The Roles of Machine Learning in Biomedical Science
KONRAD PAUL KORDING, ARI S. BENJAMIN, ROOZBEH FARHOODI
University of Pennsylvania
JOSHUA I. GLASER
Northwestern University
While the direct goal of biological modeling is to describe data, it ultimately aims to find ways of fixing systems and enhancing understanding of system objectives, algorithms, and mechanisms. Thanks to engineering applications, machine learning is making it possible to model data extremely well, without using strong assumptions about the modeled system. Machine learning can usually better describe data than biomedical models and thus provides both engineering solutions and an essential benchmark. It can also be a tool to advance understanding.
Using examples from neuroscience, we highlight the contributions, both realized and potential, of machine learning, which is becoming easy to use and should be adopted as a critical tool across the full spectrum of biomedical questions.
INTRODUCTION
The goal of nearly all of computational biology is to numerically describe a system, which is often quantified as the explained variance. In some cases, only the explained variance is of interest—for example, to make predictions. But in most cases, just describing the data successfully is not sufficient. There has been much discussion of objectives in the neuroscience community (e.g., Dayan and Abbott 2001; Marr 1982).
Uses of Models
The typical model is designed not only to numerically describe data but also to meet other objectives of the researcher. In some cases it is used to inform how to fix things—to predict what would happen based on certain interventions. In
others, the goal is to determine whether the system optimizes some objective; for example, whether the intricate folds of the brain minimize wiring length (Van Essen 1997). Or the aim may be to understand the system as an algorithm—for example, which algorithms the brain uses to learn (Marblestone et al. 2016). Probably most commonly, a model is used to understand underlying mechanisms—for example, how action potentials are enabled by interactions between voltage-dependent ion channels (Hodgkin and Huxley 1952).
So far, progress in the modeling field comes mostly from human insights into systems. People think about the involved components, conceptualize the system’s behavior, and then build a model based on their intuitive insights. This has been done for neurons (Dayan and Abbott 2001), molecules (Leszczynski 1999), and the immune system (Petrovsky and Brusic 2002).
Biomedical researchers are starting to use computational models both to describe data and to specify the underlying principles. However, understanding such complex systems is extremely difficult, and human intuition is bound to be incomplete in systems with many nonlinearly interacting pieces (Jonas and Kording 2017).
What Is Machine Learning?
The vast field of machine learning is a radically different way of approaching modeling that relies on minimal human insight (Bishop 2006). We focus here on the most popular subdiscipline, supervised learning, which assumes that the relationship between the measured variables and those to be predicted is in some sense simple (Wolpert 2012), with characteristics such as smoothness, sparseness, or invariance.
Supervised algorithms receive vectors of features as inputs and produce predictions as outputs. Machine learning techniques mostly differ by the nature of the function they use for predicting (Schölkopf and Smola 2002). Rather than assuming an explicit model about the relationship of variables, ML techniques assume a generic notion of simplicity.
The field of machine learning is undergoing a revolution. It has moved from a niche discipline to a major driver of economic activity over the past couple of decades as progress revolutionizes web searching, speech to text, and countless other areas of economic importance. The influx of talent into this field has led to massive improvements in algorithm performance, allowing computers to outperform humans at tasks such as image recognition (He et al. 2015) and playing Go (Silver et al. 2016). These developments in machine learning promise to make it an important tool in biomedical research. Indeed, the number of ML-related papers and patents in biomedical research has grown exponentially (Figure 1).
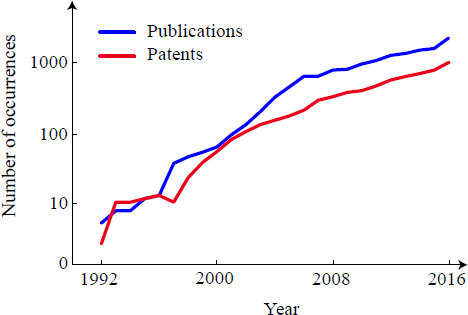
USES OF MACHINE LEARNING FOR BIOMEDICAL RESEARCH
Many kinds of questions can be answered using machine learning techniques. In some cases they are useful for predictions, such as whether a drug will cure a particular cancer. In others they set a benchmark—for example, what are the shortcomings of the human-thought-out model relative to what may be possible? In yet other cases machine learning may enhance understanding of a system by revealing which variables are shared between components of a system.
Description and Prediction
The standard use for machine learning is to make a prediction based on something that can be measured. For example, in psychiatric medicine, studies have used smartphone recordings of everyday behaviors (e.g., when patients wake up or how much they exercise) to predict mood using machine learning (Wang et al. 2014).
A typical problem in neuroscience is the decoding of neural activity (Velliste et al. 2008) to infer intentions from brain measurements. This application is useful for developing interactive prosthetic devices, in which one uses measurements from the brain of a paralyzed subject to enable a robot to execute the movement. Many algorithms have been developed to solve such problems (Corbett et al. 2012; Yu et al. 2007); for this application, general purpose machine learning tends to do extremely well (Glaser et al. 2017).
Computationally similar problems exist throughout biomedical research, in areas such as cancer (Kourou et al. 2015), preventive medicine (Albert et al.
2012), and medical diagnostics (Foster et al. 2014). In these areas only the quality of the predictions is of interest. Similarly, many engineering problems are mainly concerned with the error size of predictions. When the main goal is to obtain accurate predictions, it is best to first try machine learning methods.
Benchmarking
Often the goal is not only to describe and predict data but also to produce models that can be readily understood and taught. Machine learning can be extremely useful by providing a benchmark.
One problem when evaluating a model is that it is hard to know how much its errors are due to noise versus the insufficiency of the model. Because machine learning is a useful tool for making predictions, it may provide close to an upper bound for human-produced models. If a human-generated model produces results that are very different from the ML benchmark, it may be because important principles are missing or because the modeling is misguided. If, on the other hand, a model based on human intuition is very close to the ML benchmark, it is more likely that the posited concepts are, indeed, meaningful.
But how is it possible to know whether a model is missing important aspects? We argue that ML benchmarking can help answer those questions (Benjamin et al. 2017).
Understanding
Machine learning can also directly help understanding. One important question is whether a system carries information about some variables (for example, whether neural activity contains information about an external stimulus), but it may not be clear whether the relation between the variables is linear or nonlinear. With machine learning it is possible to determine whether information is contained in a signal without having to specify the exact nature of the relationship.
Another important question concerns the information shared between two parts of a system. For example, which aspects of the world (high dimensional) are shared with which aspects of the brain (also high dimensional)? Machine learning makes it possible to ask such questions in a well-defined way (Andrew et al. 2013; Hardoon et al. 2004).
For many questions in biology, machine learning promises to enable new approaches to enhance understanding.
MACHINE LEARNING: A NECESSITY FOR EVER-GROWING DATASETS
Datasets are rapidly growing and becoming more and more complex as they become multimodal and multifaceted (Glaser and Kording 2016). In neuroscience,
the number of simultaneously recorded neurons is increasing exponentially (Stevenson and Kording 2011), as is the amount of electronic health record data (Shortliffe 1998).
Challenges in Modeling for Complex Datasets
There are several ways in which these changes in datasets will create new problems for modeling. First, we humans are not very good at thinking about complex datasets. We can only consider a small hypothesis space. But in biology, as opposed to physics, there are good reasons to assume that truly meaningful models must be fairly complex (O’Leary et al. 2015). While humans will correctly see some structure in the data, they will miss much of the actual structure. It could be argued that it is nearly impossible for humans to intuit models of complex biological systems.
Second, nonlinearity and recurrence make it much more difficult to model complex systems (O’Leary et al. 2015), which require complex models. It can be hard to falsify models that are very expressive or have many free parameters. One needs to both explain complexity and ensure that the model will fail if the causal structure is dissimilar to the model. For full-cell interactions or full-brain modeling, the design of models that strike this delicate balance seems implausible.
Finally, in the case of the large complex systems that are characteristic of biology, a major problem is the lack of understanding of how many different models could in principle describe the data. Models can explain some portion of the variance, but not necessarily the mechanism (Lazebnik 2002). Comparing models is pointless if they are not good at describing the relevant mechanisms.
Given all these arguments, it may be that physics-based non-ML-based approaches can only partially succeed. Any reasonably small number of principles can describe only part of the overall variance (and potentially a relatively small part). It is unclear how far the typical approach in biomedical research, drawing on concepts of necessity and sufficiency, can help to enhance understanding of the bulk of activity in complex interacting systems (Gomez-Marin 2017). Machine learning has the potential to describe a very large part of the variance.
A Note about Model Simplicity and Complexity
Machine learning also changes the objectives of data collection. In traditional approaches, measuring many variables is unattractive as, through multiple comparison testing corrections, it is not possible to say much about each of them. But with machine learning, using many variables improves predictions—even if it is not clear which variables contribute—making it attractive to record many variables.
This is not just a vacuous statement about information processing. It reflects the fact that the brain and other biological systems are not simple, with few
interactions, but highly recurrent and nonlinear. The assumption of simplicity in biology is largely a fanciful, if highly convenient, illusion. And if the systems subject to machine learning are not simple, then biases toward simple models will not do much good.
Based on their intuitions, researchers are starting to fit rather complex models to biological data, and those models usually fit the data better than simpler models. However, a complex model based on a wrong idea may fit the data extremely well and thus negate the advantage of an interpretable model.
A good fit does not mean that the model is right. For example, Lamarckian evolution explains a lot of data about species, but it was based on a fundamentally misleading concept of causal transmission of traits. The problem of apparent fit affects human intuition–based models, but not ML models, which, by design, do not produce a meaningful causal interpretation.
SPECIALIST KNOWLEDGE NOT NECESSARY FOR MACHINE LEARNING
There are countless approaches in machine learning, certainly more than most biomedical researchers have time to learn. Kernel-based systems such as support vector machines are built on the idea of regulating model complexity (Schölkopf and Smola 2002). Neural networks are built on the idea of hierarchical representations (Goodfellow et al. 2016). Random forests are built on the idea of having many weak learners (Breiman 2001). One could easily fill books with all the knowledge about machine learning techniques.
Yet the use of ML techniques has actually become very simple. At application time, one requires a matrix of training features and a vector of the known labels. And given the availability of the right software packages (Pedregosa et al. 2011), generally only a few lines of code are needed to train any ML system.
Moreover, ensemble methods obviate the need to choose a single machine learning technique (Dietterich 2000). The idea is that a system can run all techniques and then combine their predictions using yet another ML technique. Such approaches often win ML competitions (e.g., kaggle.com).
Furthermore, a new trend has developed rapidly in the past few years: automatic machine learning (Guyon et al. 2015). The idea is that most ML experts do similar things: they choose one of a number of methods (or all of them if they use ensembling) and then optimize the hyperparameters of those techniques. They may also optimize the feature representation. Although this can take a significant amount of time via trial and error, the process is relatively standard and several new packages allow automation of some or all of it.1
___________________
1 Examples of these packages are available at https://github.com/automl/auto-sklearn, www.cs.ubc.ca/labs/beta/Projects/autoweka/, https://github.com/KordingLab/spykesML, and https://github.com/KordingLab/Neural_Decoding.
These developments are likely to pick up speed in the next year or two, making it less necessary for biomedical scientists to know the details of the individual methods and freeing them to focus on the scientific questions that machine learning can answer.
EXAMPLES OF STATE-OF-THE-ART MACHINE LEARNING IN NEUROSCIENCE
With examples from neuroscience we illustrate two uses of ML approaches, predictions and benchmarking.
Neural Decoding
In neural decoding the aim is to estimate subjects’ intentions based on brain activity—for example, to predict intended movements so that they can move an exoskeleton with their thoughts. A standard approach in the field is still the use of simple linear techniques such as those used in the Wiener filter, in which all previous signals during a given time period are linearly combined to predict the output.
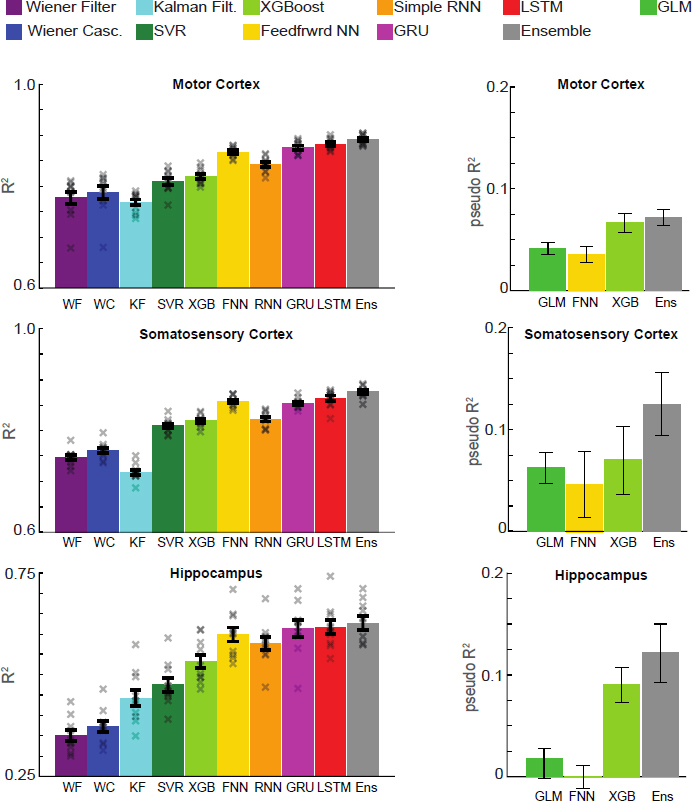
There has recently been a lot of interest in improving this and similar approaches using modern machine learning. For many applications the goal is simply good performance. To analyze the advantages of using standard machine learning, we implemented many approaches: the linear Wiener filter, the nonlinear extension called the Wiener cascade, the Kalman filter, nonlinear support vector machines, extreme gradient–boosted trees, and various neural networks (Figure 2).
The modern neural network–based techniques did very well (Glaser et al. 2017), and a combination of all the techniques, using ensemble methods, performed even better. The same phenomenon was seen when decoding from different brain regions. Thus we conclude that, to solve biomedical engineering problems, use of standard machine learning should be the starting point.
In this sense, machine learning also sets a benchmark for other decoding approaches. When neuroscientists write decoding algorithms they are often based on their insights into the way the brain works (Corbett et al. 2012). However, without a comparison to modern machine learning, it is not possible to know whether or to what extent these insights are appropriate.
As machine learning becomes automatic and easy to use, we argue that it should always be used as a benchmark for engineering applications.
Neural Encoding
Neural encoding, or tuning curve analysis, involves the study of signals from a neuron or a brain region to understand how they relate to external variables.
Such a characterization can yield insights into the role of a neuron in computation (Jonas and Kording 2017).
Typically, the neuroscientist chooses a model (often implicitly) and the average signal is plotted as a function of external variables such as visual stimuli or movements. This approach generally assumes a simple model. Would machine learning give better results?
For such applications it is impossible to know whether poor model performance is due to external variables unrelated to neural activity or to the choice of model form. In principle, input variables may affect the neuron’s activity in highly nonlinear ways. This hypothesis can be tested with machine learning.
When we compared the generalized linear model (GLM; Pillow et al. 2008), it performed considerably worse than neural networks or extreme gradient–boosted trees (Figure 2). And again, the combination of all the methods using ensemble techniques yielded the best results. It can be difficult to guess features that relate to neural activity in exactly the form specified by the GLM.
Interestingly, despite the fact that the space was rather low dimensional, GLMs performed poorly relative to modern machine learning. This may suggest that the tuning curves measured by neuroscientists are rather poor at describing neurons in real-world settings.
In this context, machine learning can conceptually contribute in the following ways:
- It can detect that a variable is represented, even if there is no linear correlation.
- It can set a benchmark that humans can strive for.
- It offers a possibility of replacing the common cartoon model of neural computation with a complex (although admittedly hard to interpret) alternative.
CONCLUSION
The incorporation of machine learning has profound implications for neuroscience and biomedical science.
For biomedical modeling, traditional modeling and machine learning cover opposite corners. Traditional modeling leads to models that can be compactly communicated and taught, while explaining only a limited amount of variance. Machine learning modeling explains a lot of variance, but is difficult to communicate. The two types of modeling can inform one another, and both should be used to their maximal possibility.
REFERENCES
Albert MV, Kording K, Herrmann M, Jayaraman A. 2012. Fall classification by machine learning using mobile phones. PLoS One 7:e36556.
Andrew G, Arora R, Bilmes J, Livescu K. 2013. Deep canonical correlation analysis. Proceedings of Machine Learning Research 28(3):1247–1255.
Benjamin AS, Fernandes HL, Tomlinson T, Ramkumar P, VerSteeg C, Miller L, Kording KP. 2017. Modern machine learning far outperforms GLMS at predicting spikes. bioRxiv 111450.
Bishop C. 2006. Pattern Recognition and Machine Learning. New York: Springer.
Breiman L. 2001. Random forests. Machine Learning 45:5–32.
Corbett EA, Perreault EJ, Kording KP. 2012. Decoding with limited neural data: A mixture of time-warped trajectory models for directional reaches. Journal of Neural Engineering 9:036002.
Dayan P, Abbott LF. 2001. Theoretical Neuroscience: Computational and Mathematical Modeling of Neural Systems. Cambridge: MIT Press.
Dietterich TG. 2000. Ensemble methods in machine learning. Proceedings of the First International Workshop on Multiple Classifier Systems, June 21–23, Cagliari, Italy. Berlin: Springer-Verlag.
Foster KR, Koprowski R, Skufca JD. 2014. Machine learning, medical diagnosis, and biomedical engineering research: Commentary. BioMedical Engineering OnLine 13:94.
Glaser JI, Kording KP. 2016. The development and analysis of integrated neuroscience data. Frontiers in Computational Neuroscience 10:11.
Glaser JI, Chowdhury RH, Perich MG, Miller LE, Kording KP. 2017. Machine learning for neural decoding. arXiv preprint 1708.00909.
Gomez-Marin A. 2017. Causal circuit explanations of behavior: Are necessity and sufficiency necessary and sufficient? In: Decoding Neural Circuit Structure and Function, eds. Çelik A, Wernet MF. Cham: Springer.
Goodfellow I, Bengio Y, Courville A. 2016. Deep Learning. Cambridge: MIT Press.
Guyon I, Bennett K, Cawley G, Escalante HJ, Escalera S, Ho TK, Macià N, Ray B, Saeed M, Statnikov A, Viegas E. 2015. Design of the 2015 ChaLearn autoML challenge. 2015 International Joint Conference on Neural Networks (IJCNN), July 12–17, Killarney.
Hardoon DR, Szedmak S, Shawe-Taylor J. 2004. Canonical correlation analysis: An overview with application to learning methods. Neural Computation 16:2639–2664.
He K, Zhang X, Ren S, Sun J. 2015. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. Proceedings of the IEEE International Conference on Computer Vision, December 7–13, Santiago.
Hodgkin AL, Huxley AF. 1952. A quantitative description of membrane current and its application to conduction and excitation in nerve. Journal of Physiology 117:500–544.
Jonas E, Kording KP. 2017. Could a neuroscientist understand a microprocessor? PLoS Computational Biology 13:e1005268.
Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. 2015. Machine learning applications in cancer prognosis and prediction. Computational and Structural Biotechnology Journal 13:8–17.
Lazebnik Y. 2002. Can a biologist fix a radio?—Or, what I learned while studying apoptosis. Cancer Cell 2(3):179–182.
Leszczynski JE. 1999. Computational Molecular Biology, vol 8. Amsterdam: Elsevier.
Marblestone AH, Wayne G, Kording KP. 2016. Toward an integration of deep learning and neuroscience. Frontiers in Computational Neuroscience 10:94.
Marr D. 1982. Vision: A Computational Approach. San Francisco: Freeman & Co.
O’Leary T, Sutton AC, Marder E. 2015. Computational models in the age of large datasets. Current Opinion in Neurobiology 32:87–94.
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, and 6 others. 2011. Scikit-learn: Machine learning in python. Journal of Machine Learning Research 12:2825–2830.
Petrovsky N, Brusic V. 2002. Computational immunology: The coming of age. Immunology and Cell Biology 80(3):248–254.
Pillow JW, Shlens J, Paninski L, Sher A, Litke AM, Chichilnisky EJ, Simoncelli EP. 2008. Spatiotemporal correlations and visual signalling in a complete neuronal population. Nature 454:995–999.
Schölkopf B, Smola AJ. 2002. Learning with Kernels. Cambridge: MIT Press.
Shortliffe EH. 1998. The evolution of health-care records in the era of the Internet. Medinfo 98:8–14.
Silver D, Huang A, Maddison CJ, Guez A, Sifre L, Van Den Driessche G, Schrittwieser J, Antonoglou I, Panneershelvam V, Lanctot M, and 10 others. 2016. Mastering the game of go with deep neural networks and tree search. Nature 529:484–489.
Stevenson IH, Kording KP. 2011. How advances in neural recording affect data analysis. Nature Neuroscience 14:139–142.
Van Essen DC. 1997. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature 385(6614):313–318.
Velliste M, Perel S, Spalding MC, Whitford AS, Schwartz AB. 2008. Cortical control of a prosthetic arm for self-feeding. Nature 453:1098–1101.
Wang R, Chen F, Chen Z, Li T, Harari G, Tignor S, Zhou X, Ben-Zeev D, Campbell AT. 2014. StudentLife: Assessing mental health, academic performance and behavioral trends of college students using smartphones. Proceedings of the 2014 ACM International Joint Conference on Pervasive and Ubiquitous Computing, September 13–17, Seattle.
Wolpert DH. 2012. What the no free lunch theorems really mean: How to improve search algorithms. SFI Working Paper 2012-10-017. Santa Fe Institute.
Yu BM, Kemere C, Santhanam G, Afshar A, Ryu SI, Meng TH, Sahani M, Shenoy KV. 2007. Mixture of trajectory models for neural decoding of goal-directed movements. Journal of Neurophysiology 97:3763–3780.
This page intentionally left blank.












