3
Non-Rodent Models for Microbiome Research
The microbiome research community has focused most of its efforts on mouse models, but as with other branches of preclinical research, exploring the microbiomes of other species could complement mouse studies and generate new knowledge relevant to humans. The four speakers in the workshop’s first panel session provided perspectives on the benefits and limitations of animal models beyond mice. The four speakers were Buck Samuel, assistant professor in the Alkek Center for Metagenomics and Microbiome Research and the Department of Molecular Virology and Microbiology at Baylor College of Medicine; Angela Douglas, the Daljit S. and Elaine Sarkaria Professor of insect physiology and toxicology at Cornell University; Karen Guillemin, the Alec and Kay Keith Professor in the Department of Biology and the Institute of Molecular Biology at the University of Oregon; and Jeff Gordon, the Dr. Robert J. Glaser Distinguished University Professor and Director of the Center for Genome Sciences and Systems Biology at Washington University in St. Louis. In a second session, three speakers discussed in vitro systems for studying microbiomes. The three speakers on the second panel were Robert Britton, professor in the Department of Molecular Virology and Microbiology at Baylor College of Medicine; Vincent Young, associate professor in the Department of Internal Medicine/Division of Infectious Diseases and Department of Microbiology and Immunology at the University of Michigan; and Donald Ingber, founding director of the Wyss Institute for Biologically Inspired Engineering at Harvard University and Judah Folkman Professor of Vascular Biology at Harvard Medical School.
EUKARYOTIC MODELS
Caenorhabditis elegans (C. elegans)
The strength of C. elegans as a model organism for microbiome research lies in the ability to conduct high-throughput experiments with a gnotobiotic organism and explore the complex cause-or-effect relationship between the presence or absence of a microbial species and a specific state of health or disease. For Samuel, his goal is to identify the pathways that are open to microbial influence and the molecules mediating that influence.
In the laboratory, C. elegans will grow for multiple generations on a nutrient-rich, chemically defined, organism-free medium (Szewczyk et al., 2003). Under these conditions, C. elegans grows 35 times slower and lives twice as long as in the wild, modeling how it and other organisms grow under starvation conditions. Beyond that observation, said Samuel, researchers know little about the artificial germ-free state in C. elegans, though it is clear that peptide uptake and intestinal metabolism are impaired in the germ-free state, while uptake of complex lipids is not. Despite being able to take up lipids, germ-free C. elegans are devoid of fat according to unpublished work from Samuel’s group.
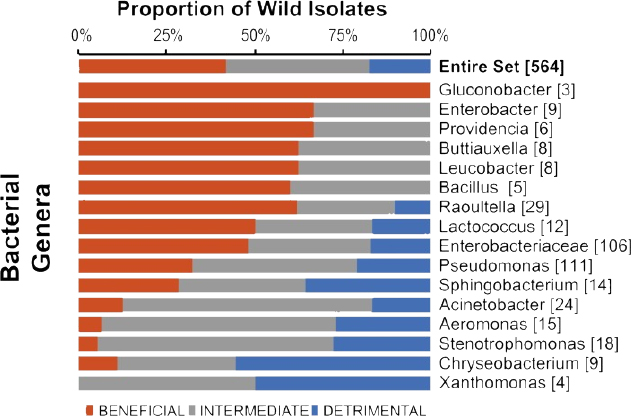
The C. elegans microbiome is relatively simple, comprising 5-15 microbial strains that support approximately 10,000 colony-forming units per healthy animal (Berg et al., 2016; Dirksen et al., 2016; Samuel et al., 2016), with most of the colonization occurring early in adulthood. So far, Samuel and his colleagues have cultivated 564 different organisms from natural C. elegans populations and can now recapitulate communities representing 80 percent of the core operational taxonomic units and 75 percent of the microbial abundance (see Figure 3-1). “We still have some missing taxa that we are interested in, but we definitely have all of the big ones,” said Samuel. In one set of knockdown experiments, Samuel and his collaborators identified new signaling pathways that C. elegans uses to regulate microbiome form and function. Approximately 40 of the actors involved have direct human orthologs, raising the possibility that this simple system will provide new insights into the basic mechanisms that hosts use to regulate their microbiomes.
Drosophila melanogaster
Like C. elegans, Drosophila is experimentally amenable to manipulation and easy to maintain in a germ-free state. The drosophila gut is not anoxic, explained Douglas, and the organisms that live there—predominantly bacteria—are tolerant of oxygen and readily cultured. Recolonizing axenic, or germ-free, flies involves adding a bacterium or collection of microorganisms to the food on which the larvae or adults feed.
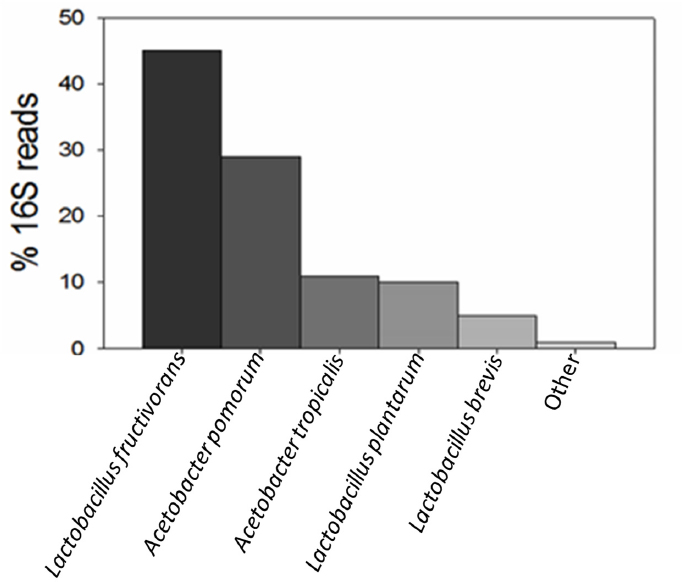
The natural drosophila microbiome is an order of magnitude less diverse than that of mammals, said Douglas, with Acetobacteraceae, Lactobacillales, and γ-Protobacteria being the dominating strains. However, the drosophila microbiome, like that of mice and humans, is inconstant—identical lines of flies grown under the same conditions in two different laboratories will have different microbiome compositions (Chaston et al., 2015; Wong et al., 2013). Characterization of the microbiota in drosophila reared in her laboratory and aggregated across different developmental stages identified five major isolates (see Figure 3-2) that can recapitulate the conventional drosophila phenotype (Wong et al., 2011).
Douglas described an elaborate set of feeding experiments with axenic drosophila as an example of how axenic insects can provide insights on nutritional interactions in the host digestive system. These experiments showed that the gut microbiota, and specifically the lactobacilli, spares drosophila’s dietary requirements for B vitamins, especially riboflavin (Wong et al., 2014). Other experiments found that axenic drosophila are inordinately fat, hyperglycemic, and hyperlipidemic (Ridley et al., 2012). Further investigation showed that one genus in the Acetobacteraceae family, Komagataeibacter, was present in normal drosophila and not in the obese flies, and that these bacteria protected against hyperlipidemia by competing with the host for dietary sugars (Huang and Douglas, 2015). In other recent unpublished work, Douglas and her colleagues have identified a few bacterial taxa and microbial communities that fail to protect against hyperglycemia.
One advantage of working with Drosophila is the ability to harness the wealth of genomic and genetic resources available. “We have tremendous panels of mutants, RNAi lines and so on, readily available from stock centers, and we can make use of the UAS-GAL4 system to exert very precise spatiotemporal control over gene expression,” said Douglas. In addition, she noted, CRISPR tools for genetic manipulation are becoming increasingly sophisticated. Two specific resources her laboratory uses are the Drosophila Genetic Resource Panel (DGRP) of 200 inbred lines with sequenced genomes (Huang et al., 2014; Mackay et al., 2012) and the Drosophila Global Diversity Panel of 84 inbred lines from five continents with sequenced genomes (Grenier et al., 2015). Using DGRP lines, she and her colleagues have found that eliminating the microorganisms in these flies causes some genotypes to become obese, others slightly overweight, and still others become even leaner than normal (Dobson et al., 2015). “We see this genetic variation as an opportunity, not as a problem,” said
Douglas. “It indicates the importance of host genotype as a determinant of microbiota-dependent traits and enables us to apply genome-wide association studies to identify candidate genetic determinants and then, because we have mutants readily available for many of these genes, to validate those genes.” Many of the candidate genes her group identified are expressed in the gut or in neurons; have homologs across the animal kingdom, including in humans and other mammals; and belong to pathways that are highly conserved across the animal kingdom. “I think this reflects the fact that the foundation of the microbiome in animals is very ancient,” said Douglas.
Comparing the transcriptome of axenic and gnotobiotic flies from 17 Drosophila lines, Douglas and her colleagues found that transcriptome-wide co-expression is significantly weaker in the axenic flies than in gnotobiotic flies (Dobson et al., 2016). They also observed the co-expression of pairs of genes differed significantly between axenic and gnotobiotic flies, leading Douglas to conclude that the microbiome promotes co-expression of specific transcriptional modules. “This work is very recent and there is more about this that we do not understand than that we do,” said Douglas. Questions she posed included whether this is a general effect across species and if reduced co-expression is associated with microbiomes that fail to support health.
Douglas said that using invertebrate models such as Drosophila and C. elegans could substantially reduce the need to conduct experiments using rodents while enhancing their scientific quality, given that researchers can use Drosophila, C. elegans, and other non-vertebrate systems to more quickly understand fundamental principles of animal microbial associations and then use mammalian models and human data to verify the relevance to humans. Toward that end, Douglas recommended strengthening the framework and infrastructure for integrating model systems with biomedical and clinical science.
Zebrafish
Zebrafish studies benefit from an abundance of genetic and genomic tools and high-throughput functional assays. As vertebrates, zebrafish offer additional complexity in terms of the kinds of microbial communities they harbor, and their optical transparency provides some unique opportunities to observe dynamic microbial communities in the gut of a living animal. There are challenges to working with zebrafish, including the difficulty of rearing germ-free animals to adulthood, the lack of standardized methods of screening for pathogens, and the lack of knowledge about the nutritional requirements of juvenile and adult animals. Maintaining water quality is an issue, too, because microbes in the natural ecosystem normally play a large part in detoxifying urea and other waste products. Guillemin explained that, while her group and others have accumulated large, curated collections of bacterial isolates and a few species of fungi isolated from zebrafish, there is no defined inoculum for them as there is for mice.
Zebrafish harbor hundreds of different bacterial species and other microorganisms at a level of complexity similar to mammals. Because it is rather straightforward to derive them under germ-free conditions, researchers can build complex microbial communities starting with mono- or di-associations. In one set of experiments, Guillemin and her collaborators found that wild-type and immune-deficient fish raised in isolation had vastly different microbiomes, but the microbial communities converged when the two types of fish were raised in the same tank or when each type of fish was raised in the same tank with others of its genotype. Guillemin said these results suggest that co-housing hosts selects for bacterial members that are transmissible, while raising a host in isolation leads to extinction of the bacterial lineages that have the ecological strategy of moving between hosts. “This has large implications for thinking about how one designs experiments profiling microbiota,” said Guillemin. “The housing conditions can have very profound effects.”
In another study, Guillemin and her colleagues found that Aeromonas introduced into the gut of an axenic zebrafish forms clumps that are readily visible in the transparent fish. When a member of the genus Vibrio is then added to the gut, this highly motile bacterium rapidly displaces the Aeromonas (Wiles et al., 2016). However, in a mutant zebrafish with defective gut peristalsis, the Aeromonas population persisted when Vibrio was introduced. “This is telling us
that the host environment, in this case peristalsis, contributes to the bacteria-bacteria competition,” said Guillemin.
Guillemin has also determined how the microbiome alters zebrafish development (Hill et al., 2016). When zebrafish hatch at three days, they are supported by a single functional islet of pancreatic beta cells. These beta cells double over the next three days, concurrently with the colonization of the patent gut tube by bacteria. In germ-free fish, this doubling does not occur, and consequently, these fish have higher circulating glucose levels relative to the conventionally reared fish. Subsequent experiments identified several Aeromonas strains and a Shewenalla strain that could reverse this effect and that a specific secreted protein—beta cell expansion factor A, or BefA—produced by the bacteria was responsible for stimulating beta cell production. Treating axenic fish with this protein, which has homologs in human-associated bacteria, triggered expansion of beta cells. Guillemin noted in closing that beta cell expansion in humans occurs during the first year of life, concurrent with the establishment of an infant’s gut microbiota.
Piglets
Studying a suspected link between human malnutrition and human gut microbes, Gordon and his collaborators found evidence that immature microbiota are causally related to undernutrition (Blanton et al., 2016; Subramanian et al., 2014, 2015). They also found that Malawian mothers with severely stunted 6-month-old infants had lower levels of sialylated milk oligosaccharides. When tested in mice, these molecules interact with gut microbes to influence growth phenotypes, including lean body mass gain, bone biology, and metabolism. To determine whether these observations in mice apply to a second species whose physiological and metabolic properties are more similar to those of humans, Gordon and his colleagues spent 2.5 years developing a protocol for birthing germ-free piglets and repeating those experiments in this piglet model.
Birthing germ-free piglets requires delivering the piglet directly from the uterus through a sterile plastic tube. Before they take their first breath, the piglets are submersed in a sterile 2 percent chlorhexidine bath and then placed into a sterile, flexible-film nursery isolator, where they are revived and kept on a heated pad until the remaining piglets in the litter are delivered. Within 24 hours, all of the piglets are transferred from these nursery isolators to larger gnotobiotic isolator tubs, with three to four piglets per isolator, in a room that can be thoroughly disinfected prior to the initiation of any experiments. Initially, the piglets are bottle-fed irradiated sow’s milk replacement, and starting at day 4 or 5, they transition to a pelleted diet. The piglets are provided with environmental enrichment, and if they are to be colonized with microbes, that occurs four days after birth by suspending an inoculum in the irradiated sow’s milk replacement.
Using the same sialylated milk oligosaccharide-supplemented diet and the same microbial culture they administered to mice, Gordon and his colleagues were able to replicate in the piglets the growth and metabolic effects they ob-
served in mice (Charbonneau et al., 2016). Gordon noted that the germ-free piglet model provides the opportunity to characterize postnatal assembly of the microbiota during the suckling and weaning transition of a mammal with a rapid growth phenotype. The piglet model also allows for studying microbial ecology and microbiota development relative to features such as biogeography that are challenging to characterize in smaller vertebrate or invertebrate models. “We think this model provides opportunities to develop technologies that are relevant for studying human communities, as well as interactions between microbiota and hosts,” said Gordon. As examples of such technologies, he listed autonomously functioning devices for remote sampling of communities along the length of the gut and implantable devices for measuring metabolism.
There are challenges of working with such a technically demanding, expensive, and time-consuming model. Experiments are limited to the first 30 days after birth, after which time the animals become too large to manage in a controlled environment; the transition to the weaned state requires careful monitoring and husbandry; and the fact that gut function is compromised during weaning in piglets limits studies with communities containing pathogens. There are also fewer analytic reagents available for the pig. Nonetheless, Gordon said he is enthusiastic about this model for select purposes, and particularly as a second species in a translational medicine pipeline for both proof of concept and mechanistic studies.
IN VITRO SYSTEMS FOR CHARACTERIZING MICROBIAL CONSORTIA
As a means of studying a microbiome’s complex microbial community at both the structural and the functional level, investigators are developing a variety of in vitro systems, including the miniature bioreactor arrays that Britton and his colleagues have created to study how microbial communities resist invasion by pathogens without the need to use mice. Other investigators have developed complex three-vessel bioreactor systems (Freeman et al., 2003; Macfarlane et al., 1998), five-vessel systems such as the Simulator of the Human Intestinal Microbial Ecosystem (Molly et al., 1993), and the ROBOGUT system, used to produce defined microbial communities to treat people with recurrent Clostridium difficile infection (Petrof et al., 2013).
Britton’s miniaturized bioreactor system uses reaction chambers crafted from commercially available plastic blocks that can be autoclaved, are small enough to fit in an anaerobic chamber, and can be combined in an array of up to 96 bioreactors. Peristaltic pumps feed media into and force waste out of the continuously stirred chambers. Tests using fecal matter from three donors showed that this apparatus could produce stable, distinct microbial communities within three to seven days (Auchtung et al., 2015). The relative proportions of the dominant phyla in the bioreactor-produced communities were similar to those of the original fecal samples. While certain phyla significantly recede and others be-
come more abundant than in fecal samples, Britton believes this system captures approximately half of the species that initially go into the bioreactors.
To test if this system could model what happens in the human gut, Britton and his collaborators conducted an experiment in which they treated some of the reactors with water (controls) and others with clindamycin, an antibiotic clinically associated with C. difficile infection in hospitals, followed by inoculation with C. difficile. In the water-treated reactors C. difficile could not compete and was washed away. It was able, however, to establish a stable invasion in the clindamycin-treated bioreactors, and by day 14 this pathogen was producing toxins and spores. Subsequent experiments showed that as few as 150 cells of C. difficile would produce a stable invasion. Britton noted that treatment with clindamycin does not affect the total mass of bacteria growing in the reactors, only the community composition. His group is now trying to determine if they can introduce specific bacteria or bacterial communities to reverse the C. difficile invasion.
Other uses for the bioreactor array include studying microbiota-driven drug metabolism, microbiota production of beneficial and detrimental metabolites, and how defined microbial consortia form from purified strains of bacteria. Britton and his collaborators are also using the bioreactor array to establish microbial communities from body sites other than the gut and to grow hard-to-cultivate microbes. Going forward, he plans to develop an interface between this device and human enteroids, grown from autopsy tissue, and organoids, produced from induced human pluripotent stem cells or embryonic stem cells, as an approach to introducing a host component into the system and to explore ways of establishing niches inside the bioreactors.
Organoids
Young and others are using human intestinal organoids to study the relationship between pathogen and host. Intestinal organoids grown from either human induced pluripotent or embryonic stem cells have both mesenchyme and epithelium (Wells and Spence, 2014), explained Young, whereas enteroids produced from autopsy tissue only have epithelium (Sato et al., 2011). Intestinal organoids have a brush border, microvilli, endocrine cells, lysozyme-producing cells, cells that resemble Paneth cells, and goblet cells that produce mucus, all in a stable matrigel environment (Spence et al., 2011). These organoids are sterile, and they have a functional epithelial barrier.
Using organoids produced from human embryonic stem cells, Young and his colleagues have shown that the C. difficile toxin disrupts the endothelial barrier within six to eight hours after injection into the interior of the organoid by disrupting the cytoskeleton of the endothelial cells (Leslie et al., 2015). Repeating this experiment with C. difficile itself produced the same results over 12 hours, whereas introducing a strain that does not produce toxin had no effect on barrier function. What was surprising about these experiments, said Young, was that C. difficile, an anaerobe, was able to grow in what he assumed was an aero-
bic environment, and further examination uncovered the reason. It turned out that there was an E. coli contaminant in one of the pieces of equipment, and E. coli reduced the percentage of oxygen in the organoid from 21 percent to approximately 8 percent, low enough to allow C. difficile to grow. Additional experiments with E. coli alone showed that it induced increases in mucus expression and changes in epithelial cell gene expression corresponding to changes in the types of complex carbohydrates these cells produce (Finkbeiner et al., 2015), suggesting that these organoids can be used to study the molecular details of host-microbe interactions. Young noted that the enhanced mucus production is similar to what happens when a human fetal small intestine is first exposed to bacteria.
Young and his collaborators are now looking at ways of monitoring and manipulating the oxygen level in intestinal organoids to facilitate the study of other anaerobic bacteria that may be even more sensitive than C. difficile to oxygen. He believes that, while most of the research conducted so far by his group and others has focused on pathogens, such as Helicobacter pylori (Huang et al., 2015; Sigal et al., 2015), Salmonella (Forbester et al., 2015; Höner zu Bentrup et al., 2006), and rotavirus (Finkbeiner et al., 2012; Yin et al., 2015), organoids offer the opportunity to examine how mutualistic organisms interact with intestinal tissues.
Other avenues of future research will include introducing increasing complexity to the system. Young and his collaborators, for example, have run some experiments in which they observed immune cells homing in on organoids with bacteria but not to those in the same system that have not been colonized. Recent papers from other groups have reported success at triggering development of an enteric nervous system as part of the organoids (Schlieve and Grikscheit, 2017; Workman et al., 2017), though Young questioned how much additional complexity will prove to be too much. “At which point are we trying to build a mouse or a person?” he asked. “What we need to figure out with these organoid model systems is where they actually fit.”
Human Organs on Microfluidic Chips
A major issue affecting the drug development enterprise, said Ingber, is that most animal studies do not predict results in human clinical trials, at least in part because animal models lack the human microbiome. To address this problem, he and his colleagues at the Wyss Institute are engineering microchips containing living human cells that reconstitute organ-level functions to accelerate drug development and replace animal testing.
Manufacturing microchips using well-developed photolithographic etching allows control of various features in biocompatible materials at the size scale of living cells (Chen et al., 1997; Singhvi et al., 1994). Ingber and his collaborators’ first major success with this approach involved using a functional alveolus on a microchip to observe the human inflammatory response to bacteria at high resolution (Huh et al., 2010) and study pulmonary edema and drug toxicity. They have
since built a small airway on a chip, complete with differentiated bronchiolar epithelial cells and beating cilia (Benam et al., 2016a,b), and are using it to study influenza virus infection, chronic obstructive pulmonary disease using cells from affected patients, and the effect of cigarette smoke on lung tissues.
Ingber’s team has also created what he calls a peristaltic human gut-on-a-chip that re-creates the human intestine, complete with fully developed intestinal villi with mucus-producing cells, endocrine cells, Paneth cells, and cytochrome P450-based drug metabolism (Kim and Ingber, 2013). They have used this system to culture a probiotic Lactobacillus found in human intestines and have confirmed that it improves barrier function. They have also cultured a commercial probiotic formulation containing eight different microbes. Gene microarray data showed that this mixture totally changes the phenotype of the human gut epithelium to resemble that of distal human ileum, the one place in the small intestine where microbes are found (Kim et al., 2016a). In contrast, a pathogenic strain of E. coli completely overgrows the villi instead of merely growing in the spaces between the villi, which is what the probiotic species do. In addition, flowing peripheral blood mononuclear cells through the vascular channel underlying the intestinal tissue triggers the type of injury response associated with pathogenic E. coli and inflammatory bowel disease (Kim et al., 2016b). These studies have identified four combinatorial therapeutic targets and shown that the commercial probiotic could partially protect against injury induced by invasive E. coli.
A new project in Ingber’s laboratory uses human, mouse, pig, and Xenopus gut-on-a-chip devices to study host tolerance to infection. His group has also developed a method for creating primary human small and large intestines and colons on a chip, as well as microfluidic chip-based models of the skin, liver, heart, kidney, brain, and blood-brain barriers. These organs and chip-based models create integrated human body-on-chips that remain coupled and functioning for up to three weeks.
In closing, Ingber said he believes organs-on-chips have the potential to gradually replace animal testing in drug development. He noted that the U.S. Food and Drug Administration, which has provided substantial funding for this work, has said it will accept data from these systems as long as Ingber and his collaborators can demonstrate that the data are as good or better than the data from animal models. His group has already demonstrated the robustness of the organs-on-chips, and therefore Ingber’s next step would be to obtain primary and induced pluripotent stem cells and microbiome samples from individual patients as a means of creating personalized medicine approaches to treating disease.










