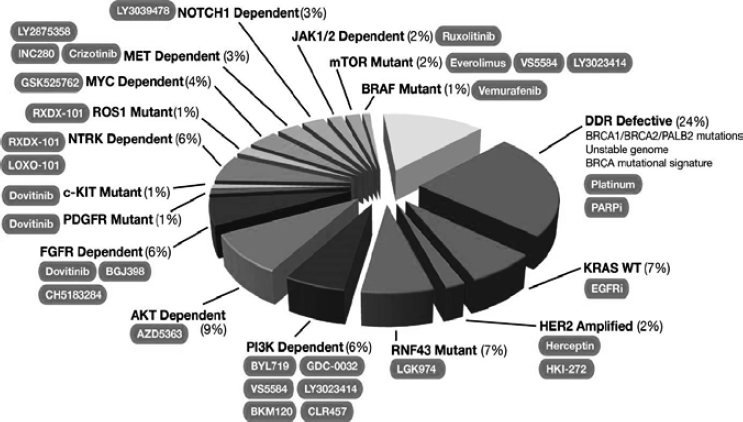
NOTE: Figure was developed by David Chang, Peter Bailey, and Andrew Biankin, University of Glasgow, and is based on data from 457 patients in the International Cancer Genome Consortium (ICGC PACA-AU) cohort. Percentages correspond to the percent of total pancreatic cancers with mutations and/or copy number alterations in the indicated gene or pathway, and shaded ovals indicate possible therapies based on the molecular aberrations.
SOURCE: Lynn Matrisian, National Academies of Sciences, Engineering, and Medicine workshop presentation, March 22, 2016.
PanCan also maintains a comprehensive clinical trial database on all pancreatic cancer trials that are currently open in the United States, Matrisian said.1 By using the Clinical Trial Finder, patients get personalized information on trials that they may be eligible for. It is estimated that in 2014 only 4.2 percent of newly diagnosed pancreatic cancer patients entered a clinical trial, Matrisian said. However, among the subset of patients who contacted PanCan for information, 15.5 percent entered clinical trials. In addition to its role in providing information for patients, the organization’s database can be used by health care professionals to easily survey the landscape of clinical trials in order to identify gaps and plan new trials. Trial sponsors can also receive information on how many times information about their trial is given out.
PanCan has contact with more pancreatic cancer patients than any other organization and is now collaborating with Genetic Alliance on its Platform for Engaging Everyone Responsibly (PEER) registry system. On the PEER registry, patients can record their experiences and outcomes, and can manage via the PEER portal their preferred privacy settings and access to data they have provided, Matrisian said. Depending on the desires of the patients, certain data are made available to researchers. This system allows researchers to access another layer of information that supplements clinical records and thus creates opportunities for novel discoveries, Matrisian said.
COLLABORATIVE APPROACHES TO GENOMIC DRUG DISCOVERY
Utilizing EHR-Linked Biobanks for Drug Discovery
Previous efforts to assess the impact of genetic variation on clinical phenotypes have relied on population studies performed through genetic analysis consortiums, said Meg Ehm, the director of external strategic alliances for genetics at GlaxoSmithKline (GSK). However, the phenotypes used in these studies have often been reduced to a “common denominator,” she said, and the studies rarely have produced longitudinal information. Only a few of these collections have included information on drug usage or drug response, and many conditions have not yet been studied, Ehm said.
__________________
1 For more information on the Pancreatic Cancer Action Network’s clinical trial database, see https://clinicaltrials.pancan.org (accessed June 8, 2016).
To address these issues, GSK is focusing its efforts on building a new entity, the Genomic Resources for Drug Discovery Consortium, which will utilize electronic health record (EHR)-linked biobanks more effectively for drug discovery purposes. The new consortium is designed to support target identification, clinical development, patient stratification, and pharmacogenetics. The overarching goal of the consortium is to realize the value in integrated medical and genomic resources in order to identify and prioritize targets and to better understand drug response, Ehm said. The consortium has five key features: comprehensive and diverse health data, comprehensive genetic data, access to biological samples, informatics, and the ability to recontact patients.
The consortium was designed with several key tenets in mind, according to Ehm. The first is the idea that it is important to develop a research-enabled environment that facilitates the precompetitive space, because accelerating drug discovery will require more resources than any one company can provide. The second is that engaging users early in the design and development of genomic and EHR-linked bioresources is critical if one is to leverage unique insights into drug discovery and development questions. Finally, consortium developers recognize that the harmonization of multiple resources, such as data collected from many types of health settings and from participants of all ages and ethnicities, will be needed to realize the full value of the partnership.
As an example of how the consortium could help research to progress more efficiently and effectively, Ehm cited work on the identification of a rare missense variant in the glucagon-like peptide 1 receptor (GLP1R) gene. GLP1R agonists, which are used in the treatment of type 2 diabetes, act as mimics of the incretin hormone GLP-1 and increase insulin levels in response to orally-consumed glucose (Scott et al., 2016). The rare missense variant that was discovered in GLP1R was found to mimic the effects of GLP1R agonists, so researchers combined genetic and phenotypic data using in silico approaches to test the association of the variant with disease outcomes. This analysis revealed that the variant was associated not only with a reduced risk of type 2 diabetes, but also with a reduced risk of coronary heart disease, thus providing supportive evidence that GLP1R agonists are not likely to be associated with an unacceptable increase in cardiovascular risk.
The Genomic Resources for Drug Discovery Consortium grew out of enthusiasm about associating genetic variants with clinical outcomes, but participating companies also intend to work toward developing evidence that links potential drug targets to disease progression, predicting out-
comes of target modulation, and using systems biology and systems pharmacology to facilitate these goals.
A Public–Private Partnership to Accelerate Genomic Medicines
The Foundation for the National Institutes of Health (FNIH) is a nongovernmental organization established by an act of Congress to develop public–private partnerships that support the mission of the National Institutes of Health (NIH). One project managed by the foundation is the Accelerating Medicines Partnership (AMP), which brings high-level government, industry, and nonprofit foundation partners together to identify and validate the most promising biological targets for therapeutics. The specific goals of AMP are to discover and validate new drug targets that companies can incorporate into their therapeutic development programs; to provide new insights into known, existing targets; to enable a significant increase in knowledge of tractable disease biology and disease pathways; and to create a rich, comprehensive, integrated knowledge base that is easy to use and available to the entire global research community, said David Wholley, the director of research partnerships for FNIH. Launched in early 2014, AMP brings together about 20 companies, NIH, and multiple nonprofit organizations to do precompetitive research and share data broadly and quickly, with the funding split between the public and private sectors. To ensure the broadest possible opportunity for commercialization, preemptive patenting is not allowed, and data become freely available as soon as quality control is finished, Wholley said.
AMP is focused on three main areas: Alzheimer’s disease, type 2 diabetes, and immune-related disorders such as rheumatoid arthritis and systemic lupus erythematosis. Within each of these disease areas, the stakeholders involved in the partnership set timelines for deliverables. AMP is governed by steering committees that are co-chaired by a representative from the NIH and an industry partner. The steering committees “are really designed to make sure that the research is continuing on track and according to our milestones and goals,” Wholley said. Steering committee members are personally committed to the goals of the program and make it a point to join each and every meeting, Wholley continued.
As an example of how AMP is using genetic and genomic data, Wholley described efforts within the program to find new medicines to treat type 2 diabetes. In order to identify potential drug targets for type 2
diabetes, AMP has mapped out a 5-year program that links human genetic data on disease risk to phenotypic data. Similar to the Genomic Resources for Drug Discovery Consortium, researchers in the AMP diabetes program realized that they could accelerate progress by aggregating their data, as opposed to working separately, Wholley said. Though funding procedures and workflows have been complicated at times, AMP has succeeded in building a knowledge portal that provides aggregated human genetic data from more than 150,000 individuals across a wide range of ages and ethnicities, and has created tools within the portal to allow easy, integrated interrogation across multiple datasets while maintaining individual-level data privacy, he said.
The challenges for this type of collaboration include both incentivizing investigators to share data and managing data restrictions, such as consent and regulations on exporting data out of countries, Wholley said. Another difficulty has been data integration, which is challenging because of the heterogeneity of the data, analytical needs, funding sources, legacy support, and data platforms. It has been critical to address concerns about publications, authorship, and acknowledgment, he said, although intellectual property issues have not been a concern because of the well-defined precompetitive agreement that all partners must agree to before joining the collaboration. Members also debate about how to prioritize spending. Finally, aligning interests and perspectives between government, academic, and industry scientists can present difficulties, Wholley said.
The Structural Genomics Consortium: Pooling Resources and Sharing Risks
Three major challenges confront researchers working on drug discovery and development both in the public and private sectors, said Chas Bountra, a professor of translational medicine at the University of Oxford. The first is target discovery—the identification of molecules (e.g., proteins) that can be modulated to treat diseases in a subset of patients. The second is the amount of duplication occurring in biomedicine across all sectors. Many groups are working on the same targets, and, unfortunately, many of those targets are destined for failure, he said. The third challenge is the development of high-quality drugs to alter disease pathways.
The Structural Genomics Consortium (SGC)2 has tried to address these challenges in several ways, Bountra said. First, members of the consortium have pooled resources and shared risk by assembling a network of government agencies, philanthropies, and eight large pharmaceutical companies. Second, through collaborations with hospitals, clinicians, patient organizations, and individual patients, the SGC can easily access such resources as primary human tissues for target validation studies and can maintain focus on patient needs within the context of specific diseases. Third, the consortium is making all of its reagents and tools for drug discovery freely and immediately available. This has generated great enthusiasm for collaborations, so that the consortium is now working with more than 300 academic labs all over the world for free, Bountra said. Finally, the consortium has used the expertise, chemical compound collections, and other resources made available through commercial partnerships to work on proteins or protein families that have been previously viewed as intractable or unusable in a drug development program.
As an example of this approach, Bountra cited work on a family of proteins with a special motif called bromodomains, which were thought to be undruggable due to their complex biology and protein structures. However, in collaboration with researchers at GSK and Harvard University, Bountra and his colleagues identified a novel inhibitor of bromodomain-containing proteins that reduced proliferation in tumors (Filippakopoulos et al., 2010). Since then, Bountra said, the molecule has been given to more than 1,000 laboratories around the world and has been found to be effective not only for a range of cancers but for sepsis, cardiac hypertrophy, male contraception, fibrosis, and chronic obstructive pulmonary disease, as reported in more than 300 publications. Pharmaceutical companies that partnered with the SGC began proprietary programs with the molecule as a starting point. To date, six companies have developed six unique investigational drugs based on the original molecule and are currently testing them in clinical trials, Bountra said.
More recently, with funding from the Wellcome Trust, the SGC has been working with clinicians, geneticists, and disease experts to identify high-priority genes that are most likely to be therapeutic targets. The group then generates the purified proteins, related active molecules, inactive control molecules, and other tools that can facilitate the development
__________________
2 For more information on the Structural Genomics Consortium, see http://www.thesgc.org (accessed July 7, 2016).
of drugs for patients. Twenty-five of these target-enabling packages are being developed in the areas of neuropsychiatry, metabolic disease, cancer, and inflammatory disease. The reagents are peer reviewed by external academic scientists to assess their quality before being released, Bountra said.
With the work of the SGC, “we are trying to generate de-risked targets which industry can then take to generate proprietary molecules, fast track them through large-scale clinical studies, and take them all the way to the marketplace,” Bountra said. “We believe this is the right thing to do for patients. It’s the right thing to do for society. It’s the right thing to do for industry.” There is hope that the creation of a new collaborative research ecosystem that involves the pharmaceutical industry, academic researchers, patients, government, and private funders, will generate many more novel medicines, Bountra continued.
THE SUSTAINABILITY OF BUSINESS MODELS
How can collaborative business models for drug discovery be sustainable over time? Successfully meeting short-term program goals is, of course, one key to sustainability, Matrisian said, because it keeps all the partners invested and moving forward. The SGC has been sustainable because of its ability to evolve and take on new research projects once the network was in place and results were being generated, Bountra said, and that evolution has allowed for additional funding. Clearly defining deliverables early on was a significant part of the respective business plans for AMP and the Genomic Resources for Drug Discovery Consortium, according to Wholley and Ehm. Data also need to remain relevant to important research questions for an initiative to be sustainable, Ehm said.
The availability of data generated from large-scale genetic bioresources tends to create opportunities for incentivized challenges, which can democratize the discovery process, Wholley said. Generating large datasets and making them broadly available can attract innovative researchers with an entrepreneurial spirit, he said. Data harmonization and the ability to associate genetic variants with longitudinal outcomes are key practical markers of efficient data accessibility and utility, Ehm said. Making the needs of patients a priority during the design of research studies is also important, Wholly said. One way to ensure that research is focused on the goals of patients is to issue direct challenges and competitions for specific funding opportunities, Matrisian said.
One shortcoming of public–private partnerships, Wholley said, is that few of them have clearly defined metrics for when enough work has been done on a topic or outline plans for how the partnership will prioritize and advance projects. Looking forward to future steps to ensure consortium relevance for the long term is also important.










