11
Talking with Molecules: Marine Bacteria and Microalgae
INTRODUCTION
For the past century, the investigation of microbes has primarily focused on laboratory-isolated strains grown in nutrient-rich cultures. This approach has been very fruitful, and has delivered invaluable insights into bacterial characteristics like metabolism, genetics, behavior, proliferation, and motility. However, it hardly represents how microbes grow and evolve in the wild. In almost all natural environments, such as soil or the human gut, bacteria are surrounded by a plethora of other microbes in a constant battle for nutrients and survival. Given how important microbial interactions are for human health and the environment, it is imperative that we understand the molecular principles that drive these multilateral interactions. It is, at the same time, an exceedingly difficult task, and appropriate model systems that can inform on more complex interactions are necessary. We have been working on one such model system, which involves a naturally occurring symbiosis in the marine environment between microalgae and bacteria. This commentary will focus on the chemical language that these two symbiotic partners use to communicate with each other, including the chemicals that are exchanged, as well as their functions.
One of the microalgae we have been investigating is Emiliania huxleyi, a single-celled microorganism, 5-10 μm across, and abundant in all the world’s oceans. It forms massive seasonal blooms that are easily visible from outer space. Because of the sheer size of these blooms, which can cover areas as large as 3×105 km2, E. huxleyi significantly contributes to global biogeochemical cycles, for example, by generating O2 during photosynthesis or synthesizing large amounts of the climatically active nutrient dimethylsulfoniopropionate (DMSP) (Holligan et al., 1983, 1993; Balch et al., 1991, Wolfe et al., 1997). Another obvious and spectacular manifestation of the global environmental consequences of E. huxleyi blooms are the White Cliffs of Dover at the southern tip of England, a mountain range that largely consists of algal coccoliths and calcium carbonate shells, that have been deposited by microalgae, including E. huxleyi, over millions of years.
In the laboratory, E. huxleyi has been shown to interact with members of the roseobacter clade of marine bacteria (Segev et al., 2015, 2016). The roseobacter are a well-defined clade within the alpha-proteobacteria (Buchan et al., 2005; Wagner-Döbler and Biebl, 2006; Geng and Belas, 2010). They are abundant in all the oceans, especially in coastal areas where they constitute nearly 20% of bacterial communities, increasing to 60% during algal blooms. They have diverse primary and secondary metabolisms as well as host-associated lifestyles; studies
___________________
a Departments of Chemistry and Molecular Biology, Princeton University.
* Corresponding Author: mrseyed@princeton.edu.
SOURCE: Wang et al., 2016.
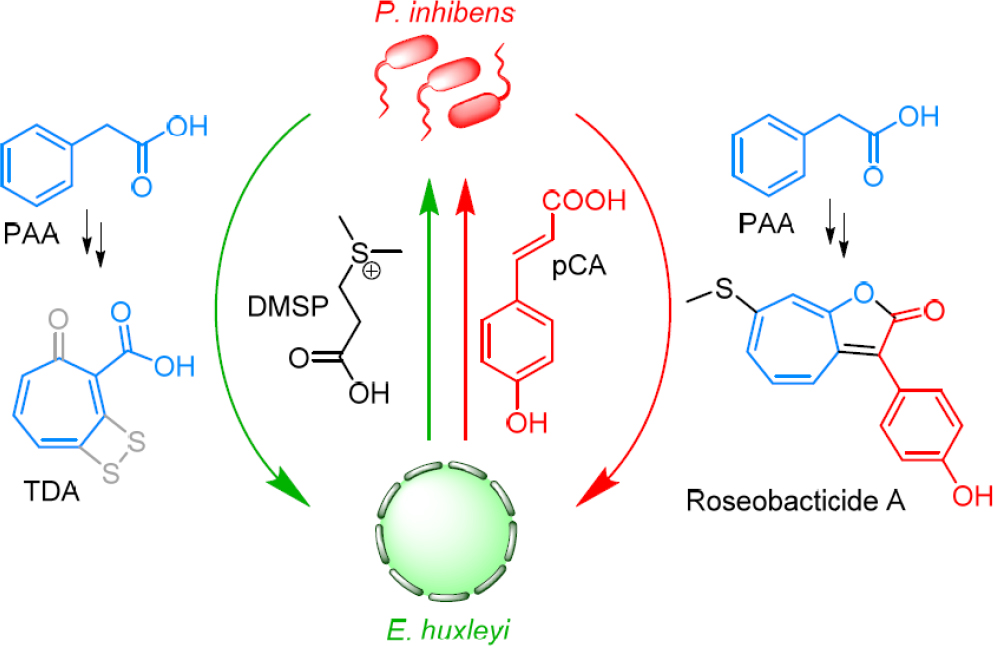
over a number of years have shown that E. huxleyi and roseobacter produce molecules that facilitate a mutualistic symbiosis. In this model, E. huxleyi provides DMSP, which the bacteria can use as a sole source of carbon and sulfur (Gonzalez et al., 1999; Newton et al., 2010), and an attachment surface. In return, Phaeobacter inhibens, a representative roseobacter, produces auxin phenylacetic acid, which promotes algal growth and health, and the antibiotic tropodithietic acid (TDA), which protects the microalgal–bacterial assembly from unwanted marine pathogens (Brinkhoff et al., 2004; Thiel et al., 2010; Seyedsayamdost et al., 2011b; Wilson et al., 2016; see Figure 11-1, green arrows).
A BACTERIAL MUTUALIST-TO-PARASITE SWITCH
More recently, we have shown that this mutualistic symbiosis model is incomplete, and that there is a parasitic phase during which the bacteria turn on the algal host (Seyedsayamdost et al., 2011b). E. huxleyi cultures in the lab produce p-coumaric acid (pCA) with increasing cell densities; the function of this molecule to algal growth is unknown, although brown and green algal strains have also been shown to secrete phenylpropanoids, molecules that are similar to pCA (Delwiche et al., 1989; Martone et al., 2009; Espineira et al., 2011). We imagined that pCA might signify an aging algal host to the bacteria, and that they had perhaps evolved to identify and respond to this molecule. To test the hypothesis, we cultured P. inhibens in the presence and absence of pCA, and found that pCA
stimulated the production of a family of compounds characterized by a broad 430 nm peak in the UV-vis spectrum (Seyedsayamdost et al., 2011b). In the absence of pCA, these compounds were not produced by P. inhibens under a variety of conditions examined. These results showed that the new metabolites, which we call roseobacticides (RSBs), were induced only in response to algal pCA and that their production was tightly regulated.
The most abundant RSB, RSB A, was isolated and its structure elucidated via a combination of 1D/2D NMR and X-ray crystallography. We found that RSB A represents a new class of small molecules with a substituted 1-oxaazulan-2-one substructure previously unobserved (Thiel et al., 2010; see Figure 11-1). While troponoloids are common, troponoid natural products are rare with less than a dozen identified to date. RSB A and TDA are both tropone natural products produced by the same bacterial strain (Bentley, 2008; Greer et al., 2008; see Figure 11-1).
What, if any, is the role of RSB A in algal–bacterial symbioses? A series of bioassays showed that RSB A lacked antibacterial, antifungal, and antifouling activities. It did, however, exhibit potent and somewhat specific algaecidal activities (Seyedsayamdost et al., 2011b). Notably, RSB A killed the host E. huxleyi and another algal strain, the cryptomonad Rhodomonas salina, at nM to μM concentrations, but did not display growth-inhibitory effects against three other microalgal strains tested.
That RSB A is a specific algaecide carries important implications for the symbiosis model. We have proposed a mutualist-to-parasite switch to explain RSB production in response to the putative senescence signal, pCA (see Figure 11-1, red arrows). The mode of the interaction may depend on the health of the algal host. When the algae are healthy, beneficial molecules are exchanged between the host and its bacterial symbiont. However, when the host senesces, the interaction changes. Under these conditions, it produces pCA, which triggers the biosynthesis of RSBs that kill the microalgae. This kind of lifestyle switch has been observed in medical microbiology before but is uncommon in environmental microbial interactions.
ROSEOBACTICIDE DIVERSITY
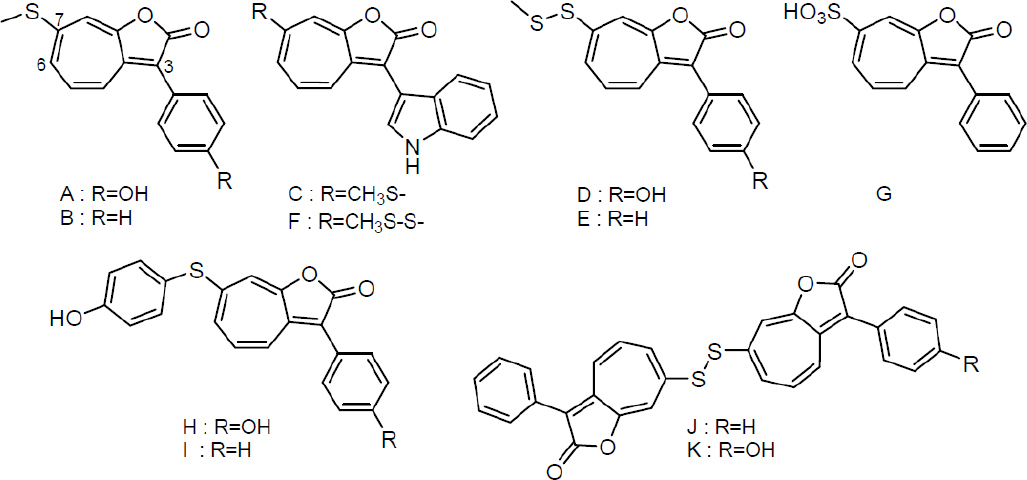
Roseobacter are opportunistic symbionts. P. inhibens interacts with E. huxleyi, but likely associates with other algal cells as well, as has been demonstrated for other members of this genera, and the roseobacter clade as a whole (Buchan et al., 2005; Wagner-Döbler and Biebl, 2006; Geng and Belas, 2010; Wang et al., 2014). Because different phenylpropanoids are produced by varying algal hosts, we examined the effect of sinapic acid, ferulic acid, cinnamic acid, and caffeic acid on RSB production in P. inhibens. Only small effects on the secondary metabolome were observed with the latter two, while sinapic acid and ferulic acid led to the production of a large diversity of roseobacticides (Seyedsayamdost et al., 2011a); we were able to elucidate the structures of 10 additional analogs (see Figure 11-2). Interestingly, we found that the set of RSBs secreted was dependent on the nature of the elicitor: pCA resulted predominantly in production of RSBs A and H, while sinapic acid elicited mostly indole-containing roseobacticides (Seyedsayamdost et al., 2011a). The most abundant variant with cinnamic acid was RSB B. These results suggest that roseobacter may match their RSB output to the host that they are interacting with. That is, phenylpropanoids might represent calling cards specific to an algal host.
METABOLIC ECONOMY IN ROSEOBACTICIDE BIOSYNTHESIS
The large diversity of discovered RSBs also provides clues regarding their biosynthesis. The diversity comes from variable substitutions at the 3 and 7 positions. The substituents at the 3 position—either phenol, phenyl, or indole—suggest that RSBs are aromatic amino acid derived. The substituents at the 7 position are due to variable thiol insertion and/or modification chemistries (see Figure 11-2). All RSBs have the 1-oxaazulan-2-one core, suggesting a specific pathway for its formation.
A series of isotope labeling studies were carried out to identify the precursors for RSB biosynthesis. These experiments surprisingly revealed the auxin, PAA, fragments of the food molecule DMSP, and even the signal pCA—for the phenol-containing derivatives—as RSB precursors (Seyedsayamdost et al., 2014). Accordingly, beneficial molecules in the mutualistic phase are converted into toxins in the parasitic phase of the interaction, a remarkable example of metabolic economy (Seyedsayamdost et al., 2014; see Figure 11-1, note color coding).
SOURCE: Seyedsayamdost et al., 2011a.
This economy likely reflects both the nutrient-poor environment in which the symbiosis occurs and the necessity for a switch-like conversion of bacterial lifestyles, from mutualistic to parasitic in the presence of pCA.
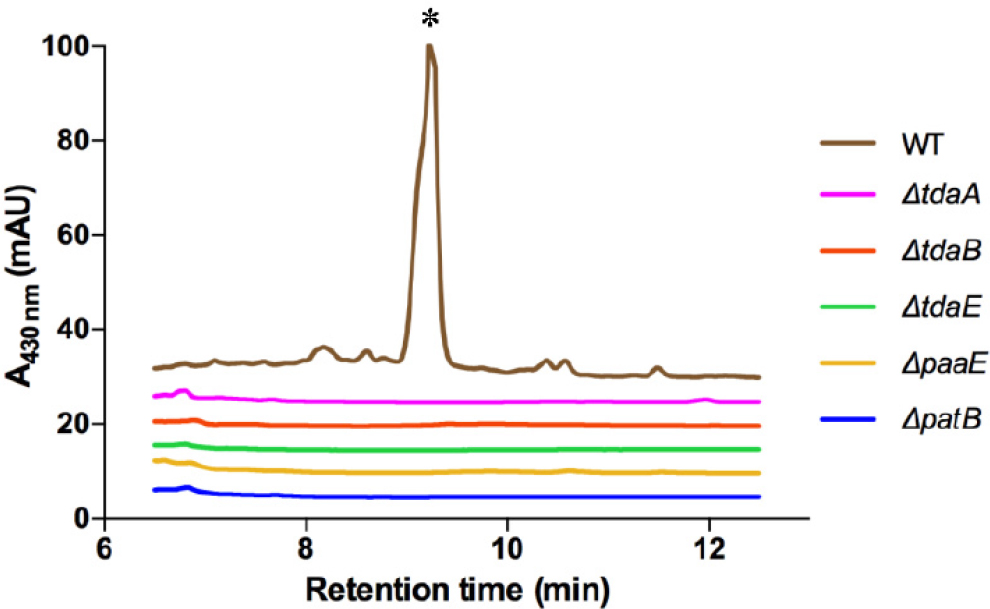
What are the biosynthetic genes responsible for RSB production? This question is usually answered using bio-informatic methods. However, RSBs represent a new chemotype, and, as such, such methods were not sufficient in identifying the responsible genes. To elucidate the RSB biosynthetic operon(s), we instead used a genetic approach (Wang et al., 2016). A transposon mutant library of P. inhibens was generated and arrayed into 96 well plates. The mutants were then tested for their ability to synthesize RSBs using a fluorescence assay. With this approach, 48 unique genes from a library of ~8,500 mutants could be identified (Wang et al., 2016). These genes fall into three operons. The first contains patB, a gene known to be involved in sulfur insertion in TDA biosynthesis (Brock et al., 2014). The second operon is the paa catabolon, which also provides the seven-membered ring precursor for TDA biosynthesis (Berger et al., 2012; Teufel et al., 2012). And lastly, and most intriguingly, the third operon involved is tda, the set of genes responsible for TDA production (Geng et al., 2008). Subsequent experiments have verified that the tda cluster can give rise to two different molecules, TDA and the RSBs (Geng et al., 2008; Wang et al., 2016; see Figure 11-3). Deletion mutants of patB, and paaE—involved in phenylacetate degradation—and tdaA, tdaB, and tdaE—all involved in TDA synthesis—abolish RSB production. To the best of our knowledge, this is the first example of a biosynthetic gene cluster capable of generating two distinct metabolites. Note that RSB and TDA are not mere analogs of each other; they are different molecules with disparate structures and bioactivities produced in different phases of the symbiotic interaction.
With the RSB biosynthetic genes identified, we can now provide an answer regarding the underlying molecular framework that enables the observed metabolic economy, which is the ability to repurpose beneficial molecules to generate RSBs (see Figure 11-1). The key molecular feature is production of two different molecules from the same set of biosynthetic genes. By intertwining RSB and TDA biosyntheses, the bacterium conserves precious nutrients and precursors. Rather than inducing a new pathway with its own set of molecules, P. inhibens diverts an already existing pathway into production of RSBs. This allows the bacteria to be efficient, with respect to precursors, and facilitates a rapid lifestyle switch in response to algal senescence. Our current hypothesis is that TDA
SOURCE: Wang et al., 2016.
and RSB biosynthesis share a very late common intermediate (Wang et al., 2016). Mutualistic conditions result in TDA production from this pathway, while during the parasitic phase, it is diverted to generate RSBs.
QUORUM SENSING–REGULATED ROSEOBACTICIDE PRODUCTION
TDA production is known to be quorum sensing (QS) regulated via the N-acyl homoserine lactone (AHL) signal 3-hydroxdecanoyl homoserine lactone. Given that TDA and RSBs largely share the same set of biosynthetic genes, we hypothesized that production of the latter would also rely on AHL signals. Indeed, an AHL-deletion mutant, in which the signal synthase was removed, failed to synthesize RSBs in response to pCA (Wang et al., 2016). However, when the mutant was supplemented with the AHL signal and pCA, RSB production was restored. These experiments show that RSB synthesis is QS regulated; a sufficient quorum and signal pCA must be present for the bacteria to initiate a lifestyle switch. The requirement of two signals for induction of RSB synthesis in part explains the tight regulation of this process and why no RSBs are observed under numerous conditions examined that lack pCA (Seyedsayamdost et al., 2011b). Future studies will further address the regulatory pathways that are activated in response to pCA.
LESSONS LEARNED FOR COMPLEX SYMBIOSES
With the insights gained into the biology and chemistry of this algal–bacterial symbiosis, we may begin to enumerate the key principles that are relevant to this and perhaps other interspecies and interkingdom interac-
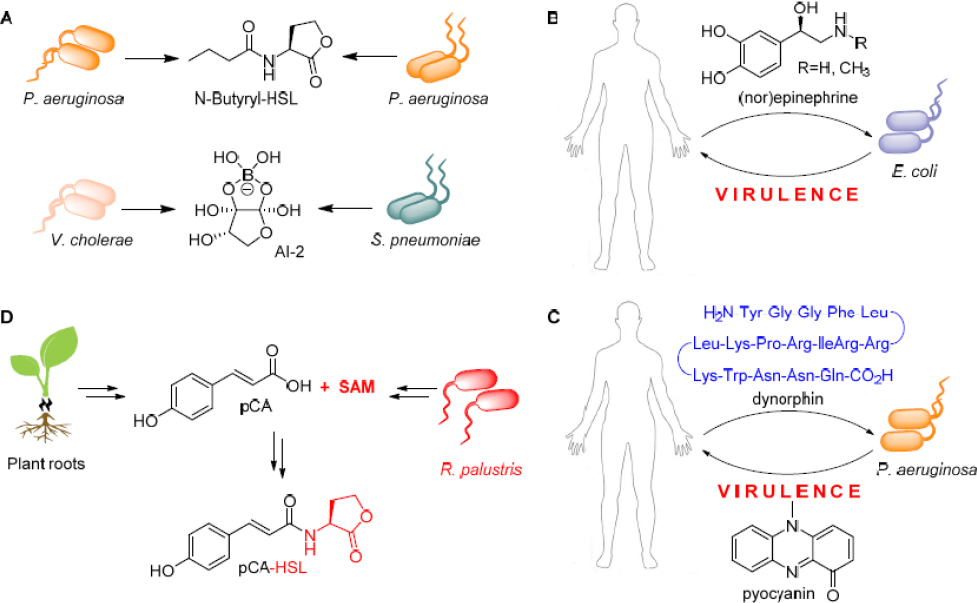
tions. First, the studies on this system show that the exchange of small molecules mediates and modulates the interaction. This is going to be, and perhaps already is, a universal feature of microbial symbioses. Numerous previous studies have already established that bacteria can communicate with their own kind using various diffusible and small signaling molecules. Proteobacteria, for example, commonly use AHLs as QS signals (Fuqua and Greenberg, 2002; Bassler and Losick, 2006; see Figure 11-4A). These signals are not limited to intraspecies communication, they, and other molecules, can mediate interspecies and inter-genus interactions. Even Gram-positive and Gram-negative bacteria have a lexicon, in the molecule AI-2, by which they communicate (Chen et al., 2002; see Figure 11-4A). A major consequence of the use of small molecules as a chemical language is that the study of symbiotic interactions necessitates biological and chemical methods. In the absence of chemical analyses, the “words” cannot be discerned.
A second key aspect of this interaction is its dynamic nature: Multiple phases are involved, each governed by a set of small molecules. Again, we can draw parallels with other systems. It has been shown that some bacteria in the human body can respond to stress signals by mounting an infection. Enterohemorraghic Escherichia coli, for example, turn on virulence and cell-attachment pathways, when they sense the fight-or-flight hormones (nor) epinephrine (Sperandio et al., 2003; see Figure 11-4B). Similarly, Pseudomonas aeruginosa activates virulence factor production in response to the stress peptide dynorphin (Zaborina et al., 2007; see Figure 11-4C). Intermittent or dynamic interactions are likely to be found in other symbioses as well.
A third feature that is not well appreciated is exemplified by RSB biosynthesis, which is synthesized by algal and bacterial precursor molecules. Lifestyle switches in other symbioses may also involve repurposed metabolites and hybrid biosynthetic pathways. We can draw parallels with the plant Rhodopseudomonas palustris interaction, where this kind of phenomenon has been observed (Schaefer et al., 2008). In this case, plant-derived pCA is used to synthesize the bacterial QS signal pCA-HSL, which allows R. palustris to sense its own cell density and the presence of plant roots at the same time (see Figure 11-4D).
Lastly, bacterial QS will likely govern production of chemical signals in other symbiotic interactions as well. Thus, understanding these regulatory processes will also aid the study of microbial symbioses.
REFERENCES
Balch, W. M., P. M. Holligan, S. G. Ackleson, and K. J. Voss. 1991. Biological and optical properties of mesoscale coccolithophore blooms in Gulf of Main. Limnol Oceanogr 36:629.
Bassler, B. L., and R. Losick. 2006. Bacterially speaking. Cell 125:237.
Bentley, R. 2008. A fresh look at natural tropolonoids. Nat Prod Rep 25:118.
Berger, M., N. L. Brock, H. Liesegang, M. Dogs, I. Preuth, M. Simon, J. S. Dickschat, and T. Brinkoff. 2012. Genetic analysis of the upper phenylacetate catabolic pathway in the production of tropodithietic acid by Phaeobacter gallaeciensis. Appl Environ Microbiol 78:3539.
Brinkhoff, T., G. Bach, T. Heidorn, L. Liang, A. Schlingloff, and M. Simon. 2004. Antibiotic production by a Roseobacter clade-affiliated species from the German Wadden Sea and its antagonistic effects on indigenous isolates. Appl Environ Microbiol 70:2560.
Brock, N. L., A. Nikolay, and J. S. Dickschat. 2014. Biosynthesis of the antibiotic tropodithietic acid by the marine bacterium Phaeobacter inhibens. Chem Commun 50:5487.
Buchan, A., J. M. Gonzalez, and M. A. Moran. 2005. Overview of the marine roseobacter lineage. Appl Environ Microbiol 71:5665.
Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545.
Delwiche, C. F., L. E. Graham, and N. Thomson. 1989. Lignin-like compounds and sporopollenin coleochaete, an algal model for land plant ancestry. Science 245:399.
Espineira, J. M., E. Novo Uzal, L. V. Gómez Ros, J. S. Carrión, F. Merino, A. Ros Barceló, and F. Pomar. 2011. Distribution of lignin monomers and the evolution of lignification among lower plants. Plant Biol 13:59.
Fuqua, C., and E. P. Greenberg. 2002. Listening in on bacteria: Acyl-homoserine lactone signaling. Nat Rev Mol Cell Biol 3:685.
Geng, H., and R. Belas. 2010. Molecular mechanisms underlying roseobacter-phytoplankton symbioses. Curr Opin Biotechnol 21:332.
Geng, H., J. B. Bruhn, K. F. Nielsen, L. Gram, and R. Belas. 2008. Genetic dissection of tropodithietic acid biosynthesis by marine roseobacters. Appl Environ Microbiol 74:1535.
Gonzalez, J. M., R. P. Kiene, and M. A. Moran. 1999. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the alpha-subclass of the class Proteobacteria. Appl Environ Microbiol 65:3810.
Greer, E. M., D. Aebisher, A. Greer, and R. Bentley. 2008. Computational studies of the tropone natural products, thiotropocin, tropodithietic acid, and troposulfenin. Significance of thiocarbonyl-enol tautomerism. J Org Chem 73:280.
Holligan, P. M., M. Viollier, D. S. Harbour, P. Camus, and M. Champagne-Philippe. 1983. Satellite and ship studies of cocclithophore production along a continental shelf edge. Nature 304:339.
Holligan, P. M., E. Fernadez, J. Aiken, W. M. Balch, P. Boyd, P. H. Burkill, M. Finch, S. B. Groom, G. Malin, K. Muller, D. A. Purdie, C. Robinson, C. C. Trees, S. M. Turner, and P. van der Wal. 1993. A biogeochemical study of the coccolithophore, Emiliania huxleyi, in the North Atlantic. Global Biogeochem Cycles 7:879.
Martone, P. T., J. M. Estevez, F. Lu, K. Ruel, M. W. Denny, C. Somerville, and J. Ralph. 2009. Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Curr Biol 19:169.
Newton, R. J., L. E. Griffin, K. M. Bowles, C. Meile, S. Gifford, C. E. Givens, E. C. Howard, E. King, C. A. Oakley, C. R. Reisch, J. M. Rinta-Kanto, S. Sharma, S. Sun, V. Varaljay, M. Vila-Costa, J. R. Westrich, and M. A. Moran. 2010. Genome characteristics of a generalist marine bacterial lineage. ISME J 4:784.
Schaefer, A. L., E. P. Greenberg, C. M. Oliver, Y. Oda, J. J. Huang, G. Bittan-Banin, C. M. Peres, S. Schmidt, K. Juhaszova, J. R. Sufrin, and C. S. Harwood. 2008. A new class of homoserine lactone quorum-sensing signals. Nature 454:595.
Segev, E., A. Tellez, H. Vlamakis, and R. Kolter. 2015. Morphological heterogeneity and attachment of Phaeobacter inhibens. PLoS One 10:e0141300.
Segev, E., T. P. Wyche, K. H. Kim, J. Petersen, C. Ellebrandt, H. Vlamakis, N. Barteneva, J. N. Paulson, L. Chai, J. Clardy, and R. Kolter. 2016. Dynamic metabolic exchange governs a marine algal-bacterial interaction. Elife 5:e17473.
Seyedsayamdost, M. R., G. Carr, R. Kolter, and J. Clardy. 2011a. Roseobacticides: Small molecule modulators of an algal-bacterial symbiosis. J Am Chem Soc 133:18343.
Seyedsayamdost, M. R., R. J. Case, R. Kolter, and J. Clardy. 2011b. The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat Chem 3:331.
Seyedsayamdost, M. R., G. Carr, R. Kolter, J. Clardy, and R. Wang. 2014. Hybrid biosynthesis of roseobacticides from algal and bacterial precursor molecules. J Am Chem Soc 136:15150.
Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: The language of hormones. Proc Natl Acad Sci USA 100:8951.
Teufel, R., T. Friedrich, and G. Fuchs. 2012. An oxygenase that forms and deoxygenates toxic epoxide. Nature 483:359.
Thiel, V., T. Brinkhoff, J. S. Dickschat, S. Wickel, J. Grunenberg, I. Wagner-Döbler, M. Simon, and S. Schulz. 2010. Identification and biosynthesis of tropone derivatives and sulfur volatiles produced by bacteria of the marine Roseobacter clade. Org Biomol Chem 8:234.
Wagner-Döbler, I., and H. Biebl. 2006. Environmental biology of the marine Roseobacter lineage. Annu Rev Microbiol 60:255.
Wang, H., J. Tomasch, M. Jarek, and I. Wagner-Döbler. 2014. A dual-species co-cultivation system to study the interactions between Roseobacters and dinoflagellates. Front Microbiol 5:311.
Wang, R., E. Gallant, and M. R. Seyedsayamdost. 2016. Investigation of the genetics and biochemistry of roseobacticide production in the Roseobacter clade bacterium Phaeobacter inhibens. M Bio 7:e02118.
Wilson, M. Z., R. Wang, Z. Gitai, and M. R. Seyedsayamdost. 2016. Mode of action and resistance studies unveil new roles for tropodithietic acid as an anticancer agent and the γ-glutamyl cycle as a proton sink. Proc Natl Acad Sci USA 113:1630.
Wolfe G. V., M. Steinke, and G. O. Kirst. 1997. Graozing-activated chemical defence in a unicellular marine alga. Nature 387:894.
Zaborina, O., F. Lepine, G. Xiao, V. Valuckaite, Y. Chen, T. Li, M. Ciancio, A. Zaborin, E. O. Petrof, J. R. Turner, L. G. Rahme, E. Chang, and J. C. Alverdy. 2007. Dynorphin activates quorum sensing quinolone signaling in Pseudomonas aeruginosa. PLoS Pathog 3:e35.








