9
Deciphering the Chemistry of the Human Gut Microbiome
INTRODUCTION
Trillions of microorganisms live in the human gut, making it one of the densest microbial habitats. These organisms play critical roles in host health, and a growing body of research has correlated changes in this microbial community to disease. Though advances in DNA sequencing have improved our knowledge of the gut microbiome’s composition, our understanding of the molecular mechanisms underlying how this community influences human biology has lagged. To gain these mechanistic insights, it is critical that we move beyond cataloging the organisms present in this environment, and gain an appreciation for the chemical transformations they carry out in the body. Thus, there is a need to uncover the molecular basis for the metabolic activities of the gut microbiome by linking metabolism to genes and enzymes. It is also essential that we develop methods for the controlled manipulation of these activities in intact communities. This paper discusses how knowledge and approaches from chemistry can play a central role in achieving these objectives.
OUR CURRENT UNDERSTANDING OF GUT MICROBIAL FUNCTIONS
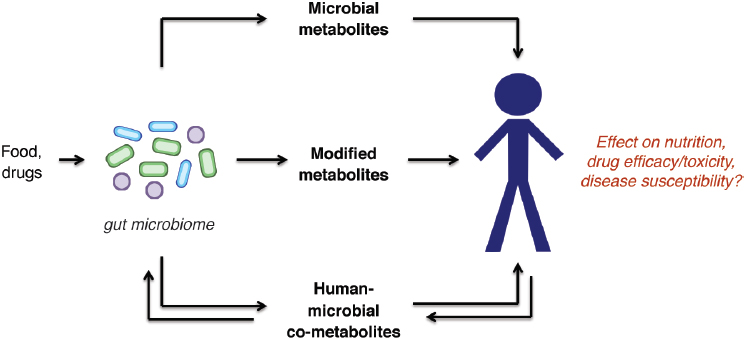
To decipher how the gut microbiome impacts human biology, it is crucial that we understand the biochemical transformations carried out by this microbial community (see Figure 9-1). Gut microbes digest otherwise intractable dietary components, providing nutrients to both the human host and other members of the microbiome. They also metabolize other foreign compounds (xenobiotics), including pharmaceuticals and environmental pollutants, altering their bioactivity and lifetimes within the body. Gut microbes can modify host-derived metabolites in unique ways, perhaps most notably, bile acids. Finally, these organisms can synthesize exclusively microbial molecules that engage host cells, including the immune system, and mediate microbe–microbe interactions. An enhanced knowledge of these activities could inform personalized nutrition and medicine, as well as reveal new strategies for treating or preventing disease.
Only a small fraction of chemistry carried out in this microbial habitat has been characterized. Half of the genes present in the human gut microbiome cannot be given any kind of annotation (Joice et al., 2014). Moreover, only about 15% of gut microbial genes can be linked to known metabolic pathways (Human Microbiome Project
___________________
a Department of Chemistry and Chemical Biology, Harvard University.
* Corresponding Author: balskus@chemistry.harvard.edu.
Consortium, 2012). We also have a limited understanding of the metabolism known to be associated with the gut microbiome; the genes and enzymes responsible for many gut microbial activities have not been identified. For example, only 6 of the nearly 50 examples of drug metabolism by human gut microbes have been linked to enzymes (Spanogiannopoulos et al., 2016). Gut microbial metabolites are also poorly characterized, as illustrated by metabolomics studies in humans and model organisms (Wikoff et al., 2009). Overall, it is clear that an enormous knowledge gap exists with regard to both the metabolites produced by this community and the enzymatic catalysts that generate them.
ELUCIDATING THE GENETIC AND BIOCHEMICAL BASIS FOR GUT MICROBIAL METABOLISM
Connecting Known Microbial Activities to Genes and Enzymes
A critical step in understanding human gut microbial metabolism is linking metabolic activities with specific microbial genes and enzymes. The genes encoding the enzymes that carry out individual transformations could be robust diagnostic markers that would predict the presence of particular functions in gut multi-omics datasets of metagenomes, metatranscriptomes, and metaproteomes. They can also be targets for genetic manipulation, helping to elucidate the roles of individual activities in microbial physiology and host health. Access to purified enzymes allows in vitro biochemical experiments that can decipher the molecular basis for activity, and, as discussed in the next section, gut microbial enzymes could also represent important targets for therapeutic development.
Multiple approaches can connect the growing number of metabolic activities associated with the human gut microbiome with microbes, genes, and enzymes. If transformations of interest can be identified in culturable strains, traditional methods such as forward genetics or activity-guided protein purification may be employed. Such approaches may be particularly useful for studying metabolic activities that do not resemble those performed by characterized enzymes. However, within the last decade, methods that incorporate DNA sequencing technology, including comparative genomics, transcriptomics, and functional metagenomics, have provided additional options for elucidating the genetic basis for gut microbial metabolism.
Knowledge of the chemistry underlying enzyme function and metabolism can greatly enable efforts to rationally mine microbial genomes for metabolic enzymes of interest. One example is the identification of microbial genes involved in anaerobic choline metabolism. Microbes in the human gut ferment choline, producing the volatile odorant trimethylamine (TMA) as an end product. TMA is further metabolized by host liver enzymes to trimethylamine-N-oxide (TMAO); both TMA and TMAO are linked to multiple human diseases. Until recently,
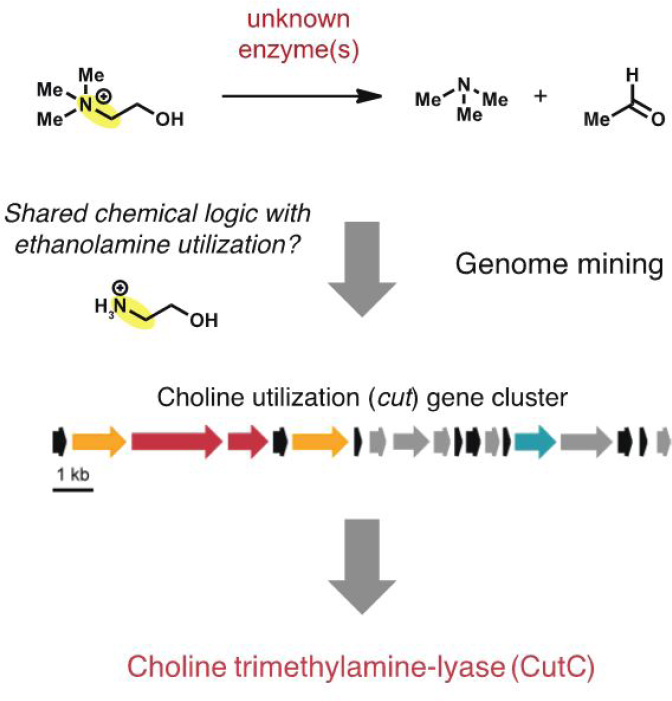
knowledge of this metabolic activity’s presence in the human gut microbiome was extremely limited because its genetic and biochemical basis was unknown. We recognized that the first step in choline fermentation, a carbon–nitrogen (C–N) bond cleavage reaction that converts choline to TMA and acetaldehyde, resembles the first reaction in bacterial ethanolamine utilization, leading us to hypothesize that these pathways shared certain transformations (Craciun and Balskus, 2012). We searched genomes of choline metabolizing bacteria for homologs of ethanolamine metabolizing enzymes and uncovered the choline utilization (cut) gene cluster (see Figure 9-2). Encoded within this gene cluster is choline TMA-lyase (CutC), a glycyl radical enzyme responsible for the key TMA-generating reaction. This discovery enabled the identification of cut genes in microbial genomes and gut metagenomes, providing information about the distribution of this activity. We anticipate that “chemically guided” genome mining may be applied more generally to uncover the genes that mediate additional gut microbial metabolic activities.
Connecting Genes of Unknown Function to Microbial Metabolism
Another major challenge facing microbiome research is how to elucidate the functions of the vast numbers of misannotated or uncharacterized genes found in these communities. Classical techniques, such as reverse chemical genetics, are limited to genetically tractable organisms. Associated phenotypes may also be difficult to observe under the artificial conditions of cultivation. Solving this enormous problem will clearly require new methods and approaches. One promising strategy is to leverage computational methods to predict the activities of uncharacterized proteins and guide experimental characterization; this approach is already widely applied to
study microbial secondary metabolism. Many classes of natural products are made using a shared biosynthetic logic, making it possible to readily identify biosynthetic gene clusters in microbial genomes and metagenomes. For certain pathways, one can even predict structural elements of the encoded natural products. Recently, Fischbach and coworkers characterized the potential for secondary metabolite biosynthesis in Human Microbiome Project metagenomes from various body sites (Donia et al., 2014). They found 3,118 putative biosynthetic gene clusters, with saccharide production dominating. Pathways for the synthesis of other structurally complex natural products like polyketides, nonribosomal peptides, terpenes, and ribosomally synthesized and post-translationally modified peptides were also located. Future challenges include identifying the secondary metabolites that are produced in these environments, which will require new techniques to monitor metabolite synthesis and exchange.
Gaining analogous predictive capabilities for other microbial metabolic pathways will require new computational and experimental tools. A promising starting point for this endeavor is to study novel members of previously characterized enzyme superfamilies. A growing number of computational methods like secondary structure prediction, protein sequence similarity networks, and genome neighborhood networks can differentiate uncharacterized family members from those with known activities (Gerlt et al., 2011). New computational and experimental strategies, including high-throughput docking of metabolite libraries and functional screening of solute binding proteins, can guide characterization.
It is substantially more challenging to study hypothetical proteins, which have no sequence or structural homology to proteins of known function. Ideally, methods for investigating these targets will be high throughput and automated, enabling many experiments to be performed in parallel; they should also be culture independent. Functional metagenomics, the expression of environmental DNA in heterologous hosts followed by screening for phenotypes of interest, may be particularly promising in this context, but is currently limited by the narrow range of available hosts, limitations in library size, and throughput of screening methods. Alternative strategies will need to incorporate high-throughput, automated methods for rapidly assessing changes in microbial phenotypes and integrate information from multi-omics measurements. Until such methods are developed, it will be important to prioritize uncharacterized proteins for further study.
CHEMICAL TOOLS FOR INVESTIGATING MICROBIOME FUNCTIONS
Chemists can also enhance our knowledge of the human gut microbiome by developing unique tools and approaches to study microbial metabolic activities. Notably, chemical tools routinely used in other areas of biology, including eukaryotic cell biology and studies of microbial pathogens, may be leveraged, adapted, and extended to understand the functions of microbial communities, including the human gut microbiome.
Chemical Genetics and the Human Gut Microbiome
Developing selective ways of manipulating gut microbial functions may hold the key not only to deciphering the contributions of individual activities to community and host health, but also to rationally altering the microbiome for therapeutic benefit. A variety of methods have been proposed for accomplishing this objective. Targeted, in situ genetic manipulations of intact microbial communities, for example, by using bacteriophage to deliver genome editing machinery to specific organisms, has generated much interest; however, it is at an early stage. Issues that may limit the utility of this approach include its current restriction to a subset of organisms and the fact that targeting single strains or species may not sufficiently alter functions that are more widely distributed in the gut microbiome.
Another option for introducing activities into communities is to add one or more probiotic organisms, either wild type or engineered (probiotics). However, addition of even a single strain alters multiple functions simultaneously. The transplantation of more complex consortia has emerged as an effective treatment for recurrent Clostridium difficile infections and is under investigation for other indications. This treatment may globally remodel microbial activities, but the mechanism underlying its efficacy is not well understood. Prebiotics and other dietary manipulations may alter microbiome activities by promoting the growth of beneficial microbial species. Though such treatments have shown benefits in clinical studies and are recommended for certain conditions, we currently
lack a predictive understanding of which organisms will respond to particular nutrients in different patients and a mechanistic knowledge of how the activities stimulated by these dietary components benefit the host.
An alternative approach to manipulating gut microbial functions is chemical genetics: the use of small molecule inhibitors—chemical probes or tool compounds—that target specific microbial processes. Originally formulated as a method to explore eukaryotic cell biology, chemical genetics provides a means of interrogating biological function in cells and organisms that complements traditional genetic approaches (Spring, 2005). This approach offers several advantages that may make it particularly suitable to application in complex microbial communities. Unlike genetic manipulation, using small molecules to modulate protein activity provides temporal and reversible control over function. The ability to “reverse” a microbiome intervention may be particularly attractive both from a research and therapeutic perspective, as the consequences of altering this community are not yet well understood. As this method does not require genome manipulation, it can be applied to organisms that are not genetically tractable, and can be used to simultaneously target activities shared between multiple bacterial species. Importantly, the small molecule inhibitors used in chemical genetics experiments are distinct from broad-spectrum antibiotics in that they do not target cellular functions that are universally essential for growth.
Such small molecule inhibitors have been used previously in microbiology, but are an underexplored strategy with respect to manipulating the human gut microbiome. Examples commonly used in environmental microbiology include compounds that target methanogenesis (bromoethane sulfonate), sulfate reduction (sodium molybdate), and nitrification (nitrapyrin) (Ormeland, 1988). Studies have also examined the impacts of inhibitor treatment on the compositions of soil and animal microbiomes. It is important to emphasize that most of the inhibitors used in these studies were identified prior to the era of modern chemical biology.
Small molecule inhibitors have also been increasingly popular tools to study microbial pathogenesis (Anthouard and DiRita, 2015). Unlike many inhibitors used in environmental microbiology, these compounds have been largely identified using strategies from modern chemical biology, including the high-throughput screening of compound libraries and structure-based inhibitor design. Such efforts have identified inhibitors of quorum sensing, additional factors influencing regulation of toxin production and virulence gene expression, secretion systems, biogenesis of pili, and other factors affecting pathogen-host interactions. Enteric pathogens that have been successfully studied using chemical genetics include Vibrio cholerae, Escherichia coli O157:H7, Salmonella typhimurium, and Clostridium difficile. In addition to gaining insights into the biology of pathogenesis, these tool compounds may also provide valuable starting points for therapeutic development.
Recent work illustrates the promise of developing small molecule inhibitors that target gut microbial metabolic activities. Redinbo and coworkers used an in vitro high-throughput screen to identify small molecules that potently inhibit E. coli β-glucuronidase while ignoring the corresponding human enzyme (Wallace et al., 2010) (see Figure 9-3). Their inhibitors were effective in modulating β-glucuronidase activity across a number of distantly related gut bacteria, highlighting the power of this strategy to target microbial functions shared between multiple strains in a community. They then investigated the ability of these compounds to prevent reactivation of SN-38G, a glucuronidated metabolite of the chemotherapeutic drug irinotecan, in the gut lumen. This microbial metabolic activity leads to severe, dose-limiting diarrhea in cancer patients, and blocking this reaction could potentially allow extension of chemotherapeutic regimens. Coadministration of their inhibitor with irinotecan prevented this side effect in mice. This work represents one of the first examples of alleviating a clinical condition by targeting a nonessential gut bacterial enzyme with a small molecule inhibitor. Although further questions remain regarding the broad effects of these inhibitors on gut microbiome composition and functions, as well as the long-term consequences of β-glucuronidase inhibition, this work provides important proof of concept for the idea of using chemical genetics to manipulate metabolism in this microbial community.
As has been the case for eukaryotic cell biology and microbial pathogens, the development of small molecule inhibitors that target additional gut microbial metabolic functions would deliver transformative tools with the potential to greatly advance our understanding of this microbial community. By combining chemical genetics with multi-omics experiments, we can begin to understand how inhibiting specific microbial metabolic activities shapes microbiome structure and function as well as host biology. Specific metabolic activities that may be interesting to target include functions associated with health, as well as disease-associated activities such as TMA production and genotoxin biosynthesis. An important challenge that will be faced in developing microbiome-targeted small
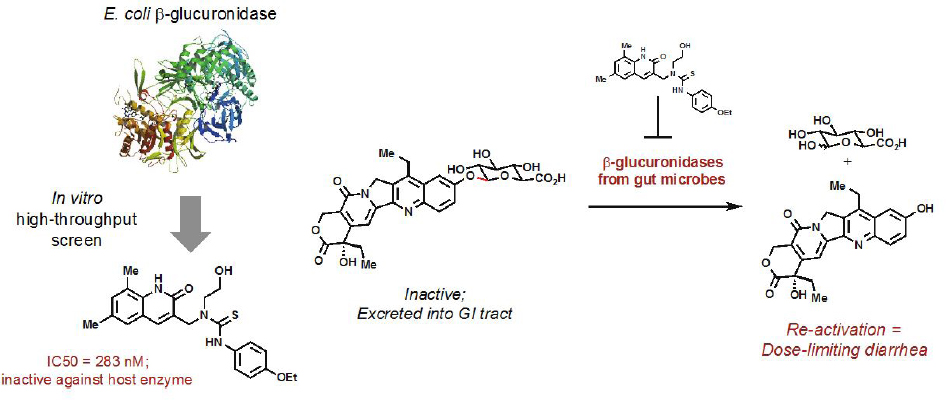
molecule inhibitors is the need to access targets located inside microbial cells, although the successful development of inhibitors that modulate intracellular β-glucuronidases illustrates that this is not an insurmountable obstacle. Moreover, many interesting microbial functions are extracellular, including excreted enzymes, receptors, and transporters. Overall, this approach to interrogating microbial functions has the potential to not only teach us about unappreciated aspects of gut microbial ecology, but also identify promising lead compounds to guide the development of microbiome-directed therapeutics.
Applying Additional Approaches from Chemical Biology to Study the Gut Microbiome
Chemical genetics is just one example of how tools developed by chemical biologists could be applied to enhance our understanding of the human gut microbiome. Many other methods are routinely used in eukaryotic cell biology and studies of microbial pathogens, but have not yet been widely applied to studies of the human microbiome and other microbial communities. Combining metabolic labeling strategies with bio-orthogonal chemistry can enable visualization of gut microbes in model hosts (Geva-Zatorsky et al., 2015). Development and application of chemical probes for imaging microbial metabolites and metabolic activities could allow us to observe gut microbial functions in real time in model organisms and even human patients. These tools could also provide information about the chemical environment of this microbial habitat. Finally, activity-based protein profiling could facilitate the discovery of microbial enzymes that are expressed and active in this community, including metabolic activities that are upregulated in patients with disease and potentially influence disease development or progression.
CONCLUSIONS
There are currently tremendous gaps in our knowledge of the metabolic activities associated with the human gut microbiome, preventing us from leveraging this community to enhance health and treat disease. Addressing this issue will require contributions from many scientific disciplines, but it is clear that knowledge and techniques from chemistry have the potential to facilitate leaps in our understanding of gut microbial functions. Microbiome research therefore provides tremendous opportunities for chemists and chemical biologists.
REFERENCES
Anthouard, R., and V. J. DiRita. 2015. Chemical biology applied to the study of bacterial pathogens. Infect Immun 83(2):456-469.
Craciun, S., and E. P. Balskus. 2012. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci USA 109(52):21307-21312.
Donia, M. S., P. Cimermancic, C. J. Schulze, L. C. Wieland Brown, J. Martin, M. Mitreva, J. Clardy, R. G. Linington, and M. A. Fischbach. 2014. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158(6):1402-1414.
Gerlt, J. A., K. N. Allen, S. C. Almo, R. N. Armstrong, P. C. Babbitt, J. E. Cronan, D. Dunaway-Mariano, H. J. Imker, M. P. Jacobson, W. Minor, C. D. Poulter, F. M. Raushel, A. Sali, B. K. Shoichet, and J. V. Sweedler. 2011. The Enzyme Function Initiative. Biochemistry 50(46):9950-9962.
Geva-Zatorsky, N., D. Alvarez, J. E. Hudak, N. C. Reading, D. Erturk-Hasdemir, S. Dasgupta, U. H. von Andrian, and D. L. Kasper. 2015. In vivo imaging and tracking of host–microbiota interactions via metabolic labeling of gut anaerobic bacteria. Nat Med 21(9):1091-1100.
Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486(7402):207-214.
Joice, R., K. Yasuda, A. Shafquat, X. C. Morgan, and C. Huttenhower. 2014. Determining microbial products and identifying molecular targets in the human microbiome. Cell Metab 20(5):731-741.
Ormeland, R. S., and D. G. Capone. 1988. Use of “specific” inhibitors in biogeochemistry and microbial ecology, in K. C. Marshall (Ed.) Advances in Microbial Ecology, Vol. 10. New York: Springer U.S. 285-383.
Spanogiannopoulos, P., E. N. Bess, R. N. Carmody, and P. J. Turnbaugh. 2016. The microbial pharmacists within us: A metagenomic view of xenobiotic metabolism. Nat Rev Microbiol 14(5):273-287.
Spring, D. R. 2005. Chemical genetics to chemical genomics: Small molecules offer big insights. Chem Soc Rev 34(6):472-482.
Wallace, B. D., H. Wang, K. T. Lane, J. E. Scott, J. Orans, J. S. Koo, M. Venkatesh, C. Jobin, L. A. Yeh, S. Mani, and M. R. Redinbo. 2010. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330(6005):831-835.
Wikoff, W. R., A. T. Anfora, J. Liu, P. G. Schultz, S. A. Lesley, E. C. Peters, and G. Siuzdak. 2009. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA 106(10):3698-3703.
This page intentionally left blank.








