6
Research Strategy
Developing a research strategy to understand the interactions between environmental chemicals and the human microbiome and the implications of the interactions for human health risk is a complex task. Understanding of how perturbations of the human microbiome might cause or contribute to the development of various diseases is in its infancy, so the task of understanding how environmental chemicals fit into the picture is even more difficult. Initially, the committee envisioned a research strategy that was similar to a flowchart or decision tree in which one or more experiments would lead naturally to a next set of experiments. However, such a straightforward approach is not feasible today given the state of the science. Thus, the committee determined that the research strategy should address broadly the three general topics highlighted in its statement of task: the effects of environmental chemicals on the human microbiome, the role of the human microbiome in modulating environmental-chemical exposure, and the importance of population variability or variation in modulating chemical–microbiome interactions. The committee addresses each of those in this chapter by describing the scientific value of the research, recommending experimental approaches for conducting the research, and identifying possible barriers specific to the research. It then describes the need for specific tool development to conduct microbiome research and finally identifies opportunities for collaboration. Because selection of chemicals for the experimental approaches is germane to all research topics, the committee first provides recommendations for selecting candidate chemicals for research. The committee emphasizes that the research strategy described in this chapter is not meant to be undertaken all at once, and the committee’s strategy will be influenced by research on the relationships between microbiome perturbations and disease. Furthermore, as discussed in this chapter, the research will be a collaboration of many agencies and organizations, each with its own priorities and interests in conducting specific research.
SELECTION OF CHEMICALS FOR EXPERIMENTAL APPROACHES
Development of research programs to study whether and how the human microbiome might modulate health risks posed by exposure to environmental chemicals requires decisions regarding the specific chemicals to be investigated and the appropriate exposure routes. The universe of chemicals that could be labeled environmental is large; it includes naturally occurring and synthetic chemicals, chemicals produced as byproducts of industrial activity and energy production, and those resulting from transformation of parent chemicals in the environment. A subset of that universe of chemicals consists of those subject to the requirements of major laws and regulations that are intended to protect human health from harmful exposures to chemicals that occur in environmental media (air, water, food, and soils), in consumer products of all types (including foods and pharmaceuticals), and in the workplace. For purposes of the present report, the committee has defined environmental chemicals as comprising the chemical subset noted above with emphasis on those regulated by the US Environmental Protection Agency (EPA).
It is clearly impossible to investigate all environmental chemicals that fit the committee’s definition. Moreover, in the absence of much more knowledge than is available now, it is impossible to specify the numbers and types of chemicals that would have to be investigated to provide an unequivocal answer to the broad question regarding the interaction between environmental chemicals and the microbiome and associated human health risks. If, for example, a clear and uniquely microbiome-mediated form of toxicity were identified for a few important chemicals, that might be sufficient to demonstrate the importance of this new branch of toxicology and the need for further study. But it is not at all clear how many failures to demonstrate such a role of the microbiome would be needed to conclude that the subject should not be further pursued.
It is important to consider criteria for selecting chemicals to be investigated carefully. In the bulleted statements below, the committee presents recommendations for appropriate criteria. Not all criteria need to be satisfied for any particular chemical to be considered suitable for study. And the need for additional criteria might become apparent as data are generated. For example, if emerging research indicates that children differ substantially from adults in their vulnerability to chemical–microbiome interactions, selecting chemicals to which children are heavily exposed or highly sensitive could be given top priority.
- Chemicals should be selected to represent the important categories of environmental chemicals regulated by EPA, such as pesticides, heavy metals, organic solvents, air and water criteria pollutants, persistent organic pollutants, consumer-product chemicals, and pharmaceuticals and veterinary drugs that have entered the environment.
- Chemicals in groups that have the highest priority for regulation because they have been shown to pose substantial health risks (substantial toxicity and widespread exposure) should be strong candidates for initial investigation.
- Chemicals that have been assessed in studies of short duration (14 and 28 days) and medium duration (90 days) would be strong candidates for initial investigation because replication of those studies to investigate chemical–microbiome interactions would be less expensive and less time-intensive than studies of longer duration. Longer-term studies will likely follow as microbiome research develops a body of knowledge and inquiry.
- Some chemicals that are known to have toxicity end points similar to health effects that have been associated with perturbed microbiomes (for example, immune-system effects, nervous-system effects, metabolic effects, and perhaps reproductive effects) should be selected.
- The candidate chemicals should include ones that have known capacity to perturb microbiomes or that can be readily studied for that property before full-scale toxicity investigations begin. This information will be important in defining doses to be used in the studies. Antibiotics that have been found in the environment could be candidates for such studies.
- The candidate chemicals should include those known to undergo transformation by the human microbiome.
- Chemicals that have produced large interindividual variability in dose–response studies are also strong candidates for investigation.
Chemicals that satisfy most of those criteria can be selected before experimental studies (animal and in vitro experiments) are conducted. In the case of observational epidemiology studies, it will not be possible to select chemicals according to the same criteria. Rather, it will be necessary to identify opportunities for fruitful studies and to make decisions about whether they involve environmental chemicals as defined in the present report. The committee recommends that, when possible, the same chemicals and methods be used for studies in whole animals, in vitro systems, and human populations to allow comparisons and integration of findings. Such an approach would maximize the possibility of reaching generalizable conclusions from the total body of research.
EFFECTS OF ENVIRONMENTAL CHEMICALS ON THE HUMAN MICROBIOME
A research priority is investigation of the effects of environmental chemicals on the human microbiome and consequent changes to human health. The question is whether environmental-chemical exposures or doses that are in the range of known or anticipated human exposures can induce microbiome perturbations that modulate adverse health effects. This section explores the scientific value of the research, recommended experimental approaches, and research barriers.
Scientific Value of the Research
As discussed in Chapter 2, the human microbiome has important effects on host biochemistry and physiology, and research over the last decade has associated disruptions in the microbiome with various disease outcomes. For example, regulation of immune-system, nervous-system, and metabolic functions occurs under the influence of gut microbiome metabolites, and alterations in gut metabolite profiles have been associated with aberrations in the functioning of these systems. Such aberrations can lead to both short- and long-term adverse health consequences. There is recent evidence that exposures to some environmental chemicals can alter microbiome composition but little evidence that those alterations have adverse effects on health status. There is, however, evidence that long-term, low-level exposures to some antibiotics alter animal microbiomes so as to increase capacity to extract energy from food and lead to obesity (Cox et al. 2014). That finding is consistent with the use of low-level antibiotic treatment to promote more rapid growth of farm animals. Thus, it is reasonable to hypothesize that some environmental chemicals might alter microbiome composition and result in aberrations in health status. Most important, assessment of whether environmental chemicals can cause microbiome disruptions has the potential to identify and prevent or ameliorate adverse health outcomes caused by such disruptions.
Full exploration of the association between environmental-chemical exposure, microbiome disruptions, and adverse health outcomes is contingent on a deeper understanding of exactly what a disrupted or “unhealthy” microbiome is—a topic that extends well beyond the scope of this research program. As understanding grows, however, determining whether environmental-chemical exposures can cause such structural or functional disruptions will become a high priority because the exposures constitute a cause that potentially can be regulated and mitigated. Ultimately, greater understanding should stimulate new toxicology concepts and testing protocols that include the effects of chemicals on the microbiome.
Experimental Approach
A research program that addresses the question of how environmental chemicals affect the microbiome and the possible consequences could consist of defining toxicity end points for the microbiome, identifying environmental chemicals that can perturb (structurally and functionally) the microbiome, and using animal and epidemiology (human) studies to demonstrate that microbiome perturbations by environmental chemicals cause or modulate a change in health. The research program will require using short-term, high-level experiments—for example, using established study protocols to screen chemicals for effects on the microbiome—and conducting more detailed followup studies that require new population cohorts or that aim to elucidate toxicity mechanisms.
Defining Toxicity End Points for Microbiomes
The dose–response relationship is central to toxicology in that it quantitatively reflects the effect that a given exposure has on a given biologic system. The dose–response relationship relies heavily on quantitative measures of health outcomes or end points, such as gene expression, enzyme activity, or alterations in cellular physiology. If a microbiome is considered the “biologic system” for which a dose–response relationship needs to be defined, the question of which end points best
reflect microbiome toxicity arises. Accordingly, end points that exhibit dose-dependent properties and act through known mechanisms will need to be established. Because no known end points of microbiome toxicity have been established, comprehensive approaches—including 16S rRNA or internal transcribed spacer (ITS) gene community profiling, metatranscriptomics, metaproteomics, metabolomics, and other measures of physiologic activity—will be needed to capture all aspects of the microbiome response to a given toxicant. For example, although 16S rRNA and ITS sequencing approaches will capture changes in community structure, measures of the microbiome stress response—both general and specific to a particular environmental chemical—will be captured best through metatransciptomic approaches. However, the committee emphasizes that an integrated approach that includes the collection of data from multiple -omics assays will be important for establishing the most comprehensive view of the microbiome response to an environmental chemical.
To establish quantifiable end points, the committee recommends studying the effects of chemicals with different mechanisms on mouse and human microbiomes by using bioreactors, such as the simulator of the human intestinal microbial ecosystem (SHIME), described in Chapter 4.1 Many antimicrobial agents are good candidates for this investigation because they have known mechanisms—for example, inhibition of DNA replication and transcription, protein synthesis, or cell wall biosynthesis—and exhibit predictable dose–response relationships. Furthermore, some antimicrobials are bacteriostatic (they restrict growth and reproduction) whereas others are bactericidal (they cause cell death). Therefore, antimicrobials should help to establish quantitative end points that ultimately could be used to understand or predict the toxic effects of environmental chemicals on a microbiome.
The feasibility of using bioreactors has been demonstrated, but several important factors must be carefully considered before these studies are undertaken. First, the source of the microbiome needs to be considered. Human stool samples and rodent fecal or cecal contents are popular microbiome sources; however, how accurately they reflect the microbiome of a particular gastrointestinal niche remains a topic of intense debate (Dantas et al. 2013), and how accurately a rodent-specific microbiome reflects what might be observed in the human microbiome is unclear. Regardless, for the purpose of establishing testable end points, those microbiome sources are ideal because they are easily collected and stored and can be collected longitudinally. Second, although variability that results from diet, age, or sex can be strictly controlled in rodents, it cannot be in human studies, so experiments will need to be designed with consideration of the variation and variability associated with the human microbiome. Third, antimicrobials could influence community structure through selection via antibiotic resistance that could be especially important during long-term incubations. Therefore, acute, short-term dosing schemes will be essential for developing signatures of microbiome toxicity, and long-term chronic dosing studies should be interpreted with caution.
Once a stable bioreactor system is established, increasing doses of antimicrobials that have different mechanisms can be added, and samples can be collected longitudinally. Use of a longitudinal study design allows comparisons of acute and chronic dosing schemes. Samples can be subjected to comprehensive analysis by a suite of -omics tools. Changes in microbial membrane potential, membrane permeability, and DNA replication can also be assessed (Maurice and Turnbaugh 2013). Next, extensive statistical and bioinformatic analyses can be applied to determine patterns in gene expression, metabolite concentrations, or other physiologic measures that are consistently altered in comparison with unexposed microbiomes and hence can serve as end points for studies of effects of environmental chemicals. Data are likely to identify specific members of the microbiome that contribute to specific end points; thus, defined culture systems (such as monocultures or cultures that are representative of the major taxa in structure and function) could provide an avenue to clarifi-
___________________
1 Although the focus here is on the gut microbiome, the experimental approaches could be adapted for skin and lung microbiomes and other body niches.
cation of the mechanistic role of specific taxa of bacteria or fungi. Having established the identities of microorganisms that are most sensitive, one can conduct more detailed studies to increase understanding of a chemical’s mechanism of action.
Once a repertoire of end points—such as changes in physiology, gene expression, protein concentrations, or metabolite concentrations—is established for antimicrobial exposure, the experimental approach can be applied to environmental chemicals of concern. However, there are several caveats to the experimental approach outlined. First, it does not take into account metabolism by the host and so might miss compounds that undergo biotransformation or bioactivation through host-dependent mechanisms before having their effects on the microbiome. Second, it assumes that environmental chemicals of concern work through mechanisms analogous to antimicrobial chemicals (that is, by affecting DNA, protein, or cell-wall biosynthesis). Third, it assumes that bioreactor systems accurately model what is present in the gut or other body niches, faithfully represent the community structure and its metabolic activity, and are capable of growing even the most fastidious organisms. Fourth, the approaches do not fully capture differences that can occur through different routes of exposure, such as inhalation and dermal. Therefore, the committee recommends that model systems that faithfully recapitulate the host–microbiome interaction of the skin and lung be considered so that all exposure routes are captured fully.
Identifying Environmental Chemicals That Perturb Microbiomes
Other high-priority research would be aimed at developing a high-throughput bioreactor system that operates under physiologically relevant conditions to screen environmental chemicals in a uniform manner for their ability to perturb microbiomes. The goal is to provide a reproducible platform for assessing dose-dependent effects of environmental chemicals on defined microbial communities and on individual microbial species within a community through measures of physiologic activity (such as metabolic activity and membrane permeability) and biologic activity (such as DNA replication and transcriptional response). Once established, the measures of the microbiome response to environmental-chemical exposure could be used to populate a database and later to inform screening programs in mice and perhaps could be cross-referenced with signature responses in human populations.
Although development of bioreactors to investigate microbiome interactions is still in the early stages, several devices described in Chapter 4 have found their way into basic and translational research. Bioreactor systems permit flexibility in study design by using single strains or defined or complex communities, can be cultivated for various periods to assess acute and chronic exposures, and can be modified to include different host components, including mucin barriers or dietary constituents that more closely resemble in situ conditions. Such bioreactor systems can be designed to incorporate surfaces for microbial attachment so that the response of mixed-species biofilms and free-swimming microorganisms can be assessed, thereby recapitulating the primary modes of microbial lifestyle in and on the human host. In the bioreactor studies, it will be essential to use doses of chemicals relevant to human exposures, including concentrations typically associated with environmental or industrial accidents. However, dose estimates might need to be re-examined to take into account interactions with the microbiome at both internal and external body sites (Silbergeld 2017). For example, although estimates of arsenic exposure via drinking water typically reflect the absorbed dose (the dose passed from the gastrointestinal environment into circulation), the dose to the microbiome could be substantially higher. Furthermore, members of the microbial community are not likely to exhibit the same dose–response relationship with an environmental chemical. Therefore, experimental systems that range from individual strains of bacteria to complex microbiomes must be considered to investigate the potential of an environmental chemical to alter the microbiome. A final consideration is whether a mechanism of action is mediated by the host or is
independent of the host. If it is host-independent, simpler bioreactors that require less investment can be developed because they do not require a host component for incorporation into the system.
A long-term goal would be to evaluate distinct microbiome configurations that are representative of different life stages or disease states, which might represent periods of increased susceptibility to environmental chemicals. Such platforms would provide important information regarding susceptible human populations and would be important in trying to capture human variation and variability.
Linking Microbiome Perturbations by Environmental Chemicals to Adverse Health Outcomes
Animal Studies
Evidence of adverse health outcomes caused by perturbations of microbiomes induced by environmental chemicals could be provided by animal experiments, especially for chemicals that require metabolism by the host. Although the bioreactor experiments can screen environmental chemicals rapidly, they cannot fully capture host-mediated processes that in many cases have been identified as key mechanistic components of environmental-chemical toxicity. The committee recommends starting with gnotobiotic animals that have a defined microbiome or standardized community (as described in Chapter 4) to reduce measurement and experimental variability. When diet and other environmental factors can be carefully controlled, it should be possible to assess the interactions of environmental chemicals with the microbiome and the host and their contribution to adverse outcomes. For example, if one observes a correlation between the environmental-chemical exposure, microbiome perturbation, and adverse outcomes, one could transfer the perturbed microbiome into germ-free mice and observe whether the adverse outcome is recapitulated in them. If so, that would be strong evidence that the microbiome perturbation induced by the environmental chemical is involved in manifestation of the observed adverse outcome. An important caveat to that approach is that only a portion of the microorganisms present in the donor community will be efficiently transferred to the germ-free host. The approach has been used in only a few experiments that use gut communities; therefore, it is unclear whether it will be an effective approach for all gut communities and for those from skin or lung.
A long-term goal is to screen environmental chemicals by using animal models to assess microbiome perturbations in inbred, transgenic, and outbred lines and established disease models. The outbred lines particularly allow assessment of the consistency of effects of chemical exposures in genetic and microbial gradients in such animals. The studies are not limited to rodents; for example, zebrafish, fruit flies, or nematodes might be best suited to studies that require high-throughput analysis. Unlike the defined gnotobiotic experiments discussed above, these studies will allow better understanding of realistic microbiome variation and consistency of effects. Ideally, the studies would also include multiple animal models, multiple animal facilities, and gnotobiotic transfers from defined communities or multiple human donors.
Epidemiology Studies
Epidemiology and population exposure studies that are already under way could be used to identify microbiome co-variation with an environmental chemical of interest. The approach could involve, for example, identifying a human population in which a chemical exposure of interest has been tracked and collecting new samples appropriate for microbiome analyses, generating new microbiome-relevant data from biobanked samples from such a cohort, or adding measurements of environmental-chemical exposures of a human population that is being followed for other purposes, including microbiome measurements. For short-term, proof-of-concept purposes, simple measures of microbiome structure might be sufficient to identify cases in which a perturbation occurs either in tandem with or after chemical exposure and manifestation of adverse health outcomes; the microbiome changes would then need
to be investigated in more detail to characterize their functional or clinical consequences (if any). In such cases, it will also be crucial to separate health effects mediated by microbial activity from those induced by direct chemical exposures of the host. That research could use existing prospective cohort infrastructure, including banked specimens and could benefit particularly from collaboration among institutions, such as environmental- and population-health scientists and funding agencies.
Barriers
Defined, validated, and quantitative measures of host–environmental-chemical interactions exist but not for chemical interactions with microbial communities, although individual microbial physiology can be robustly detailed. Thus, defining measurable and quantifiable end points that reflect toxicity to the microbiome are of paramount importance. Many of the antimicrobial experiments that the committee describes are likely to require substantial investments of time and resources, are exploratory and thus unlikely to be supported through traditional funding mechanisms, and require unique expertise not found in a single laboratory. Successful studies will require a consortium of microbiologists, toxicologists, microbiome-analysis experts (those who have expertise ranging from sequencing to metabolomics), bioinformatics experts, and persons who have other relevant expertise, as appropriate. Only after clear, quantitative measures of microbiome toxicity have been established can the approaches be applied to representative environmental chemicals of concern. Identification of exposures or doses that are in the range of known or anticipated exposures will also be important, although a range of doses should be studied. Finally, a major challenge will be capturing human microbiome variation and variability that might not be apparent on the basis of sequencing but probably would be observed with metabolic output. Thus, more functional analyses of the human microbiome that use metatranscriptomics, metaproteomics, and metabolomics will be required.
There are several barriers to development of the bioreactor platforms. First, as discussed in Chapter 4, there is the difficulty of establishing and maintaining physiologic communities in vitro. Second, perturbations of the microbiome could require host-mediated chemical metabolism from such organs as the liver or some other host-mediated process that has not been incorporated into the bioreactor platform. Third, as discussed in Chapter 4, bioreactors might not be able to capture functional diversity, including interindividual, developmental, and body-site variation. Fourth, although the research discussed above should help to identify end points to use, end points for assessing microbiome toxicity have not yet been established. Fifth, there is little understanding of how microbial-community composition and interaction depend on life stage and on the developing or aged host tissues.
Overall, an additional barrier to research to understand how environmental chemicals might affect the human microbiome is the unknown level of functional redundancy that could exist within the human microbiome. For example, many chemicals are capable of altering microbiome composition, but is the altered composition itself a response, and would one expect it to be monotonically dose-dependent? If alteration of the microbiome composition can be shown to be causally related to an adverse host response (for example, a change in the abundance of microorganisms that metabolize chemical X or in the abundance of microorganisms that produce a lipid mediator of inflammation), is it possible that the response would behave in a threshold-like manner because of the large potential for functional redundancy in the microbiome? As a hypothetical example, a detoxification product of a metabolized chemical could be generated by a broad class of enzymes represented by different genes throughout various taxa in a microbiome. In that case, a shift in the composition of the microbiome—even a robust shift—might have little consequence if sufficient redundancy in function remains in the microbiome. At high doses, where the microbiome is reduced in biomass and abundance, there could be threshold effects related to metabolism or elimination, but such high doses
might not be relevant to environmental exposures. Answering questions about associations of adverse outcomes with changes in a microbiome induced by environmental-chemical exposures will require experiments with bioreactors (investigating the microbiome only), germ-free models (investigating the host only), and conventional animals (investigating the host and the microbiome, including their interactions).
THE ROLE OF THE HUMAN MICROBIOME IN MODULATING EXPOSURES TO ENVIRONMENTAL CHEMICALS
Another high-priority research topic is the effects of the human microbiome on exposure to environmental chemicals. Specifically, what is the role of a microbiome in modulating absorption, distribution, metabolism (activation or inactivation), and elimination (ADME) of environmental chemicals? This section explores the scientific value of the research, recommended experimental approaches, and barriers.
Scientific Value of the Research
As discussed in Chapter 3, there is increasing evidence that microbiomes can modulate the relationship between external exposure and internal dose of some environmental chemicals or their active metabolites. Conceptually, interactions between a microbiome and environmental chemicals might influence all aspects of the ADME profile of a given chemical. For example, some microorganisms present in the gut microbiome can metabolize foreign chemicals in ways similar to metabolism by the liver and other organs. Because the toxic properties of many environmental chemicals are influenced or directly caused by some of their metabolic products, the creation of metabolites by a microbiome could influence toxicity outcomes. Accordingly, understanding of the specific interactions between a chemical and a microbiome is particularly important in the context of assessing risk because it provides a means of quantifying the relationship between external chemical exposure and the target-tissue dose of the parent chemical or the active metabolite associated with an adverse effect.
Scientists have little understanding today of the total capacity of microbiomes to biotransform environmental chemicals; for most cases, the specific microbial enzymes and microbial species involved have not yet been elucidated. Thus, fundamental research should be aimed at broader identification of specific microbial enzymes and microbial species that mediate chemical transformation processes. Ultimately, linking the specific microorganisms, genes, and enzymes to a particular chemical transformation process is essential if substantive progress is to be made in addressing individual susceptibility and interspecies extrapolation questions at a mechanistic level and in understanding the degree of functional redundancy within a microbiome. Furthermore, if the effect of the microbiome on chemical exposure can be quantified, models can be developed by using a compartmentalized approach that could improve exposure assessment for specific chemicals in a hypothesis-driven manner without necessarily understanding the contributions of individual microbial species.
Experimental Approach
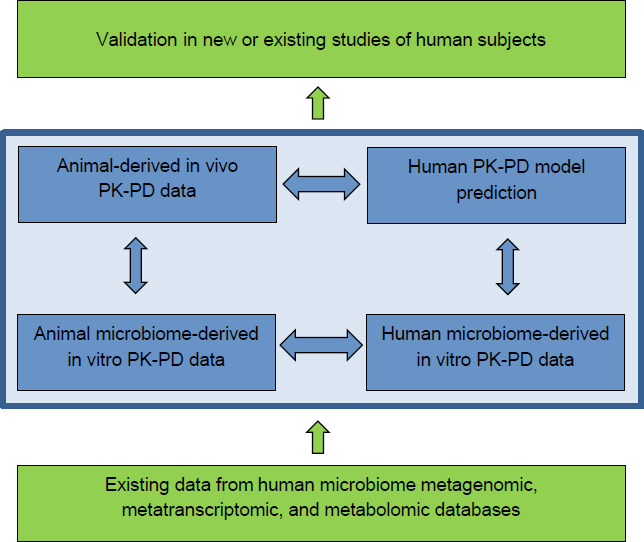
Determination of health risks associated with exposure to environmental chemicals and the potential roles of the microbiome in modulating such risks depends on an understanding of the biologic effects of the chemical, its distribution, its metabolism, and its clearance in model systems that permit analyses of the role of the microbiome in such processes. The committee has organized the experimental approach so that the data generated could feed directly into development of a microbiome component for physiologically based pharmacokinetic or pharmacodynamic (PBPK-PD) models (see Box 6-1). The goal of developing quantitative models provides a framework for guiding basic research toward outcomes that can be valuable for risk assessment in the near term. The emphasis on PBPK-PD modeling is not to imply that basic research should be a secondary part of the research
strategy. Rather, the committee recognizes that the knowledge of microbiome roles in metabolism of environmental chemicals has progressed substantially. Through thoughtful selection of chemicals for study on which there is existing knowledge, there might be an opportunity to accelerate progress in understanding how much the microbiome might influence ADME processes.
The traditional PBPK-PD modeling approach follows a data-based parallelogram strategy that incorporates in vitro cell type-specific data on both animals and humans and in vivo data generated from model animals (see blue boxes in Figure 6-1). Those three data sources feed into development of the PBPK-PD model to permit prediction of human responses to chemical exposure (Goldsmith et al. 2012). The widely used framework can be adapted to incorporate data on microorganism-specific contributions to ADME. A successful strategy for PBPK-PD modeling of human-microbiome effects on chemical exposure would be enhanced by including information from existing human-microbiome databases on microbiome gene content (metagenomes), transcription, and metabolism and by efforts to improve existing reference databases. That information could be used to infer potential chemical-metabolism pathways in a microbiome and to formulate initial models. Opportunities to validate model predictions in existing human population-based studies or those initiated specifically for the purpose of such studies should also be pursued (green boxes in Figure 6-1). The committee notes that the initial focus of this research is on the gut microbiome because a large body of literature implicates it in chemical transformation processes. However, the overall strategy could be generalizable to other tissue sites, including the oral, respiratory, and skin microbiomes.
Animal Studies to Generate Pharmacokinetic–Pharmacodynamic Data
Absorption and metabolism are two primary determinants of chemical kinetics in mammalian systems (Yoon et al. 2012). The human microbiome encodes a vast ancillary metabolic potential and plausibly plays a role in such processes. But few experimental animal studies have been designed explicitly to assess the specific role of the microbiome in ADME, and such microorganism-specific data have not been incorporated into PBPK-PD models. Integration of such data into current models could help to explain response variability within human populations and reduce uncertainties in current model predictions.
To determine initially whether the microbiome plays a role in a chemical’s kinetic behavior, comparative studies of conventional animals (ones that have an intact microbiome) and germ-free animals would allow assessment of the effects (and their magnitude) of the microbiome on ADME processes in vivo. In the simplest form, experimental
animals are exposed (via oral, dermal, inhalation, or intravenous routes), the concentrations of the parent chemical and its metabolites are assessed in several target organs and in the circulation and urine, and binding of the parent chemical or its derivatives to receptors in target organs (if known) is investigated (Yoon et al. 2012). Although germfree animal models have some caveats, as noted in Chapter 4 and in the section on barriers below, they offer a unique opportunity to consider host vs microorganism-derived chemical interactions in vivo and to some extent extricate host from microbial contributions to these processes.
To develop data that might be more directly relevant to human microbiome-derived chemical transformation, one could also consider experiments that compare germ-free animals with ones that have been colonized with a microbial inoculum derived from human feces (humanized) or colonized with specific human-derived microorganisms to study the functions of interest. Humanized animals offer an opportunity to evaluate effects by using complex microbiomes derived from heterogeneous sources that differ in their constitution, such as those from infants, adults, or people who have chronic diseases known to influence microbiome composition. Animals that have been colonized with a defined microbial community allow assessment of microorganisms that are suspected of playing a key role in chemical transformation. Both approaches offer an opportunity to generate information on the capacity of such organisms to modulate chemical exposures and influence ADME. Similarly, comparing untreated conventional animals with antimicrobial-treated animals would allow assessment of the effects of acute microbiome perturbation on ADME and toxicokinetics.
In Vitro Systems for Generating Pharmacokinetic–Pharmacodynamic Data
Once a microbiome has been implicated in modulating ADME processes in an animal model, in vitro systems, such as bioreactors and gut-on-a-chip, can be used to isolate the microbial com-
ponent and compare mechanisms among species. In vitro experiments should be used to define functional traits of the microbial community that transform the environmental chemical, to identify microorganisms and microbial interactions implicated in chemical transformations, to identify microorganism-modified metabolites, and to obtain microorganism-specific chemical transformation rates, which should be compared with those obtained by using human microbiomes for incorporation into PBPK-PD models. Environmental-chemical metabolites formed in vitro should be reintroduced into animal models to test or verify their mechanism. As shown in Figure 6-1 (bottom blue bars), microbiomes obtained from mice exposed to an environmental chemical could be used in parallel with human microbiomes from exposed populations, if available, to determine whether the same metabolites are produced after chemical exposure and through similar types of microbial interactions.
A major advantage of in vitro systems is the potential to implement high-throughput studies. Development and standardization of high-throughput in vitro systems will require careful consideration of model microbial reference communities and reference strains that broadly represent the diverse metabolic functions of the unperturbed human microbiome, which are as yet poorly defined. As discussed below, further development of microbial reference strains will require continued effort to improve functional annotation of metagenomes with emphasis on identifying the specific enzymatic pathways that act on environmental chemicals.
Identifying Specific Microbial Enzyme Functions
New chemical probes and chemical screening technologies are emerging that could reduce the experimental effort and time needed to isolate and identify specific proteins and microorganisms that interact with environmental chemicals in the microbiome. For example, chemical probes designed to target enzyme active-site chemistries have been used to profile cytochrome P450 enzyme activities and drug–protein interactions in vivo (Wright and Cravatt 2007; Wright et al. 2009; Sadler and Wright 2015) and to identify microbial glycoside hydrolases and other enzymatic activities in bacteria (Chauvigné-Hines et al. 2012). Chemical probes that mimic structures of specific classes of environmental chemicals and have reactive tags (such as biotin) could also be useful for initial screening efforts to identify direct interactions between a chemical and a microbial species within a complex microbial community. In conjunction with chemical probes, genetically engineered bacterial reporter strains could be used as sensitive indicators of microbiome perturbations. The approach has been used extensively to sense environmental chemicals in the field (Roggo and van der Meer 2017), but reporter strains have been used less commonly to sense and record specific signals in the mammalian gut microbiome (Kotula et al. 2014). However, the potential for reporter strains to affect the microbiome-community structure and function should be carefully considered.
Emerging technologies that hold promise for characterizing how environmental chemicals are metabolized in a microbial community include stable-isotope labeling, which permits tracking of labeled chemicals, and advanced mass-spectrometry methods (Berry et al. 2013). Coupling those approaches with single-cell genomics strategies should prove useful for identifying the specific microorganisms responsible for chemical interactions (Lasken 2012; Berry et al. 2013; Koppel et al. 2017). Although such discovery-based studies might have a longer time horizon, their early inclusion as part of an integrated research strategy is critical for achieving the goal of assessing personalized microbiome status as a potential risk factor for environmental-chemical interactions.
Barriers
Although the components of this research strategy for assessing the role of a microbiome in modulating ADME of environmental chemicals are based on an established framework for PBPK-PD model development, several barriers to its implementation remain to be resolved, as outlined below.
- In situ conditions that might influence ADME, such as dietary interactions and pH and oxygen gradients, are largely unknown in human populations of potentially heightened susceptibility, such as infants and patient populations that exhibit changes in microbiome diversity in association with their underlying disease. Thus, it might be difficult to recapitulate such conditions in model systems accurately.
- Germ-free animal models are known to have altered host tissue physiology compared with conventionally raised animals, including adaptive changes in expressing enzymes that are critical for metabolic transformation of drugs and environmental chemicals. The extent to which adaptive changes in normal host metabolism occur in germ-free or other gnotobiotic systems is not broadly understood and requires rigorous evaluation. Thus, for some chemicals, the use of germfree models could be problematic for measuring PBPK-PD parameters.
- To address variations in microbiome structure and function that are naturally present in human populations, large experimental design matrices might be required, whether animal models or in vitro systems are used, and might require large resource investments. In designing cost-efficient studies to identify sources of variability effectively, such statistical techniques as design of experiments could be used.
- Development of in vitro model systems, such as gut-on-a-chip, that include the microbiome is still in its infancy. There is still no consensus on microbial reference communities or strains that reflect the metabolic potentials of an unperturbed microbiome accurately. That knowledge gap might present challenges in obtaining comparable results from in vitro systems that can be directly extrapolated to the whole animal or to human systems.
- Because the human gut microbiome cannot be fully recapitulated—for example, in a germfree rodent or in vitro system—some microorganisms will be missing from such studies. That limitation reiterates the need for fundamental studies to understand what gene products (enzymes and proteins) are encoded by the microbiome and are involved in metabolism of chemicals.
THE IMPORTANCE OF MICROBIOME VARIATION AND VARIABILITY
As discussed in Chapter 2, the human microbiome structure and function vary with, for example, body site, life stage, genetics, geography, and health status. The human microbiome also differs from microbiomes of animal species. Variation and variability have important implications in assessing risk posed by environmental-chemical exposure.2 This section explores experimental approaches to examine the importance of variation and variability among humans and then between humans and laboratory animals.
Assessing the Importance of Human Microbiome Variability and Variation
As noted, microbiome variability and variation within the human population are substantial, and a question is whether knowledge of population and life-stage variation and variability in the human microbiome will improve understanding of the susceptibility to environmental chemicals and of individual health risk. The subsections that follow discuss the scientific value of the research and recommend research designed to investigate this important topic.
Scientific Value of the Research
As discussed in Chapters 1 and 2, humans and their microbiomes have co-evolved to form an ecosystem that is comprised of distinct habitats whose microbial community structure and function vary. Many factors—such as age, race, genetics, health status, physical condition, diet (including early-life nutrition), and geography—affect microbiome structure and function. Susceptibility to environmental-chemical exposure and associated health risk might be modified not only by those factors but by the variation and variability of the human microbiome structure and function (see Figure 6-2). Understanding how the variation and variability of the human microbiome might affect
___________________
2 See Chapter 1 for a discussion of the definitions of variation and variability.
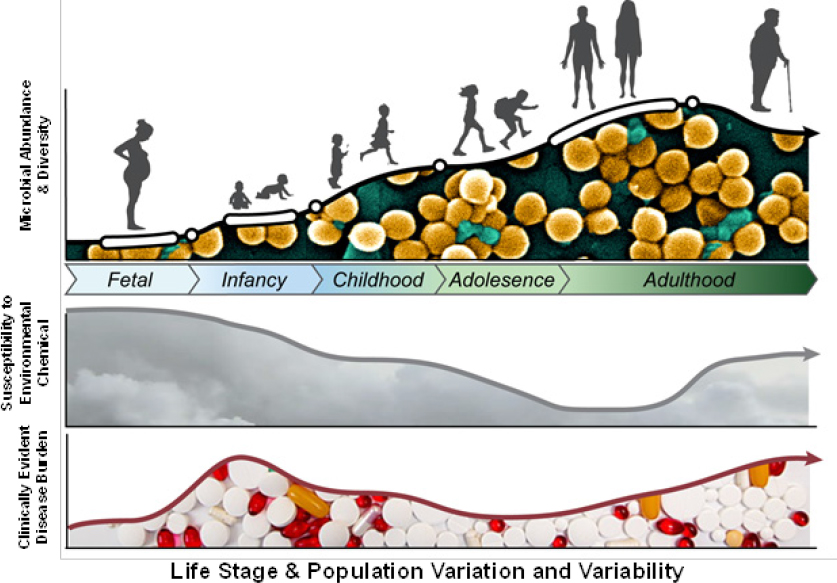
chemical-microbiome interactions will be critical in assessing microorganism-mediated risk posed by environmental-chemical exposures. For example, the microbial community and its functions are sparser and less varied in the infant than in the adult; if one considers only the adult microbiome, one could miss identifying critical windows or periods of susceptibility. Explicitly considering human microbiome variation and variability might also substantially improve our capacity for identifying at-risk populations and for developing strategies to mitigate exposures and reduce associated disease incidence in these populations.
Experimental Approach
The goals of the research described are to understand the importance of human microbiome variability and variation at any given life stage or among specific populations and ultimately to ensure that studies consider such variation and variability adequately and appropriately in assessing the health risks to human populations posed by exposure to environmental chemicals. In conducting this (and other) human microbiome research, two points need to be emphasized. First, the respiratory, gut, and skin microbiomes vary in their taxonomic composition and function, so one needs to consider the environmental exposure route when selecting the specific organ and tissue system to study. For example, although studies of one community could inform those of another, studying the response of the gut microbiome to an environmental chemical that is absorbed mainly through the skin might not be directly informative for human risk assessment. Second, community composition and its function are not the same, so examining microbial function rather than only taxonomy should be encouraged. For example, subtle variations in low-biomass communities might impart important functional differences in metabolites or small-molecule intermediates; conversely, because functional redundancy is probable in many microbial communities, variations in community composition might not necessarily impart key functional differences.
Variation and variability can be understood best by conducting comparative studies that assess functional similarities and differences of environmental exposure in the factors known or hypothesized to affect microbiome diversity. Specifically, the studies should characterize microbiome communities and their functional differences by such factors as race, sex, life stage, and health status and emphasize populations that represent key windows of potential vulnerability, such as infants, pregnant women, adolescents, and geriatric populations, and resiliency, such as healthy adults. Functions that would be strong candidates for evaluation include microbial activities and pathways for chemical metabolism, regulation of transport and barrier integrity, and modulation of factors relevant to host developmental, metabolic, immunologic and neurologic outcomes.
In the near term, large and well-characterized human population studies that are already under way could be used for conducting comparative studies. Sample collection for microbiome analysis could be added to current studies of large, longitudinally followed, and well-characterized cohorts in which toxicant exposures have been or could be readily assessed. The longitudinal component of such human studies offers an opportunity to examine short-term and long-term effects of exposure and could be particularly enlightening with respect to populations at heightened risk, for example, early-life acute or chronic toxicant exposures that have the potential to affect microbiome development in a manner that manifests as disease later in childhood. Such an effort is likely to yield valuable data at moderate cost. This approach could be enabled by developing rapid and agile funding opportunities for supplemental grants to awarded projects that are investigating chemical–microbiome interactions.
In the longer term, improved computational approaches, advances in data science, and innovative human-study design will advance understanding of the implications of variation and variability of the human microbiome. Specifically, the advent of high-throughput DNA and RNA sequencing and high-throughput untargeted protein and metabolite profiling technologies with rapidly developing computational methods will provide measures that, when integrated, will allow modeling of cause–effect relationships. When combined with detailed characteristics of the human host, a computational approach might identify modifiers (factors that affect variability and variation) that are important in manifestation of the health effect. However, tackling the integrated analysis of heterogeneous data types at the scale and complexity necessary will demand data-science innovation and computational advances that are outlined further in the section “Tool Development.”
In conjunction with the human studies, it will be important to replicate or validate the findings or observations from those studies in bioreactors or gnotobiotic-animal models described earlier in this chapter. For example, a bioreactor or gnotobiotic-animal model could be used to investigate the responses of microbiomes that were isolated from groups (defined by some host factor) that did and did not manifest a given effect that resulted from exposure to an environmental chemical.
Assessing the Importance of Microbiome Variation Between Animals and Humans
Understanding the importance of the variation between animal and human microbiomes is critical. The central question is whether the differences are so great that effects are being missed or mischaracterized by using the animal models to predict health risks associated with environmental-chemical exposure. Furthermore, do the interspecies uncertainty factors that are used to extrapolate effects in animals to humans account for the microbiome variation? The following text explores the scientific value of the research and recommends research that could be conducted to address such questions.
Scientific Value of the Research
For most health risk assessment, animal models have been the basis for determining the toxic effects of chemicals and estimating the potential for adverse health effects in humans. The extent to which toxicity studies have already accounted for mediating effects of microbiota on health effects is currently unclear. For environmental chemicals
for which there is reasonable evidence or suspicion of microbiome-mediated health effects, it is important to determine whether chemical–microbiome interactions of consequence observed in model systems are similar in human microbiomes. If not, understanding why findings in model-system microbiomes differ from those in human microbiomes is imperative. For example, are the differences the result of an inability to recapitulate a microbiome in an in vitro system (that is, key microbial-community members are not present in the in vitro system), or do they result from true variation between an animal microbiome and the human microbiome? The research described below should produce new knowledge of microbial-community function that should allow assessment of the capacity of animal models to recapitulate the activities of human microbiomes.
Experimental Approach
Like research to investigate variation and variability within the human population, this research involves conducting comparative studies that focus on functional differences rather than only taxonomy. Ultimately, the goal would be to focus on functional capacity encoded by the human microbiome to identify the animal species and study designs most appropriate for extrapolating to humans. The comparative studies should focus on evaluating functional similarities and differences between native microbiomes from humans and test animals, such as mice, zebrafish, fruit flies, pigs, and nonhuman primates; native microbiomes from laboratory-reared and wild model organisms; and native human and animal microbiomes and microbiomes resulting from transplantation of human microbiota into test animals. Functions that would be strong candidates for evaluation would be microbial activities and pathways for chemical metabolism, regulation of transport and barrier integrity, and modulation of factors relevant to host developmental, metabolic, immunologic, and neurologic outcomes. Important considerations for the experiments include use of appropriate controls that take into account effects of the vehicle or chemical form administered and use of a range of doses that include environmentally relevant exposures to evaluate effects on the shape of the dose‒response relationship. Near-term goals of the research would be the following:
- Identification of functional pathways, including chemical metabolism pathways, that are uniquely encoded by microbiomes from select model organisms and performance of multi-omics functional characterization of microbiomes from humans and animal models, including comparisons of animals from different colonies and genetic backgrounds to assess various potential microbiome compositions.
- Functional characterization of human microbiome samples in response to environmental-chemical exposure and conduct of comparative analyses of functional profiles after transplantation of microbiota into model organisms.
- Understanding of differences and similarities between model-organism and human-host responses (such as metabolism, absorption, elimination, immunity, and behavior) to environmental-chemical exposures by using defined microbial communities in in vitro or gnotobiotic models.
- Assessment of the redundancy and uniqueness of microbiomes of various model organisms and humans through comparative microbial functional genomic and metabolomic studies.
Model organisms will be essential for testing causal relationships between environmental exposures, microbiome perturbations, and health outcomes. They will enable the identification of the molecular and cellular underpinnings of observed interactions. Thus, model systems that can be used to represent the human condition faithfully are critical. Over the long term, model organisms or microbiomes that faithfully and stably encode functions relevant to human microbiomes might be engineered by using synthetic-biology or genetic-engineering strategies.
Barriers
Any model system for assessing microbiomes and even human-based studies have inherent and specific limitations. Knowledge of such limitations should inform decisions on experimental
approaches to be used and what research cannot be adequately addressed with a single approach. Potential barriers include the following:
- There could be difficulties in obtaining detailed functional characterization of some microbiomes—for example, with multi -omics approaches—because of limitations related to sample collection, sample type, sample quantity, and the preponderance of host components, such as human DNA, RNA, or protein that is usually associated with tissue samples. Such microbiomes would include those from difficult-to-access host sites or from sites that have small amounts of retrievable material, which potentially limit their study with multiple analytic approaches.
- A related barrier is the understanding of how sample collection, processing, and storage could affect multilevel functional characterization of a given microbiome or data interpretation. Research study protocols should strive to harmonize tools and methods among systems whose microbiomes are to be compared, for example, gut microbiomes from an animal model and human subject.
- Detailed functional characterization of microbiomes could be difficult because of technologic limitations in generating reliable reference databases of microbial genomic and metabolomic annotations and the poor scalability and relatively high cost of some animal models.
- Inability to reproduce findings related to chemical–microbiome interactions derived from a given experimental approach because of lack of standardization is a barrier. Investigators will need to control and disclose variables relevant to microbiome assessments, including initial characterization of microbiomes, animal-care procedures and conditions, choices of laboratory reagents, and methods for sample processing and outcome measurements. If a lack of reproducibility is observed, the extent to which such an observation is due to microbiome differences rather than other variables unique to the models or human cohorts would be important to clarify.
- Many experimental systems present important technical challenges in creating exposure conditions that appropriately mimic the human condition to be studied. Such challenges, which already exist in toxicology studies of health risks associated with environmental-chemical exposure, are amplified when microbiome-modulated influence is of central concern, and they can become even larger when variability and variation are of key interest.
TOOL DEVELOPMENT
The research strategy developed by the committee emphasizes the three main elements highlighted in the statement of task: the effect of chemical exposure on the human microbiome, the role that the human microbiome plays in environmental-chemical exposure, and the importance of population variation and variability in modulating microbiome-mediated effects of environmental-chemical exposure. While deliberating on the three elements, the committee identified several important tool-development needs that are pertinent for addressing the research described in this chapter. Those needs are relevant to a much broader set of concerns throughout the field of microbiome research and therefore are beyond what is encompassed in the charge to this committee. Consequently, progress in those matters will not be the province solely of the research strategy set forth in this chapter. Progress in the areas discussed below should be monitored and applied where appropriate to improve knowledge about the influence of the microbiome on health risks associated with exposure to environmental chemicals.
In Vitro Models
As discussed in Chapter 4, in vitro models will be used mainly for three goals: to understand biochemical transformations of environmental chemicals by different body-site microbiomes (gut, lung, and skin) by using state-of-the-art analytic tools, to identify important interactions between environmental chemicals and the microbiome and their effects on microbial-community structures (diversity) and functions that could affect host health, and to understand host transformation of environmental chemicals that might affect microbiome composition or function. In vitro model systems that faithfully model the host gut environment—
including protective mucus barriers, immune cells, and cellular architecture—have not yet been developed, despite such advances as the SHIME and mucosal-SHIME (M-SHIME) systems. As discussed, consideration of nutrient flow, oxygen tension, mechanical stress, and microbial biofilm formation are not yet captured in a single platform, so current in vitro model systems are unable to incorporate microbial communities that are fully representative of naturally occurring microbiomes—that is, ones that contain population, structural, and physiologic diversity, such as a mix of biofilm (or adherent) and free-swimming microbial populations. It is important to understand how various factors—such as nutrient and oxygen gradients, protective mucus barriers, epithelial cell types and architecture, mechanical stress, and fluid shear stress—change microbiome gene expression and metabolism, and which factors need to be recapitulated in an in vitro system. Once in vitro systems are able to incorporate complex characteristics, the effects of an environmental-chemical exposure on the microbiome can be tested with improved robustness and understanding of the chemical–microbiome interaction and its effects on the host. Beyond in vitro systems that can faithfully model the gut, there is a great need to develop systems for studying the skin, lung, and other body sites.
Microbial Reference Communities
Past initiatives of the Human Microbiome Project (HMP) have provided some initial healthy-adult reference community data. The HMP collected samples from at least 17 body sites from among 300 people who are representative of the variation in race, ethnicity, and sex of a healthy-adult cohort of the US population. Those data are being used to inform the generation of microbial reference communities and to standardize microbial populations that faithfully recapitulate the variation present in the human microbiome. However, additional work is needed to advance the microbiome field. The development and use of reference communities for in vitro and animal studies will allow comparison of results among institutions and increase reproducibility of results. It is likely that several representative reference communities beyond those informed by the HMP will be needed to account for the developmental, disease, and geographic, racial, and ethnic differences that determine the interindividual variation discussed earlier in the present report. Capturing key demographic, medical, social, and lifestyle factors that might also shape a person’s microbiome will be important for the use of reference communities because these factors might affect interpretation of results and decision-making. It will be important to consider and incorporate not only the taxonomic variation observed in the human microbiome but the functional capacity and characteristics that continue to be discovered.
Reference Information and Annotation
The functional -omics data generated from, for example, metatranscriptomic and metabolomics approaches could help to elucidate time-resolved microbiome activity in response to environmental stresses that potentially lead to changes in host health. By understanding the time-course changes with high-complexity multi-omics longitudinal datasets, one could construct better predictive models that lead to the identification of higher-confidence biomarkers and targets. For those approaches to be used for understanding microbiome dynamics, the genomic, transcriptomic, and metabolic databases and libraries need to expand their coverage of relevant strains, genes, enzymes, and metabolite identities and functions. The vastness and complexity of the microbiome have resulted in genomic databases that contain scores of unannotated genes about which we know almost nothing. Similarly, there remains much to be annotated and identified in metabolomic databases, including chemical structure, metabolite source (human vs microbe), and metabolism pathway. Enriching the databases will facilitate clear identification of the potential for interactions among host and microbial states and for biotransformation of environmental chemicals.
As human metagenomics-sequence databases continue to expand, computational modeling strategies for reconstructing metabolic pathways and identifying enzyme homologues among metage-
nomes can provide an initial framework for inferring functions and chemical interactions of specific genes in the microbiome (Saad et al. 2012; Das et al. 2016). That approach was recently used to identify over 800 bacterial genera (from 397 human metagenomes) that might express enzymes that metabolize environmental chemicals (Das et al. 2016). Such estimates could be experimentally constrained by using metatranscriptomic and metaproteomic analyses to define which of the predicted gene products are expressed and under what circumstances.
Computational Models
An overarching goal is the development of computational models that can predict chemical–microbiome interactions and their consequences. Development of such models is in its infancy and will require large-scale data generation. At the molecular level, as noted above, most biochemically relevant microbial gene products are not yet characterized and need to be cataloged. Similarly, associations between specific microorganisms at the strain level and relevant phenotypes, such as biochemical activities or health outcomes, need to be bioinformatically identified and cataloged. Having that information will allow development of single-protein and single-microorganism models that can be extended to model biochemical activities arising specifically from microorganism–microorganism and host–microorganism interactions. The ultimate aim is to develop multiscale metabolic models that incorporate many different microbial members, human cell types, and even organ systems. The large number of interacting components and the likely stochasticity of the interactions make predictive computational models of host–microorganism interactions challenging.
It is not yet possible to predict the community-wide effects of most chemical exposures on the human microbiome; that is, how will the whole community structure be affected by a particular chemical exposure and with what temporal dynamics? Conversely, it is not yet possible to predict how the microbiome affects the ADME characteristics of a particular environmental chemical. Predictive models of microbial and chemical effects on human health outcomes will need to be developed to complement the predictive host–microorganism models. For example, computational associations have not yet been made between microbial products and most health-relevant human immune pathways or systemic metabolism. Long-term effects of microbiome activity on health, such as the induction of chronic disease, will be particularly difficult to model.
OPPORTUNITIES FOR COLLABORATION AND COORDINATION
In the United States, several agencies play roles in assessing health risks associated with exposures to environmental pollutants. Similarly, microbiome-related research is being conducted by several agencies and sectors. Progress in fields related to risk assessment and in microbiome research has occurred largely independently. The segregation of research programs in those fields, historically and currently, poses a major barrier to the advancement of knowledge on interactions between environmental chemicals and human microbiomes and the potential effects of such interactions to influence human health. Funding mechanisms that promote multidisciplinary research that specifically encourages collaboration between experts in such fields as exposure science, epidemiology, toxicology, risk assessment, human health, and microbiome research are crucial for the implementation of the research strategy described in the present report. To support such efforts effectively, agencies and research entities that conduct microbiome and human-health research are encouraged to develop collaborations with their counterparts in fields related to risk assessment and vice versa. For example, collaborations between the National Institutes of Health (NIH) and EPA or state environmental and public-health agencies that have a long history of assessing the health risks posed by environmental-chemical exposures are encouraged. That type of interdisciplinary collaboration should be sought out, encouraged, and supported to make the best use of existing knowledge and resources at each agency or organization. Like-
wise, initiatives similar to the Center for Children’s Health, the Environment, the Microbiome and Metabolomics at Emory University, jointly funded by EPA and the National Institute for Environmental Health Sciences (NIEHS) could be considered as vehicles for stimulating and fostering the types of interdisciplinary research needed. Because pharmaceuticals and other products regulated by the Food and Drug Administration (FDA) enter the environment, collaboration between EPA and FDA might be valuable. The participation of experts in diverse research disciplines during the entire research cycle—planning and designing studies, conducting experiments, and analyzing data—is likely to result in studies that are better suited to addressing the research recommended by the committee. Moreover, such multidisciplinary initiatives could serve as an ideal training environment for the next generation of researchers whose expertise spans several fields.
To assist members of the various research communities, Box 6-2 lists some important resources that could serve as a starting point for identifying potential collaborators and notes where the resources could be leveraged to address the research described by the committee. The resources and related programmatic efforts present potential high-yield opportunities to advance the current understanding of the health consequences of environmental chemical–human microbiome interactions.
CONCLUDING REMARKS
The committee believes that implementation of its proposed research strategy will substantially advance understanding of whether and to what extent the human microbiome affects the nature and magnitude of adverse health effects caused by exposures to environmental chemicals. In the relatively near term (2–4 years), results from the proposed research should allow judgments to be made about whether explicit consideration of microbiome interactions in the study of environmental-chemical toxicity yields information that is not available from traditional studies, that is, ones that do not explicitly consider microbiomes. Within a similar time frame, it should also be possible to gain some understanding of whether any such new information arises from the study of the effects of chemicals on the microbiome, from the study of the effects of the microbiome on chemical exposure, or both. Near-term results from the proposed research should thus allow judgments to be made about the need for and priorities to be assigned to continued pursuit of this new field of environmental research. Those results should also provide substantial guidance on preferred study methods.
If results from the near-term research provide relatively convincing evidence that explicit consideration of the microbiome in the development of chemical toxicity yields information that has previously been absent, the committee recommends that EPA begin to incorporate that information into human health risk assessments at least on an experimental basis. The longer-term research results should provide an understanding of the nature and magnitude of the sources of variation and variability that affect chemical–microbiome interactions and their health consequences. Those results will likely have the most important effects on the conduct of risk assessments. Ultimately, both the near-term and longer-term research should lead to the type of information that is needed to assess the importance of the microbiome as a contributor to the human health risks associated with exposures to environmental chemicals and thus allow informed decisions to be made about the need for and nature of continuing research.
REFERENCES
Berry, D., B. Stecher, A. Schintlmeister, J. Reichert, S. Brugiroux, B. Wild, W. Wanek, A. Richter, I. Rauch, T. Decker, A. Loy, and M. Wagner. 2013. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc. Natl. Acad. Sci. USA 110(12):4720-4725.
CALEPA (California Environmental Protection Agency). 2016. The Mission of the Office of Environmental Health Hazards Assessment (OEHHA) [online]. Available: https://oehha.ca.gov/ [accessed August 23, 2017].
CALEPA (California Environmental Protection Agency). 2017. Consolidated Table of OEHHA/ARB Approved Risk Assessment Health Values [online]. Available: https://www.arb.ca.gov/toxics/healthval/healthval.htm [accessed August 23, 2017].
Chauvigné-Hines, L.M., L.N. Anderson, H.M. Weaver, J.N. Brown, P.K. Koech, C.D. Nicora, B.A. Hofstad, R.D. Smith, M.J. Wilkins, S.J. Callister, and A.T. Wright. 2012. A suite of activity-based probes for cellulose degrading enzymes. J. Am. Chem. Soc. 134(50):20521-20532.
Cox, L.M., S. Yamanishi, J. Sohn, A.V. Alekseyenko, J.M. Leung, I. Cho, S.G. Kim, H. Li, Z Gao, D. Mahana, J.G. Zárate Rodriguez, A.B. Rogers, N. Robine, P. Loke, and M.J. Blaser. 2014. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. 158(4):705-721.
Dantas, G., M.O.A. Sommer, P.H. Degnan, and A.L. Goodman. 2013. Experimental approaches for defining functional roles of microbes in the human gut. Annu. Rev. Microbiol. 67:459-475.
Das, A., M. Srinivasan, T.S. Ghosh, and S.S. Mande. 2016. Xenobiotic metabolism and gut microbiomes. PLoS ONE 11(1):e0163099.
EPA (U.S. Environmental Protection Agency). 2017a. Integrated Risk Information System [online]. Available: https://www.epa.gov/iris [accessed August 22, 2017].
EPA. 2017b. Regional Screening Levels (RSLs) [online]. Available: https://www.epa.gov/risk/regional-screening-levels-rsls [accessed August 23, 2017].
EPA. 2017c. Provisional Peer Reviewed Toxicity Values for Superfund (PPRTV) [online]. Available: https://hhpprtv.ornl.gov/ [accessed August 23, 2017].
Gao. J., L.B. Ellis, and L.P. Wackett. 2010. The University of Minnesota Biocatalysis/Biodegradation Database: Improving public assess. Nucleic Acids Res. 38(Database Issue):D488-D491.
Goldsmith, M.R., J.C. Johnson, D.T. Chang, R. Tornero-Velez, J.B. Knaak, and C.C. Dary. 2012. Parameters for pesticide QSAR and PBPK/PD models to inform human risk assessments. Pp. 3-15 in Parameters for Pesticide QSAR and PBPK/PD Models for Human Risk Assessment, J.B. Knaak, C. Timchalk, and R. Tornero-Velez, eds. Washington, DC: American Chemical Society.
Koppel, N., V. Maini Rekdal, and E.P. Balskus. 2017. Chemical transformation of xenobiotics by the human gut microbiota. Science 356(6344):eaag2770.
Kotula, J.W., S.J. Kerns, L.A. Shaket, L. Siraj, J.J. Collins, J.C. Way, and P.A. Silver. 2014. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc. Natl. Acad. Sci. U.S.A. 111(13):4838-4843.
Lasken, R.S. 2012. Genomic sequencing of uncultured microorganisms from single cells. Nat. Rev. Microbiol. 10(9):631-640.
Maurice, C.F., and P.J. Turnbaugh. 2013. Quantifying and identifying the active and damaged subsets of indigenous microbial communities. Methods Enzymol. 531:91-107.
MN DH (Minnesota Department of Health). 2017. Human Health-Based Water Guidance Table [online]. Available: http://www.health.state.mn.us/divs/eh/risk/guidance/gw/table.html [accessed August 23, 2017].
NASA (National Aeronautics and Space Administration). 2017. Multi-omics analysis of human microbial-metabolic cross-talk in the space ecosystem (Multi-Omics) – 08.16.17 [online]. Available: https://www.nasa.gov/mission_pages/station/research/experiments/1949.html [accessed August 22, 2017].
NCBI (National Center for Biotechnology Information). 2017. dbGaP [online]. Available: https://www.ncbi.nlm.nih.gov/gap [accessed August 22, 2017].
NICHD (National Institute of Child Health and Human Development). 2017a. Data and Specimen Hub (DASH) [online]. Available: https://dash.nichd.nih.gov/ [accessed August 22, 2017].
NICHD. 2017b. Maternal-Fetal Medicine Units (MFMU) Network [online]. Available: https://www.nichd.nih.gov/research/supported/Pages/mfmu.aspx [accessed August 22, 2017].
NICHD. 2017c. The Human Placenta Project [online]. Available: https://www.nichd.nih.gov/research/HPP/Pages/default.aspx [accessed August 22, 2017].
Roggo, C., and J.R. van der Meer. 2017. Miniaturized and integrated whole cell living bacterial sensors in field applicable autonomous devices. Curr. Opin. Biotechnol. 45:24-33.
Saad, R., M.R. Rizkallah, and R.K. Aziz. 2012. Gut Pharmacomicrobiomics: The tip of an iceberg of complex interactions between drugs and gut-associated microbes. Gut. Pathog. 4(1):16.
Sadler, N.C., and A.T. Wright. 2015. Activity-based protein profiling of microbes. Curr. Opin. Chem. Biol. 23:139-144.
Silbergeld, L.N. 2017. The microbiome. Toxicol. Pathol. 45(1):190-194.Wright, A.T., and B.F. Cravatt. 2007. Chemical proteomic probes for profiling cytochrome p450 activities and drug interactions in vivo. Chem. Biol. 14(9):1043-1051.
Wright, A.T., J.D. Song, and B.F. Cravatt. 2009. A suite of activity-based probes for human cytochrome P450 enzymes. J. Am. Chem. Soc. 131(30):10692-19700.
Wu, H., E. Esteve, V. Tremaroli, M.T. Khan, R. Caesar, L. Mannerås-Holm, M. Ståhlman, L.M. Olsson, M. Serino, M. Planas-Fèlix, G. Xifra, J.M. Mercader, D. Torrents, R. Burcelin, W. Ricart, R. Perkins, J.M. Fernàndez-Real, and F. Bäckhed. 2017. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 23(7):850-858.
Yoon, M., J.L. Campbell, M.E. Andersen, and H.J. Clewell. 2012. Quantitative in vitro to in vivo extrapolation of cell-based toxicity results. Crit. Rev. Toxicol. 42(8):633-652.






















