2
Illuminating the Microbial Dark Matter Beneath Your Feet: Microbial Catalysis in the Terrestrial Subsurface
Kelly C. Wrighton,a,* Rebecca A. Daly,aand Michael J. Wilkinsa,b
INTRODUCTION TO THE TERRESTRIAL SUBSURFACE: A RESOURCE-LADEN MICROBIAL FRONTIER
Above the Earth’s core and mantle, the crust is a solid layer that extends outward 5-10 km on oceanic plates and 30-50 km on continental plates. This continental crust, known as the terrestrial zone, is composed of a variety of layers that consist of igneous, metamorphic, and sedimentary rocks. These rocks weather and reform over geologic cycles lasting millions to billions of years. Near the Earth’s surface, these weathered minerals and organic material combine to produce a narrow lens of soil, surface, and vadose zones that cover the Earth, providing suitable habitats and niches for abundant biological diversity (Ehrlich et al., 2015). Beneath these layers, and extending to the mantle, is the subsurface, a region of the planet completely disconnected from surface light-driven reactions (see Figure 2-1).
Once thought to be relatively free of microorganisms, recent estimates report that up to 19% of the Earth’s total biological mass (10-100 Pg carbon) may be contained in the terrestrial subsurface (Whitman et al., 1998; McMahon and Parnell, 2014). Microbial life in this environment exists across a wide range of rock and habitat types (see Figure 2-1). Generally, biomass density decreases with depth in the terrestrial subsurface; however, significant cell abundance has been detected at microbial hotspots, often rock interfaces, where chemical conditions are conducive to subsurface life (Seckbach, 1999). Indigenous microorganisms have been identified from many subsurface habitats, spanning physical and chemical extremes. Life has been recovered from beneath Antarctic ice sheets (Christner et al., 2014), 2.8-km-deep gold mines (Lin et al., 2006), highly saline fluids within permafrost (Gilichinsky et al., 2003), and in cave systems (Sarbu et al., 1996). Compared to the marine subsurface (Biddle et al., 2006), only a small fraction of the microbial habitats below our feet have been sampled. Except for cave and mine environments, the accessibility of terrestrial subsurface geological materials is limited by the high cost of continental drilling and the identification of representative samples while minimizing sample contamination (Wilkins et al., 2014). Consequently, the terrestrial subsurface represents one of the least explored ecosystems on the planet. Yet, this ecosystem offers a window into novel enzymatic reactions that support life in extreme, rock-hosted habitats.
___________________
a Department of Microbiology, The Ohio State University.
b School of Earth Sciences, The Ohio State University.
* Corresponding Author: wrighton.1@osu.edu.
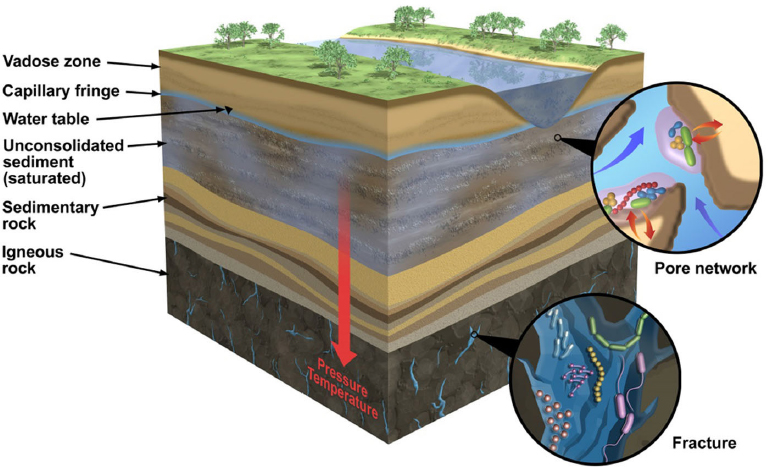
SOURCE: Ehrlich et al., 2015.
In addition to existing independent of direct light exposure, rock-hosted life has several other unique constraints from surface-adapted life. With increasing depth, microbial habitats are exposed to higher in situ temperatures and pressures (see Figure 2-1). Given that pressure increases by approximately 10 MPa per km of depth, inhibitory pressures may only be encountered at great depths within the Earth, where temperatures would be far above the maximum limit for life. Thus, it is thought that temperature, rather than pressure, is normally a greater constraint on life in the terrestrial biosphere (Zeng et al., 2009).
Primary constraints on rock-hosted life are sufficient physical space and water availability. Although water is present in huge volumes in the terrestrial subsurface—some estimates (McMahon and Parnell, 2014) put the volume at 108 km3—the availability of water is linked to both pore space and connectivity, features that vary considerably in the subsurface. For instance, the matrix of shallow saturated aquifer sediments is analogous to a saturated sponge, allowing relatively free chemical and genetic exchange. Conversely, microbial habitats in deeper sedimentary rocks are restricted by submicron pore openings and limited fluid exchange, with life likely confined to hairline fractures in bedrock or at depositional boundaries with higher porosity rocks (Krumholz et al., 1997). Extended water contact time in these environments also increases rock dissolution, so organisms often must tolerate brine-level salinities. Furthermore, in undisturbed, deep rock-hosted habitats, organisms are faced with extremely low fluxes of energy and nutrients, which restricts microbial metabolism to the slowest on the planet (Amy and Haldeman, 1997). Thus, life in the subsurface must have unique adaptations to overcome physical and geochemical stressors not found in surface habitats.
Beyond the potential for finding organisms that catalyze novel chemical reactions, the desire to manage microbial metabolisms has increased with the human dependency on subsurface resources. Humans interact with the terrestrial subsurface via a range of processes linked to groundwater extraction, energy and mineral recovery, waste disposal, and inadvertent contamination. In the shallow terrestrial subsurface, alluvial aquifers are globally important freshwater sources (Gleeson et al., 2012). Because of industrial processes like agriculture, the processing of nuclear materials, and fossil fuel exploration and development, many aquifers have been contaminated worldwide. Subsurface microorganisms, either through natural processes or by stimulation of their activities via nutrient
addition, can degrade many contaminants to harmless or less toxic products or greatly reduce their solubility and hence mobility (Lovley et al., 1989; Wilkins et al., 2009).
Deeper into the terrestrial subsurface, human interaction through waste storage or resource extraction alters the biogeochemical conditions of the subsurface, with currently unknown impacts on the engineered infrastructure of these systems. For instance, it has been proposed that CO2 generated from the combustion of fossil fuels at power plants be injected into subsurface reservoirs in an attempt to reduce anthropogenic greenhouse gas emissions to the atmosphere. The subsurface is also utilized or being considered for the sequestered storage of high-level radioactive waste from nuclear power generation and residual waste from past production of weapons-grade materials (Ehrlich et al., 2015). In terms of resources, oil and gas extracted from subsurface environments comprises a significant fraction of energy consumed in the United States. Microbially catalyzed reactions in these systems can have deleterious effects on energy yield and infrastructure, with economic costs of billions of dollars annually (Morozova et al., 2010). Therefore, as a society we have significant motivation to understand and predict microbially catalyzed reactions in these economically critical ecosystems within our planet, both before and after human interaction.
In this Proceedings of a Seminar Series, we discuss new research findings from the Earth’s terrestrial subsurface microbiome. Highlighted are microbial reactions in two subsurface regions impacted by human activity: a shallow metal-contaminated aquifer system and sedimentary shale rocks subjected to hydraulic fracturing. In the aquifer system, we demonstrate how genomic information defines new carbon cycling roles for enigmatic microorganisms, activities that further stimulate other microorganisms capable of heavy-metal contaminant removal. In fractured shales, we show how hydraulic fracturing creates a new and sustainable ecosystem, driven by methylamine metabolisms 2,500 meters below the surface. Together, these case studies showcase the phylogenetic and metabolic novelty present in the terrestrial subsurface that has only recently been uncovered. Future studies will likely reveal more insights into the chemical reactions that sustain life deep beneath our feet.
CASE STUDY FROM SATURATED UNCONSOLIDATED SEDIMENTS: GENOMICS ILLUMINATES MICROBIAL DARK MATTER IN GROUNDWATER
Until recently, the ability to cultivate microorganisms was the only protocol for providing access to microbial chemical reactions. However, the first realizations that the true extent of microbial diversity existed far beyond what was in cultivation came from analyzing microbes directly from the environment, where it was clear that on average only 1% of cells were recovered by cultivation (Solden et al., 2016). Today, we know that most of the microbial phyla on this planet lack a single cultivated representative, thus obscuring a large fraction of microbial catalyzed reactions; phyla composed exclusively of uncultured microbial representatives are referred to as candidate phyla (CP). Borrowing language from astronomy, microbiologists operationally defined these CP as microbial dark matter, as these organisms account for a large portion of the Earth’s biomass and biodiversity, yet their basic metabolic properties are unknown. Understanding the metabolic roles of this microbial dark matter presents a grand challenge to the scientific community. Without understanding the metabolic mysteries of the CP, and other phyla, our knowledge of the microbial world, and the chemical reactions they catalyze, will remain profoundly skewed. In the past 5 years, advances in genomic technologies have provided a complementary path to gaining metabolic insight from microorganisms, independent of cultivation. Microbial genomes can now be directly sequenced from the environment using metagenomics, a method where all environmental DNA is sequenced and reconstructed into individual genomes (Hedlund et al., 2014). In 2012, in conjunction with Jill Banfield and colleagues, we applied metagenomic and metaproteomic tools to groundwater biomass collected from a former uranium milling site bordering the Colorado River (Wrighton et al., 2012). While prior research focused on the bioremediation activity of metal-reducing bacteria (Wilkins et al., 2009), our meta-proteogenomic, the linkage of community-wide proteomic data to metagenomes, approached assigned metabolic roles for uncultivated bacteria that previously lacked characterized physiologies. We discovered that microbial dark matter lineages were a dominant and active fraction of the aquifer microbial community, and provided the first metabolic blueprints for five CP lineages. Soon after, more extensive genomic sampling defined these CP lineages into a single bacterial radiation (the candidate phyla radiation, CPR) that accounted for 15% of the known bacterial diversity (Brown et al., 2015; Hug et al., 2016). Today, hundreds of genomes from more than 40 phyla constitute this radiation, making many uncultivated
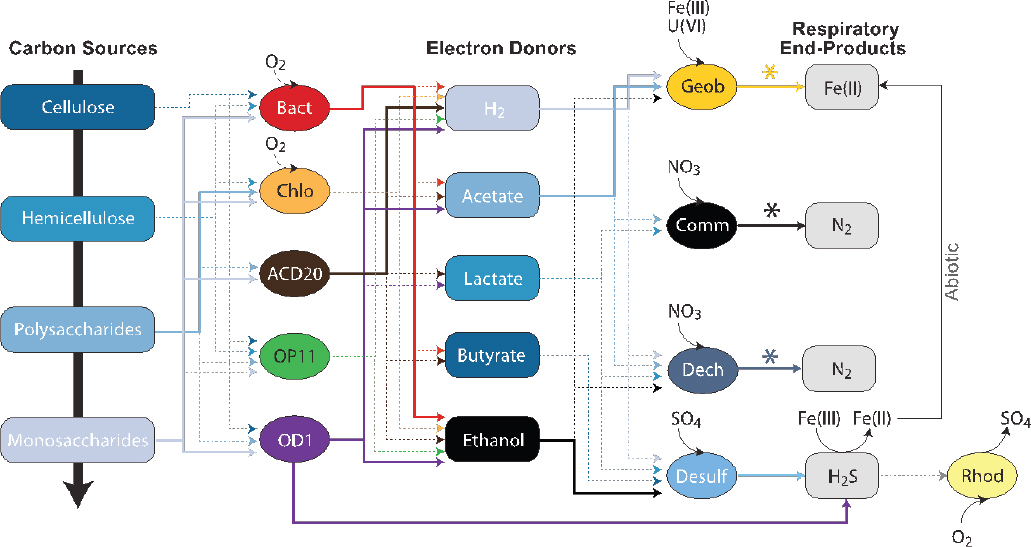
SOURCE: Wrighton et al., 2013. Reprinted by permission from Macmillan Publishers Ltd: The ISME Journal 3:873-876, copyright 2009.
lineages more genomically sampled than historically well-studied, cultivated lineages. The reactions catalyzed by the CPR, and other undescribed, uncultivated lineages, harbor diverse metabolisms.
Analyses of genomes from the CPR have led to new perspectives on microbial protein synthesis, genetic codes, ultrasmall cell volumes approaching the expected minimum of 0.009±0.002 μm3, and microbial carbon cycling in the subsurface (Solden et al., 2016). Notably, their genomes lack biosynthesis pathways for nucleotides, lipids, and most amino acids. This auxotrophy suggests that these organisms may be dependent on one or more members of the surrounding community, expanding our perspective on microbial metabolic interdependence. The CPR lineages are inferred to play critical roles in fermenting more recalcitrant carbon (see Figure 2-2), excreting hydrogen and organic acids. Thus, in the absence of oxygen, the CPR release labile substrates that facilitate other microbially catalyzed transformations of uranium, iron, and sulfur in the aquifer (Wrighton et al., 2013). These findings demonstrate how metagenomic approaches can untangle the metabolic interdependencies, shaping the potential for bioremediation within groundwater microbial communities.
Despite the application of extensive metagenomic sampling—often combined with metaproteomic and metatranscriptomic analyses—our understanding of the metabolic capabilities of CPR organisms has only just begun. Given critical differences in both the phylogenetic divergence and host environments between CPR microorganisms and well-studied bacterial strains, it is also possible these uncultivated representatives interact with the environment in new ways. Emphasizing this disconnect, less than 50% of most CPR genomes possess current functional annotations (Brown et al., 2015). Biochemically targeting the functionality of poorly annotated or novel proteins in the CPR (Wrighton et al., 2016), identifying new pathways using metabolite measurements as a guide (Johnson
et al., 2016), and developing cultivation regimes that account for metabolic codependence (Ge et al., 2016) have afforded new information on the chemical reactions catalyzed by uncultivated microbes. Moving forward, unlocking the chemical mysteries of biological dark matter will rely on efforts directed to illuminating the enigmatic metabolic reactions encoded in these cryptic genomes.
CASE STUDY FROM THE DEEP BIOSPHERE: HYDRAULIC FRACTURING CREATES A NEW METHYLAMINE-DRIVEN ECOSYSTEM 2,500 METERS BELOW THE EARTH’S SURFACE
Shale gas accounts for one-third of natural gas energy resources worldwide. In the United States, shale gas has been predicted to provide half of the natural gas annually by 2040, with the Marcellus shale in the Appalachian basin projected to produce three times more than any other formation (Daly et al., 2016). Recovery of these hydrocarbons is dependent on hydraulic fracturing technologies, where the high-pressure injection of water and chemical additives generates extensive fractures in the shale matrix. Hydrocarbons trapped in tiny pore spaces are subsequently released and collected at the surface, along with a portion of the injected fluids that have reacted with the shale formation. While attention has been paid to the economic benefits and environmental impacts of this process, the biogeochemical changes induced in the deep subsurface are poorly understood.
Microbial metabolism and growth in hydrocarbon reservoirs have both positive and negative impacts on energy recovery. Undesirable microbial activity during these processes can include oil field souring, the corrosion of wells and pipelines, and pore clogging due to biomass accumulation and biogenic mineral precipitation around wells (Morozova et al., 2010). Alternatively, some hydrocarbon industries have stimulated microbial metabolism, especially methanogens, to increase energy production (Kirk et al., 2015). We used genome-resolved metagenomics, combined with detailed metabolite analyses, to infer the consequences of microbial metabolism from two geographically distinct shale formations after hydraulic fracturing. Our findings show that hydraulic fracturing not only created the physical space for rock-hosted life, but also provided the water, nutrients, and organisms to create a new ecosystem deep beneath the Earth’s surface. With pressures hundreds of times greater than at the surface, salinities four times the ocean (see Figure 2-3), and temperatures equivalent to the hottest day in Death Valley, we were interested in the unique organisms and their adaptive mechanisms in this extreme environment.
Of the microorganisms able to persist in this environment, Halanaerobium is the most prevalent across all shale systems sampled to date (Mouser et al., 2016). Our genomic and paired metabolite analyses indicate that this organism has the capacity to ferment chemical additives (e.g., ethylene glycol; see Figure 2-3) and produce corrosive sulfide, directly impacting the engineering of this economically important resource. We also identified a new genus of bacteria, here named Candidatus Frackibacter, due to its unique recovery only from fractured shales (Daly et al., 2016). In addition to microorganisms, we also sampled hundreds of bacterial and archaeal viruses present in fractured shales. Our metagenomic and metabolite data revealed a probable role for viruses as active predators, which cause the release of cellular contents during lysis. These findings first demonstrated the intrinsic roles viruses play in controlling microbial function in the deep terrestrial biosphere. Together, genomic findings hint at the novel, and untapped, bacterial and viral diversity contained within the terrestrial subsurface biosphere, a rich source to discover extremophilic and robust enzymes for industrial applications.
Our metabolite and metagenomic findings show that members of the shale microbial community produce and utilize glycine betaine (see Figure 2-3), an organic compound that protects cellular osmolarity and, potentially, increases barotolerance (Smiddy et al., 2004). Both Halanaerobium and Candidatus Frackibacter have the capacity to degrade this microbially produced osmoprotectant, in turn producing trimethylamine (TMA). Our combined chemical and genomic data show that methyl-C1 methanogenic substrates are produced both by the microbial community (e.g., monomethylamine [MMA] and TMA) and are added exogenously during hydraulic fracturing (e.g., methanol, MMA) are substrates for biogenic methane production (see Figure 2-3). This microbial methylamine cycle of osmoprotectant synthesis–fermentation to trimethylamine—methyl-C1 methanogenesis offers a mechanism for sustaining this microbial ecosystem in fractured shales, completely independent from chemical additions in the initial fracturing. Ultimately, methylamines represent a target to increase methane recovery from these systems in the future (see Figure 2-4). Results from this study highlight the resilience of microbial life to
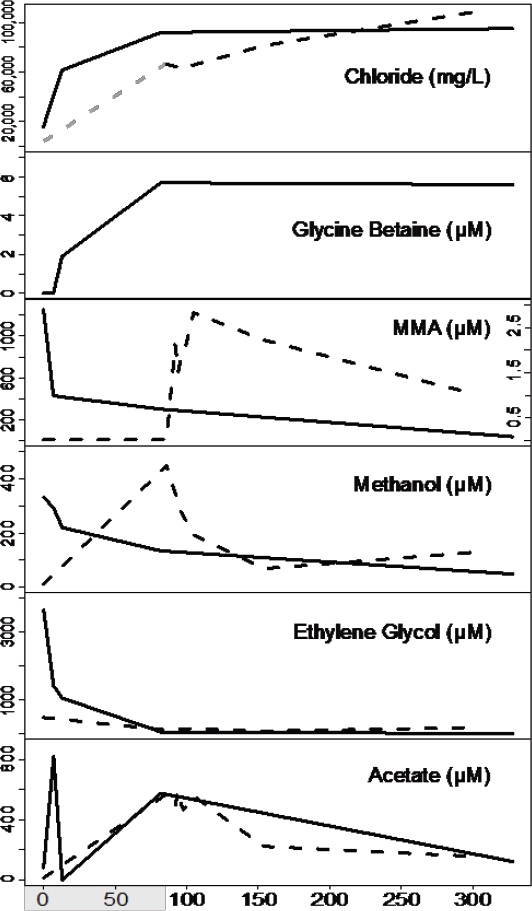
SOURCE: Daly et al., 2016.
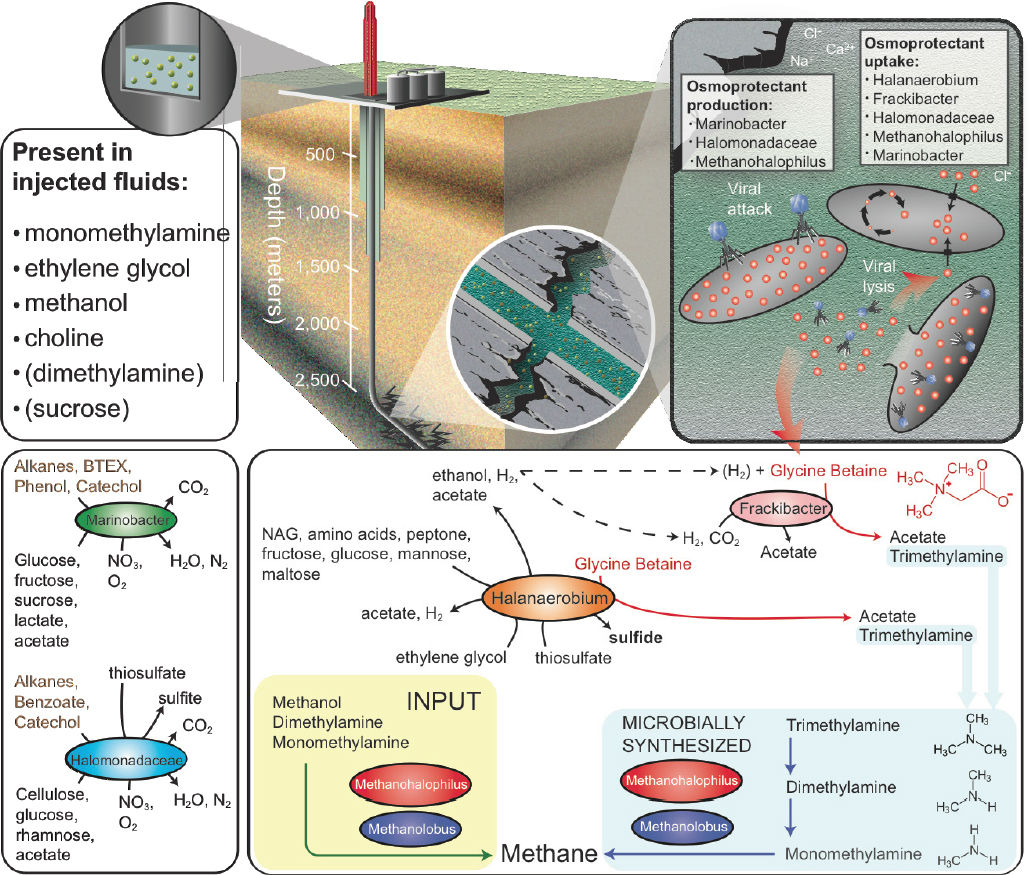
SOURCE: Daly et al., 2016.
adapt to, and colonize, a new habitat structured by physical and chemical features far different than their origin, with implications for life on this planet.
CONCLUDING REMARKS
Until recently, the ability to culture microorganisms was a prerequisite for genome sequencing, providing full access to the metabolic reactions organisms catalyze. The advent of techniques like metagenomics has opened a new window into the microbial and viral diversity within the terrestrial subsurface. Given the novelty and the abundance of high-quality genomes discovered each year, it is clear that we have only begun to appreciate the chemical potential catalyzed by these microbial communities.
A major challenge to the exploration of the terrestrial subsurface is access. Compared to surface systems, marine environments, or even our own bodies, the terrestrial biosphere is an under-sampled ecosystem and represents a new opportunity for studying biodiversity and biochemical reactions under extreme chemical and physical conditions. Diverse physiochemical conditions in these habitats often require unique adaptations for life to persist, including mechanisms for tolerance to salinity, pressure, and a lack of light-driven reactions. The ability to survive under such conditions in an environment that is buffered from changes that can impact life at the Earth’s surface have led scientists to consider the subsurface of planets and moons in our own solar system as possible refuges for microbial life. Finally, the subsurface microbiome has a major impact on the geosphere by mitigating contaminants in groundwater, the turnover of organic carbon, and the weathering of rocks and minerals. Understanding the biota present in the subsurface terrestrial ecosystems, including their interactions with each other and the environment, is critical for the management of subsurface resources. This knowledge is necessary as humans increasingly exploit the subsurface through hydrocarbon extraction, mining, and carbon dioxide sequestration activities. Future development of new chemical and biological technologies coupled to the continued sampling of subsurface environments will enhance our understanding of microbially catalyzed reactions in this critical ecosystem.
ACKNOWLEDGMENTS
Both the aquifer and hydraulically fractured shale investigations would not be possible without the support from the U.S. Department of Energy (DOE). The Rifle, Colorado, Integrated Field Research Center Project is managed by Lawrence Berkeley National Laboratory for the DOE (DE-AC02-05CH11231). Portions of this research were performed under the DOE Joint Genome Institute and Environmental Molecular Sciences Laboratory (JGI–EMSL) Collaborative Science Initiative and used resources at the JGI–EMSL, which are DOE Office of Science user facilities. Both facilities are sponsored by the Office of Biological and Environmental Research and operated under Contract Nos. DE-AC02-05CH11231 (JGI) and DE-AC05-76RL01830 (EMSL). Additional support for the hydraulically fractured shale research was made possible by the Deep Carbon Observatory’s Census of Deep Life supported by the Alfred P. Sloan Foundation award to KCW and a National Science Foundation Dimensions of Biodiversity grant awarded to MJW and KCW (Award No. 1342701).
REFERENCES
Amy, P. S., and D. L. Haldeman (Eds.). 1997. The Microbiology of the Terrestrial Deep Subsurface. Boca Raton, FL: CRC Press.
Biddle, J. F., J. S. Lipp, M. A. Lever, K. G. Lloyd, K. B. Sorensen, R. Anderson, H. F. Fredricks, M. Elvert, T. J. Kelly, D. P. Schrag, M. L. Sogin, J. E. Brenchley, A. Teske, C. H. House, and K. U. Hinrichs. 2006. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci USA 103:3846-3851.
Brown, C. T., L. A. Hug, B. C. Thomas, I. Sharon, C. J. Castelle, A. Singh, M. J. Wilkins, K. C. Wrighton, K. H. Williams, and J. F. Banfield. 2015. Unusual biology across a group comprising more than 15% of domain bacteria. Nature 523:208-211.
Christner, B. C., J. C. Priscu, A. M. Achberger, C. Barbante, S. P. Carter, K. Christianson, A. B. Michaud, J. A. Mikucki, A. C. Mitchell, M. L. Skidmore, T. J. Vick-Majors, and The WISSARD Science Team. 2014. A microbial ecosystem beneath the West Antarctic ice sheet. Nature 512:310-313.
Daly, R. A., M. A. Borton, M. J. Wilkins, D. W. Hoyt, D. J. Kountz, R. A. Wolfe, S. A. Welch, D. N. Marcus, R. V. Trexler, J. D. MacRae, J. A. Krzycki, D. R. Cole, P. J. Mouser, and K. C. Wrighton. 2016. Microbial metabolisms in a 2.5-km-deep ecosystem created by hydraulic fracturing in shales. Nat Microbiol 16146.
Ehrlich, H. L., D. K. Newman, and A. Kappler (Eds.). 2015. Erlich’s Geomicrobiology, 6th ed. Boca Raton, FL: CRC Press. Ge, Z., P. R. Girguis, and C. R. Buie. 2016. Nanoporous microscale microbial incubators. Lab Chip 16:480.
Gilichinsky, D. E. Rivkina, V. Shcherbakova, K. Laurinavichuis, and J. Tiedje. 2003. Supercooled water brines within permafrost—an unknown ecological niche for microorganisms: A model for astrobiology. Astrobiology 3:331-341.
Gleeson, T., Y. Wada, M. Bierkens, and L. Beek. 2012. Water balance of global aquifers revealed by groundwater footprint. Nature 488:197-200.
Hedlund, B. P., J. A. Dodsworth, S. K. Murugapiran, C. Rinke, and T. Woyke. 2014. Impact of single-cell genomics and metagenomics on the emerging view of extremophile “microbial dark matter.” Extremophiles 18:865-875.
Hug, L. A., B. J. Baker, K. Anantharaman, C. T. Brown, A. J. Probst, C. J. Castelle, C. N. Butterfield, A. W. Hernsdorf, Y. Amano, K. Ise, Y. Suzuki, N. Dudek, D. A. Relman, K. M. Finstad, R. Amundson, B. C. Thomas, and J. F. Banfield. 2016. A new view of the tree of life. Nat Microbiol 16048.
Johnson, W. M., M. C. Kido Soule, and E. B. Kujawinski. 2016. Evidence for quorum sensing and differential metabolite production by a marine bacterium in response to DMSP. ISME J 10:2304-2316.
Kirk, M. F., B. H. Wilson, K. A. Maruart, L. H. Zeglin, D. S. Vinson, and T. M. Flynn. 2015. Solute concentrations influence microbial methanogenesis in coal-bearing strats of the Cherokee Basin, USA. Front Microbiol 6:1287.
Krumholz, L. R., J. P. McKinley, G. A. Ulrich, and J. M. Suflita. 1997. Confined subsurface microbial communities in Cretaceous rock. Nature 386:64-66.
Lin, L. H., P. L. Wang, D. Rumble, J. Lippmann-Pipke, E. Boice, L. M. Pratt, B. S. Lollar, E. L. Brodie, T. C. Hazen, G. L. Andersen, T. Z. Desantis, D. P. Moser, D. Kershaw, and T. C. Onstott. 2006. Long-term sustainability of a high-energy, low-diversity crustal biome. Science 314:479-482.
Lovley, D. R., M. J. Baedecker, D. J. Lonergan, I. M. Cozzarelli, E. J. P. Phillips, and D. I. Siegel. 1989. Oxidation of aromatic contaminants coupled to microbial iron reduction. Nature 339:297-299.
McMahon, S., and J. Parnell. 2014. Weighing the deep continental biosphere. FEMS Microbiol Ecol 87:113-120.
Morozova, D., M. Wandrey, M. Alawi, M. Zimmer, A. Vieth, M. Zettlitzer, and H. Würdemann. 2010. Monitoring of the microbial community composition in saline aquifers during CO2 storage by fluorescence in situ hybridisation. Int J Greenh Gas Con 4:981-989.
Mouser, P. J., M. Borton, T. H. Darrah, A. Hartsock, and K. C. Wrighton. 2016. Hydraulic fracturing offers view of microbial life in the deep terrestrial subsurface. FEMS Microbiol Ecol 92(11).
Sarbu, S. M., T. C. Kane, and B. K. Kinkle. 1996. A chemoautotrophically based cave ecosystem. Science 272:1953-1955.
Seckbach, J. (Ed.). 1999. Enigmatic Microorganisms and Life in Extreme Environments. Dordecht, Netherlands: Springer.
Smiddy, M., R. D. Sleator, M. F. Patterson, C. Hill, and A. L. Kelly. 2004. Role for compatible solutes glycine betaine and L-carnitine in Listerial barotolerance. Appl Environ Microbiol 70(12):7555-7557.
Solden, L., K. Lloyd, and K. Wrighton. 2016. The bright side of microbial dark matter: Lessons learned from the uncultivated majority. Curr Opin Microbiol 31:217-226.
Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: The unseen majority. Proc Natl Acad Sci USA 95:6578-6583.
Wilkins, M. J., N. C. VerBerkmoes, K. H. Williams, S. J. Callister, P. J. Mouser, H. Elifantz, A. L. N’Guessan, B. C. Thomas, C. D. Nicora, M. B. Shah, P. Abraham, M. S. Lipton, D. R. Lovley, R. L. Hettich, P. E. Long, and J. F. Banfield. 2009. Proteogenomic monitoring of Geobacter physiology during stimulated uranium bioremediation. Appl Environ Microbiol 75:6591-6599.
Wilkins, M. J., R. A. Daly, P. J. Mouser, R. Trexler, S. Sharma, D. R. Cole, K. C. Wrighton, J. F. Biddle, E. Denis, J. K. Fredrickson, T. L. Kieft, T. C. Onstott, L. Petersen, S. M. Pfiffner, T. J. Phelps, and M. O. Schrenk. 2014. Trends and future challenges in sampling the deep terrestrial biosphere. Front Microbiol 5:481.
Wrighton, K. C., B. C. Thomas, I. Sharon, C. S. Miller, C. J. Castelle, N. C. VerBerkmoes, M. J. Wilkins, R. L. Hettich, M. S. Lipton, K. H. Williams, P. E. Long, and J. F. Banfield. 2012. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337:1661-1665.
Wrighton, K. C., C. J. Castelle, M. J. Wilkins, L. A. Hug, I. Sharon, B. C. Thomas, K. M. Handley, S. W. Mullin, C. D. Nicora, A. Singh, M. S. Lipton, P. E. Long, K. H. Williams, and J. F. Banfield. 2013. Metabolic interdependencies between phylogenetically novel fermenters and respiratory organisms in an unconfined aquifer. ISME J 8:1452-1463.
Wrighton, K. C., C. J. Castelle, C. A. Varaljay, S. Satagopan, C. T. Brown, M. J. Wilkins, B. C. Thomas, I. Sharon, K. H. Williams, F. R. Tabita, and J. F. Banfield. 2016. RubisCO of a nucleoside pathway known from Archaea is found in diverse uncultivated phyla in bacteria. ISME J 10:2702-2714.
Zeng, X., J. L. Birrien, Y. Fouquet, G. Cherkashov, M. Jebbar, J. Querellou, P. Oger, M. A. Cambon-Bonavita, X. Xiao, and D. Prieur. 2009. Pyrococcus CH1, an obligate piezophilic hyperthermophile: Extending the upper pressure-temperature limits for life. ISME J 3:873-876.










