3
Developing Effective Biological Detection Systems
OVERVIEW: TECHNOLOGY-FOCUSED OUTLOOK FOR FUTURE BIOLOGICAL DETECTION SYSTEMS
This section draws on a paper commissioned by the Planning Committee, “Technology-Focused Outlook for Future Biological Detection Systems,” by Duncan MacCannell and Toby Merlin (see Appendix H).1 Merlin, director of the Division of Preparedness and Emerging Infections at the Centers for Disease Control and Prevention (CDC), began his review of the invited paper by explaining the scope of the paper, which was to look specifically at technology to update those discussed at a 2013 Institute of Medicine and National Research Council workshop on technologies to enable autonomous detection for BioWatch (IOM and NRC, 2014). He noted that he and MacCannell did not, by design, look at the important aspects of BioWatch dealing with specimen collection or aerosol identification. Rather, they focused their review on detection and characterization of biological agents, with an emphasis on nucleic acid amplification and sequencing technologies that would be ready for deployment in 5 to 10 years. For a BioWatch system improvement to be ready for deployment, he explained, it has to be hardened, scaled, standardized, deployed, and validated across a network. “If we picked a technology right now, it would take quite a bit of investment and quite a bit of time to do that scaling, validation, and deployment,” said Merlin. Given this requirement, any technology they considered had to be mature. In the parlance of technical readiness, where Level 1 technology is aspirational and Level 9 is something a consumer could purchase off the shelf, technologies they deemed mature were those between Levels 7 and 9.
Any acquisition program needs requirements, and BioWatch requires technologies that are highly sensitive, are exquisitely specific, have a low cost per sample, have fast turnaround time, have the flexibility to incorporate new targets, have the ability to be standardized and perform reliably across a network, and
___________________
1 Although the authors of the paper mentioned specific products and tradenames, doing so does not imply endorsements by CDC, the Department of Health and Human Services, or the National Academies of Sciences, Engineering, and Medicine.
might be incorporated into an autonomous system. Any new technology should also have practical bioinformatics requirements and robust quality control and quality assurance. The challenge, said Merlin, is that no system will meet all of these requirements, so it is necessary to set priorities, which for BioWatch have to include high specificity to preclude false positives. “There is little tolerance for false positives in the operating world,” said Merlin.
The first set of technologies Merlin discussed were those that detect nucleic acid signatures, and, in his opinion, there is a real opportunity to upgrade and expand the current primary polymerase chain reaction (PCR) process. In the category of commercial, real-time, multiplex PCR systems, he started with the Luminex xMAP system, a highly flexible solution array technology that uses probes based on fluorescent color-coded microspheres. This system is approved by the Food and Drug Administration (FDA) for clinical assays and is easy to adapt to accept new primer/probe sets. Its primary weaknesses are that it requires a high level of expertise to operate and troubleshoot, it may be difficult to keep the instrument working consistently, and it is not considered cost effective for BioWatch. When asked if requiring a high level of expertise is an actual weakness if a technology meets all other requirements, Merlin replied that it is not unreasonable for BioWatch to expect a higher level of expertise to operate a technology, but that should not have to apply to knowing how to troubleshoot or repair an instrument on a regular basis.
Two other multiplex real-time PCR systems are ThermoFisher’s TaqMan Array and newer OpenArray. A strength of these systems is that the sample is aliquoted into a fixed array of multiple reaction wells on a single card containing 382 wells for TaqMan and more than 3,000 wells for the OpenArray system. Another strength is that this technology is used widely in laboratories and is familiar to those who do nucleic acid analysis. A major weakness, however, is that it dilutes the sample by taking a small sample (with a small amount of DNA) and divides it into hundreds or thousands of aliquots, which Merlin said leads to lower sensitivity than with other methods.
The Wafergen SmartChip is a relatively new version of a multiplex real-time PCR system that uses 5,184 extremely small volume reaction wells on a single chip. A strength of this system is that users can print their own arrays, which enables incorporating a new target quickly. Merlin characterized it as the most flexible of the multiplex real-time PCR systems, but it, too, suffers from low sensitivity associated with low reaction volumes. In addition, the instrumentation is proprietary and does not have an extensive track record.
The LexaGene Digital Droplet system is not on the market yet, but the manufacturer claims the system can analyze 22 targets simultaneously at a low cost per test. This technology converts a sample into tens of thousands of separate droplets, each in its own oil immersion that serves as the reaction vessel for PCR amplification. A strength of this approach, said Merlin, is that it enables detection of small amounts of a target that inhibitors might obscure. Another strength is that it can handle a large volume of samples. Merlin said the LexaGene technology originated from a BioWatch-funded project.
The BioFire Film Array is a commercially available system that has been around for years and for which there are many FDA-approved diagnostic panels, said Merlin. In his opinion, BioFire is an example of what can be done to simplify and automate real-time PCR. The user injects a sample into a disposable pouch containing freeze-dried reagents for multiplex real-time PCR. The user then inserts the pouch into the BioFire machine, which produces results in an hour. The Department of Defense (DoD) worked with BioFire to create the Warrior Panel, a pouch that can detect 16 agents and 26 total targets using its Joint U.S. Forces Korea Portal and Integrated Threat Recognition System (JUPITR) system to detect aerosolized biological and chemical agents at U.S. military installations. Strengths of the BioFire Film Array are that it can handle a large sample size and it is a simple-to-use, walkaway instrument. Although this system’s throughput is too slow and its cost per sample is too high for BioWatch’s purposes, Merlin said it does illustrate what is possible with real-time PCR.
Turning to next-generation sequencing, Merlin said it has exciting possibilities for BioWatch. Technologies from Illumina and ThermoFisher IonTorrent use different approaches to producing short reads, which provide higher fidelity and lower error rates than with longer reads. Pacific Biosciences produces a long-read, single-molecule sequencer that can produce from 3,000 to 10,000 base pair reads. The advantage of longer reads is that they reduce the amount of bioinformatic analysis required after sequencing.
Oxford Nanopore’s MinION system, another long-read sequencing, is small enough to fit in the palm of a hand, has a low cost per sample, and provides real-time data on the molecule it is sequencing. However, said Merlin, it requires a great deal of sample preparation before sequencing. “The amount of time for all of this is not something you would do as a routine on BioWatch samples currently or in the foreseeable future,” he explained. He added that Roche and Genia are working on a similar system, though few details are available. In summary, Merlin said that next-generation sequencing is rapidly evolving, but BioWatch is not likely to be deploying it in its next technological iteration.
Metagenomics—sequencing many targets at the same time in a single sample—encompasses a range of developing sets of technologies. Highly multiplexed amplicon sequencing looks at species-specific and strain-specific target amplicons using what Merlin called targeted sequencing. This approach first requires performing PCR on a sample and then sequencing the amplicons. Although the presequencing steps are rate limiting for this technology, it does afford the opportunity to look for multiple targets simultaneously and to amplify targets that are present in small amounts. The ThermoFisher IonAmpliSeq and Ion Chef systems, used in conjunction with IonTorrent sequencers, are highly sensitive and good for complex or difficult samples, said Merlin, but the minimum turnaround time is currently around 10 hours. Illumina’s TruSeq system, which feeds into Illumina’s sequencers, has similar advantages with a turnaround time of 6 to 27 hours. Fluidigm’s Targeted Sequencing Library Preparation System is compatible
with both ThermoFisher and Illumina sequences. It uses microfluidic chips to amplify multiple samples against thousands of primers with a turnaround time of 12 to 14 hours, not including sequencing time.
A second metagenomics approach uses shotgun sequencing, which involves extracting nucleic acids from a sample, sequencing everything present, and using bioinformatics to identify what is most likely present. Shotgun sequencing is useful for microbial ecology studies that aim to identify predominant organisms, but it is not good for finding organisms present in small amounts. Additional issues with bioinformatic analysis and turnaround time make it unlikely that shotgun sequencing will be feasible for large-scale surveillance applications in the next 5 to 10 years, said Merlin.
One final technology Merlin and MacCannell included in their invited review was noted in the 2013 technology update (IOM and NRC, 2014) and has since come to market. The Quanterix SIMOA SR-PLEX is used primarily for immunoassays. However, the manufacturer claims it can be used to detect nucleic acids, is sensitive enough to detect one molecule of the target, and is 1,000 times more sensitive than standard immunoassay technologies. This technology is capable of limited multiplex testing, and it has the potential to serve as an orthogonal test method that could be used to verify a positive result.
In summary, said Merlin, there are newer mature technologies available that offer multiplex testing, amplicon sequencing, flexibility in selection of targets, rapid testing, and “the Holy Grail of walkaway testing, where you can load the sample, push a button, come back in an hour, and have a result.” He added that none of them, in his opinion, are perfect fits for BioWatch as they currently exist. “I wanted to emphasize, again, that whatever the technology, the scaling, standardizing, hardening, deploying, and validating of any new technology across a network is both essential and time consuming,” said Merlin. As an illustration, he recounted developing a test for the Laboratory Response Network, getting to the point of deployment, and finding out that production of the reagents needed for this assay was not reliable from lot to lot, and therefore not usable in practice.
ENHANCEMENTS TO EXISTING SYSTEMS
This session focused on enhancements to existing BioWatch systems rather than new technologies. To provide context for this panel session, moderator Bruce Budowle explained that it is important to appreciate that BioWatch’s purpose is to detect a substantial release rather than a single particle. The system has to be sufficiently robust to deal with the types of environmental insults to which the targets are going to be exposed, but it is not necessary to have exquisite sensitivity. He then introduced the three panelists and the topics of their presentations: Dana Kadavy, director of biological sciences at Signature Science, addressed capture and extraction; Henry Erlich, senior scientist at Children’s Hospital Oakland Research Institute, discussed improvements to PCR technologies; and C. Titus
Brown, University of California, Davis, spoke about approaches to improve informatics, data flow, and connectivity. An open discussion followed the three presentations.
Capture and Extraction
BioWatch’s current collection and capture system (see Figure 3-1) is designed for an airflow rate of 100 liters per minute over a 24-hour collection cycle. As Dana Kadavy explained, a 3-micron polytetrafluoroethylene filter sits in a filter holder inside the aerosol collector, which extends upward from the mechanical portion of the device that is plugged into a power source. A field staff member removes the filter holder, never touching the filter itself, and replaces it with a new filter preloaded offsite into a new filter holder. The field staff member places the filter and holder in appropriate primary, secondary, and tertiary containment vessels and documents and maintains chain of custody until they deliver to the laboratory for analysis. Typically, collection occurs early in the morning.

At the BioWatch laboratory, a technician removes the filter from its holder, cuts it into quadrants using a sterile scalpel or single-use scissors, and places one quadrant into a screw-top tube containing glass beads.2 The sample then undergoes a cell disruption step involving extreme agitation for several minutes in a buffered lysis solution followed by immediate cooling. The resulting mixture is extracted in a two-stage filtration process using one of two methods, depending on how many samples a laboratory processes, followed by a heat inactivation step to ensure all DNA-degrading enzymes are inactivated. The initial step in either process removes large particulates and any precipitates in the system, Kadavy explained, while the second step traps the target signatures on a filter that is then washed extensively. The target signatures are then eluted from the filter with PCR grade water and placed in a 96-well master plate for analysis.
These steps, said Kadavy, require a great deal of manual manipulation following a detailed protocol that includes alerts to the laboratory staff of potential pitfalls along the way. For example, the protocol contains alerts about not leaving the sample in the lysis solution for too long or not letting the filters become too dry. The protocol, she added, requires the analysts to be vigilant throughout the process.
Aspects of this process that Kadavy said could be improved included collection and capture efficiency, coverage, sampling errors, agent stability and viability, extraction efficiency, laboratory contamination and exposure, and automation to reduce manual manipulation and increase throughput and reduce cost. “What can we do to lop off some hours in the analytical phase, especially in the front end, in the sample processing, which is always a bottleneck?” she asked. “The endgame here is to make the process more effective and decrease the time to answer.” She also wondered if BioWatch might be missing an opportunity to include targets beyond biologicals, even though the BioWatch mission is exclusive to biological agents.
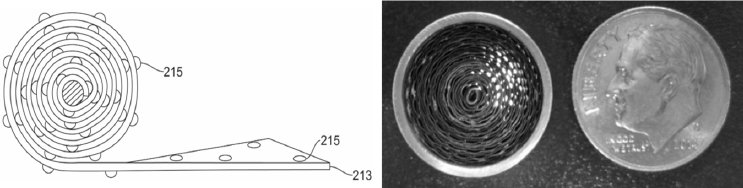
Kadavy also wondered if it would be useful to evaluate alternative materials for the filters in the portable sampling units. One challenge with other filter materials is that although they may be better at capturing biological particulates, releasing the particles back into solution tends to be difficult. An alternative might be to use a filterless collection system or something radically different, such as the dime-sized, battery-powered, coated coiled aluminum ribbon collector originally developed by the Defense Advanced Research Projects Agency for chemical detection (see Figure 3-2). This device, which operates at an airflow of 20 liters per minute for 5 hours and can be mounted almost anywhere with a magnet, could be used to augment the main collector in an indoor environment, said Kadavy. She noted that some of the BioWatch Generation 3 technologies that were tested included filterless capture devices and she asked if it would be possible to leverage some of those technologies going forward. In the same vein, she thought it worth exploring whether increasing the amount of air pulled through the collectors would increase the amount of material collected on a filter.
___________________
2 This is a condensed version of the sample processing, which in practice follows an extensive standard operating procedure.
To address sampling errors associated with uneven distribution of particles on a collection filter, Kadavy suggested it might be a good idea to collect duplicate filters and archive one of the two filters instead of quartering one filter. Another approach would be to extract the entire filter and store a portion of the extract for subsequent analysis, although this would require BioWatch laboratories to have freezer space for those samples. With regard to agent stability and viability, she suggested modifying the concept of operations to include some shorter-period sampling points to reduce the problem of severe sample desiccation on the filter, which decreases the odds of having a viable sample. Another approach would be to use a gentler air sampling system, such as a wetted-wall cyclone particulate collector. The Department of Homeland Security (DHS) has funded many studies looking at filter materials and extraction methods with the goal of increasing extraction efficiency, said Kadavy, and she thought there were small changes that could be made to increase extraction efficiency from filters. “I really think there are some optimizations that can occur here,” she said.
One way to decrease analysis time is by automating many of the steps that currently require manual manipulation, and Kadavy noted that there are several commercial robotic systems for nucleic acid extraction. Some of these are small enough to fit in more space-constrained BioWatch laboratories. She wondered if there were modularized technologies evaluated during the Generation 3 testing process that could be leveraged to enhance the current process.
In her opinion, minor modifications to the capture and extraction modes of the current system have the potential to improve efficiency, especially with regard to the likelihood of a positive detection if a biological threat agent is present. She suggested that BioWatch should examine more closely some of the work DHS, other federal agencies, the national laboratories, and industry have funded to see if there are proven technologies that BioWatch can leverage.
Improvements in PCR
Before addressing approaches for improving the PCR step in the BioWatch process, Henry Erlich, session moderator, reviewed some of the relevant performance metrics used to characterize PCR performance. One obvious metric, he
said, is sensitivity as it relates to the limit of detection. While it is possible to generate a signal from a single template molecule, the limit of detection in the real world is defined by the nature of the extract and the instruments used in the analysis. Other metrics include specificity, which is determined by the primers and probes; robustness, or the ability to perform in a variety of circumstances; the potential for quantification; and target breadth, or the number of targets detectable in a single reaction. Erlich noted there are two approaches to increasing target breadth: multiplex PCR and amplification of a universal target, such as the bacterial 16s RNA gene, followed by further analysis with probes or sequencing.
Erlich said various characteristics of the several types of DNA polymerase have the biggest effects on PCR performance. Some polymerases, for example, have greater fidelity or are more resistant to inhibitors than others. It is possible today to screen thermophilic bacteria or genetically engineer a polymerase to find or produce a polymerase with specific performance characteristics, possibly including the ability to function in the presence of PCR inhibitors. The thermal profile of a polymerase and the number of amplification cycles can have a significant effect on specificity, he said. For example, increasing the annealing temperature and elongation reaction temperature can increase specificity. Reaction conditions can have a major effect on both sensitivity and specificity, too, as can the instrument platform and the choice between using probes or sequencing as the analytic method.
The most significant change in PCR performance occurred almost 30 years ago when the heat-stable Taq polymerase was discovered in an organism growing in a Yellowstone hot spring. This polymerase made PCR more specific, efficient, and automatable with a simple thermal cycling instrument, said Erlich. Multiplex and real-time PCR have both led to improvements in PCR performance. One of the advantages of real-time PCR is that it enables quantitative measurements.
There are a number of steps that could make multiplex PCR more robust, said Erlich. Designing amplicons to be of similar size can minimize primer artifacts that reduce sensitivity, such as the formation of what are called primer dimers. Adjusting primer concentrations to balance efficiency among different primers, and designing primers with universal sequences, can also improve the performance of multiplex PCR, as can using real-time PCR instruments with the capacity to detect multiple fluorescent probes.
There is a real need, said Erlich, for reference collection standards to measure inclusivity and exclusivity with respect to targets, near neighbors, and more distant, nontargeted organisms. Inclusivity panels would include the set of pathogen strains the assay should detect, while exclusivity panels would include a set of closely related species and strains the assay should not detect. Reference collection standard backgrounds are needed to determine how environmental background properties affect PCR performance, and should include a panel of other organisms in the environment the assay should not detect, as well as background substances such as dust and pollen that might interfere with an assay.
In closing, Erlich said the PCR performance standards for BioWatch should establish that an assay has sufficient sensitivity to detect the release of a tested biothreat agent at a program-relevant concentration above the baseline of environmental background. Performance standards should also establish that an assay has sufficient specificity to detect pathogenic strains of concern but not to cross-react with other strains and organisms, within acceptable program false-positive and false-negative rates, and that it is sufficiently robust for routine operational use.
Enhancing Informatics, Data Flow, and Connectivity
One issue BioWatch needs to think about, said C. Titus Brown, is the increasing wealth of available genomic data. The reasons are twofold. First, he said, “our awareness of what is in the environment and likely to be in various samples is growing at an astronomical rate, along with microbial databases.” There are now some 100,000 microbial genomes that are finished, close to finished, and mostly annotated within public repositories, with 10,000 to 20,000 genomes being added per year, many of which are for strain variants of existing genomes.
The second reason why BioWatch needs to think about genomic data and databases is that researchers are depositing a great deal of data from human symbionts, that is, organisms that live on and in the human body, something that Brown had not considered until preparing for this workshop. In general, these sequences are not in GenBank, but are in the publicly accessible but uncurated Sequence Read Archive. “I do not think anybody is taking a close look at those in terms of how they might share sequence with some of the things that we are concerned about,” said Brown.
All of this is relevant to improving BioWatch’s current methodology because some primer sets may be less pathogen specific than previously thought and new data could help develop more specific probes. Public databases, said Brown, could be screened against private databases to determine which portions of the private databases are not public and can be used for primer design by those who are trying to design biological weapons. In addition, he said, public databases can be used to expand at relatively low cost the secondary “reflex” panel screening for distinguishing between pathogens and nonpathogenic microorganisms.
The last subject Brown addressed was what he called the pangenome problem. “Increasingly, we are finding that many bacteria in the environment exist in vast clouds of sequence diversity, the so-called pangenomes,” he said. “This complicates genomic analysis immensely because genomic elements previously thought to be signatures of specific bacteria seem to be widely shared among strain variants of those bacteria in the environment.” In other words, he explained, no single genotype seems to exist.
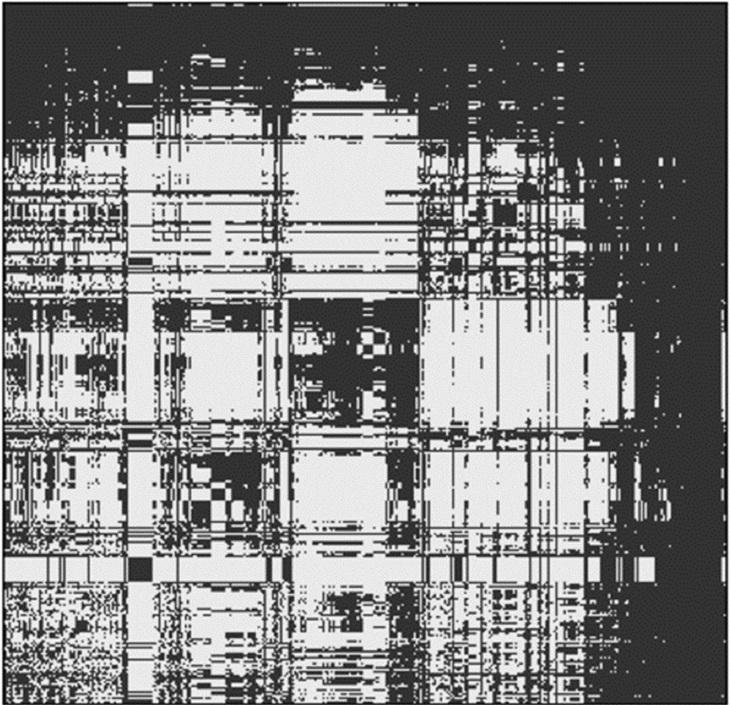
To illustrate this problem, Brown showed an analysis he conducted in which he randomly selected Escherichia coli genomes and asked which genetic elements within those 50 genomes correlate with only one other genome (see Figure 3-3).
Choosing a unique element to identify one specific strain requires finding a column of the resulting matrix that was present only on the diagonal, which turns out be essentially impossible with a single primer pair, he said. This could be a significant problem given that most bacteria in the environment have not been sequenced. His suggestion was to consider developing multiple primer pairs.
One action Brown suggested that BioWatch might take is to use informatics tools to postprocess multiple false-positive events to distinguish endemic organisms from signals. In his opinion, the type of modeling that David Brown spoke about earlier would be well suited to integrating this type of data over multiple detectors or to determine if there were signatures for specific seasons or wind directions. It may be necessary, he said, to use more expensive techniques, such as shotgun metagenomics at great depth, to better understand some of the false-positive events that occur as a means of better shaping the analysis pipeline.
He noted that the MinION sequencer may show promise in BioWatch given that it has been deployed in the field looking for Ebola virus. “It may not be high throughput or particularly rugged yet, but it does show more promise than any of the other large sequencers because you can actually carry it out into the field and you do not need much of a lab to work with it,” said Brown. In closing, he said he believes that there will be dramatic improvements in the sequencing technology that may be included in future BioWatch technologies.
Discussion
An unidentified participant began the discussion by noting that a technology enhancement working group that met last year emerged with a priority list that the three speakers did a good job summarizing. The participant then asked if the current BioWatch assays could be multiplexed to include several agents in a single reaction given that there are now numerous fluorescent detectors available for use with TaqMan PCR. Erlich replied that he could not answer the question specifically because he was not sure what the current reactions are, but he has great confidence that it is possible to develop robust multiplex PCR assays for almost any targets. The limiting factor, he said, is the availability of instruments that can detect multiple fluors.
The same participant then noted that because of BioWatch’s outstanding quality assurance program, it should be possible to develop spiking standards of known concentration that would enable real-time PCR to provide quantitative or near-quantitative results that would be useful when dealing with advisory committees. Erlich agreed with that idea.
Willy Valdivia commented that there needs to be an examination of signature stability and erosion that could be used to develop models regarding signature stability. “I think that is something we need to discuss in the future if we want to design better PCR assays and to develop algorithms for processing metagenomic information or information from any sequencing platform,” he said. The goal would be to identify specific genomic regions in the pathogens of interest that would be stable enough over time, independent of new sequences in the databases, to serve as representative of the taxonomic diversity of the organism of interest. Brown responded that what Valdivia proposed is a basic research problem, and he predicted that it will be possible in the next 10 to 20 years to get the full genome of every cell collected on a filter. “We still would not know what 99.999 percent of those bacteria did and we would not know which of them were virulent, which of them were pathogens, and which of them were harmless,” said Brown. “We simply do not have the information to characterize them.”
The challenge over the next 5 years, Brown said, is to optimize the current process to preferentially return positives only when known pathogens are present. He added that, beyond that time frame, a new set of technologies will deliver whole genomes. “At that point, we want to integrate more in the way of pathway modeling and other approaches to figure out whether or not these newly isolated
genomes, which are probably going to be bioengineered by bad actors at that point, are actually dangerous,” said Brown. “That is going to be a different kind of question than genomic identification.” Valdivia agreed and said the problem with developing primers today is the error rate in the public databases, an assessment with which Brown agreed.
Budowle wondered if it would be possible to develop a PCR assay for antibiotic resistance genes in the known target organisms that would be used after a first signal or a second panel signal, rather than having to culture the identified organism. Everyone on the panel thought that was a good idea. Budowle then raised the idea of spiking samples with an internal control to determine if a negative assay resulted from PCR inhibitors that were present on the original filter. Mark Scheckelhoff, BioWatch’s director of laboratory operations, said that is done currently, though he could not provide details.
Scheckelhoff then reiterated a comment Michael Walter made earlier, which is that BioWatch does not have the ability to do its own research and development. “That is something that really hampers the program’s ability to make some of these straightforward improvements,” he said. BioWatch, he explains, depends on DoD for its assays and the CDC’s Laboratory Resource Network (LRN) reagents for its verification work. “Not only is there a large cost associated with doing a new validation or verification for each one of these signatures, you have to remember that there is not necessarily the same priority or impetus to spend that money, in terms of where the LRN stands or where the DoD stands,” said Scheckelhoff. “While we are a big customer, we are not driving their individual policies or priorities.”
When Budowle asked if the program collects data on dirty filters to see if there is some seasonal relationship or other information that might be gleaned from those data, Scheckelhoff replied that the program does collect such data, but how dirty samples look in one jurisdiction is different than in another jurisdiction. In addition, the look of a filter is not indicative of whether there will be inhibitors present. This is a location-specific and season-specific issue that requires making adjustments to the concept of operations to address and mitigate this issue.
Budowle then brought up the idea of looking at a cost-benefit analysis for BioWatch in terms of its mission. Walter replied that the program did have an independent cost-benefit analysis done recently. This calculation, which was based on a number of assumptions, found that if there was one attack in 50 years and BioWatch saved 1,100 lives, it will have paid for itself given its current budget. Walter likened BioWatch to fire insurance—nobody wants to pay for it, until they are glad they have it when a fire destroys their home. The issue BioWatch faces, he said, is that since the anthrax letters were sent to Congress there has not been a bioterrorism attack, and so the threat has receded from the public’s eye and Congress’s attention. At the same time, when the BioWatch program was considering the costs of deploying what at the time was to serve as the Generation 3 system, Congress did not balk at spending $6.1 billion over 23 years.
NOVEL TECHNOLOGIES TO EXPAND CAPABILITIES
As an introduction to the panel that would discuss some up-and-coming technology options, session moderator Tom Slezak said he asked the panel to think more broadly than about the primary detection issue and to consider issues such as the challenges associated with indoor detection and determining how far an agent has spread after an initial attack. The three panelists in this session were Wayne Bryden, president of Zeteo Tech, who discussed the use of mass spectrometry as a detection and identification technology; Lyle Probst, chief executive officer of Excite PCR Corporation, who spoke about a novel potential use of PCR technology; and Sam Reed, president of DNA Electronics, who addressed the possible use of next-generation sequencing as a BioWatch technology. An open discussion followed the three presentations.
Mass Spectrometry
A mass spectrum, explained Wayne Bryden, is the pattern of molecular fragments produced when a source of energy, such an electron beam, ion beam, or laser, strikes a molecule, ionizes it, and causes it to break into smaller fragments. This process occurs in the vacuum of a mass spectrometer, which typically measures how long it takes for the fragments to travel through the vacuum and reach a detector, with the time of flight (TOF) relating to the mass-to-charge ratio of each fragment. In 2002, the Nobel Prize in chemistry was awarded for the development of two forms of mass spectrometry—matrix-assisted laser desorption/ionization (MALDI) and electrospray ionization—that are particularly useful for characterizing biological molecules.
With “whole cell” MALDI-TOF mass spectrometry, it is possible to analyze an entire microorganism, and in the late 1990s, the Defense Advanced Research Projects Agency funded work by Bryden and his colleagues Catherine Fenselau and Robert Cotter, who were then at the Johns Hopkins University Applied Physics Laboratory, to develop a completely autonomous MALDI-TOF tool for biological detection. The resulting machine could analyze a sample aerosol every 5 minutes, archive portions of each sample, analyze the sample, and run a detection algorithm. The instrument performed well in building protection tests, said Bryden, when the target-to-clutter ratio was high. “The virtue of mass spectrometry is that you can detect anything,” he said, “but the curse of mass spectrometry is that you detect everything, so success depends on separating the anything from the everything.”
Bryden said there are four main strategies for reducing clutter in mass spectrometry, several of which are commercially available. One strategy, useful for clinical work, involves culturing whatever is in a sample overnight, taking the resulting cultures, and running them through the MALDI-TOF process (see Figure 3-4). The commercial systems, Biotyper and VitekMS, work well in the clinic,
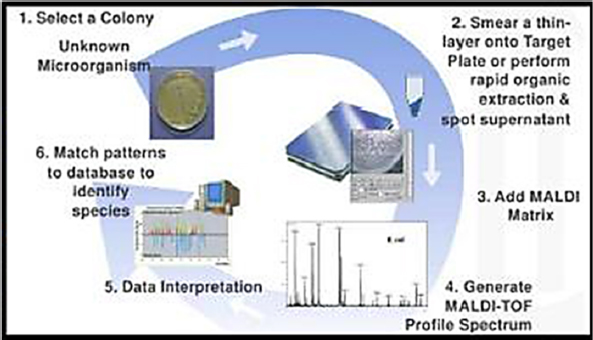
and though the instruments are expensive at approximately $350,000, accuracy is greater than 99 percent, and the cost per sample is somewhere between 25 cents and one dollar. In addition, research has begun identifying strain-level markers and markers for antibiotic susceptibility. Both instruments have been approved for use in clinical diagnostics by the FDA and the relevant European regulatory agency. Bryden estimated that the two instruments have analyzed close to 100 million samples. He noted that the data the instruments generate go back to the manufacturers, who continue to modify and improve the algorithms used to identify organisms, and as a result it is now considered the gold standard assay in the clinical setting.
A second approach looks for a trigger that signals the mass spectrometer to begin sampling when the target is present. In most cases, the trigger is an optical signal such as fluorescence or changes in light polarization. Bryden’s company has developed a briefcase-sized MALDI-TOF instrument that uses polarization measurements as the trigger. The technical challenge with this approach is making sure the trigger can sense the presence of the targeted agent in the outdoor environment, though Bryden said it does work well in the indoor environment. The combined trigger-MALDI-TOF system can analyze a sample in a few minutes.
The Army Research Office has funded work by Bryden’s company and Fenselau, who is now at the University of Maryland, to build a field-deployable MALDI-TOF assay for peptide biological toxins such as ricin, abrin, and botulinum toxin. This approach, he explained, takes advantage of the existence of carbohydrates that will capture these types of toxins and bind them strongly while the sample is washed to remove clutter. Treating the washed sample with hot acetic or formic acid for approximately 1 hour fragments the toxins, and the resulting fragments are then analyzed by the mass spectrometer. All of the laboratory steps have been incorporated into a single-use microfluidics cartridge that can be plugged into a portable MALDI-TOF mass spectrometer. Detection algorithms,
Bryden said, rely on publicly accessible peptide sequence information. Every peptide in the resulting map can be further sequenced in seconds to provide an additional level of certainty, and a mass spectrometry–based activity assay that the CDC developed can provide a third level of certainty (Kalb et al., 2015).
The final approach for clutter reduction that Bryden discussed, which he called digital molding, coats every particle in an aerosol stream with the matrix material that enables the MALDI process and then analyzes each matrix-coated particle. Laboratory tests have shown this approach has single-particle sensitivity and good specificity, but issues with the matrix-coating process need to be solved for this technology to be useful in the field. Bryden’s company is working on improving that process, adding optical measurements to the system, and producing an instrument capable of real-time detection in the field.
Point-of-Need PCR
Lyle Probst, who was part of the team that developed the first-generation BioWatch system, discussed his company’s work developing a battery-powered microfluidic system with an integrated and automated sample preparation module that performs point-of-need PCR in 30 minutes. This system, which the company has named FireflyDx, can detect a variety of pathogens, including those of interest to BioWatch, without the need for highly trained personnel. Probst noted that the same device can be used to process environmental, clinical, and even agricultural samples. Because the company is developing applications for multiple markets, he expects that economies of scale will drive down the cost per sample. The current portable device, about the size of a toaster, produces results that are equivalent to those generated by industry-standard benchtop clinical instruments. Among the many uses Probst envisions for the device, particularly for a smaller handheld device the company hopes to have available in 2 years or so, is to test a space for biological contamination as a safeguard for first responders.
The analytical process, explained Probst, starts with placing a liquid sample into the FireflyDx cartridge and inserting the cartridge into the Firefly instrument. Samples can be any biological fluid or an extract of a swab off a surface. Once the cartridge is inserted, the instrument performs sample purification, nucleic acid extraction, and real-time PCR. The device cartridge is programmed for a specific assay. For example, a Bacillus anthracis cartridge runs a specific protocol designed to crack open this organism’s tough spore coating, while an influenza virus cartridge is programmed to use a much less harsh process to preserve the integrity of the fragile virus particle. “The end user does not have to know any of that,” said Probst. “They just grab the influenza cartridge, put it in the machine, and because of the chip that is in there, it is already preprogrammed with a protocol for an RNA virus.”
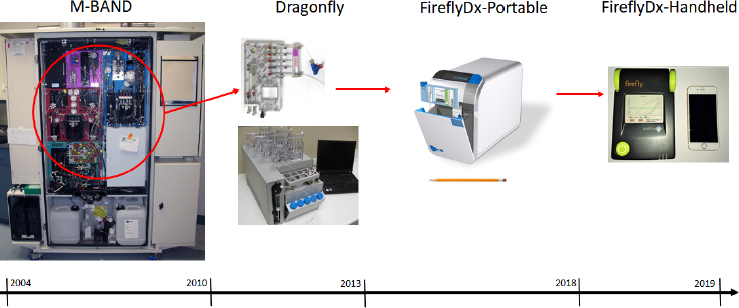
The FireflyDx instrument evolved from m-BAND, a cabinet-sized, BioWatch Generation 3 instrument designed to process airflow of 400 liters per minute for 30 days unattended and produce results every 3 hours for 17 signatures
across six organisms and three toxins. Taking what he and his colleagues learned developing this instrument, they worked at shrinking each module and reducing the cost of the overall instrument (see Figure 3-5) while retaining the performance equivalent to an industry-standard laboratory diagnostic instrument. The goal, explained Probst, was to avoid having to take a sample back to the laboratory to confirm the results from the portable device. He noted that the sample-preparation cartridge would fit in both the portable and handheld versions. The current cartridge runs four PCR assays simultaneously.
Early in the development process, Probst and his colleagues decided to keep real-time PCR as the core technology, as opposed to designing around it, and to focus on making it faster and less expensive. One benefit of this decision is that it is straightforward to take any available PCR assay and port it directly onto the device at industry-standard volumes. “We intentionally designed this to be assay agnostic,” said Probst. “We do not want to be an assay development company.” Noting that the plastic cartridge is the key to the successful development of the FireflyDX system, he explained that it is inexpensive and the reagents it contains are stable at room temperature for over a year. It contains an RFID chip to enable automated chain of custody, and it can be autoclaved before disposal. The company projects the instrument will cost between $20,000 and $30,000, with the cartridges running around $20 apiece.
Probst said he sees FireflyDX serving as a confirmatory device and explained that it has been designated to serve in that role in DHS’s SenseNet program. In one field test not connected to the SenseNet program, FireflyDX, running an assay developed by GenArraytion, was able to accurately and rapidly detect Zika virus. In a separate test program, FireflyDx was able to detect a marker for genetically modified organisms, which is relevant to the corn and soybean export market, with the same performance as the industry-standard laboratory PCR system.
Rapid Targeted Next-Generation Sequencing
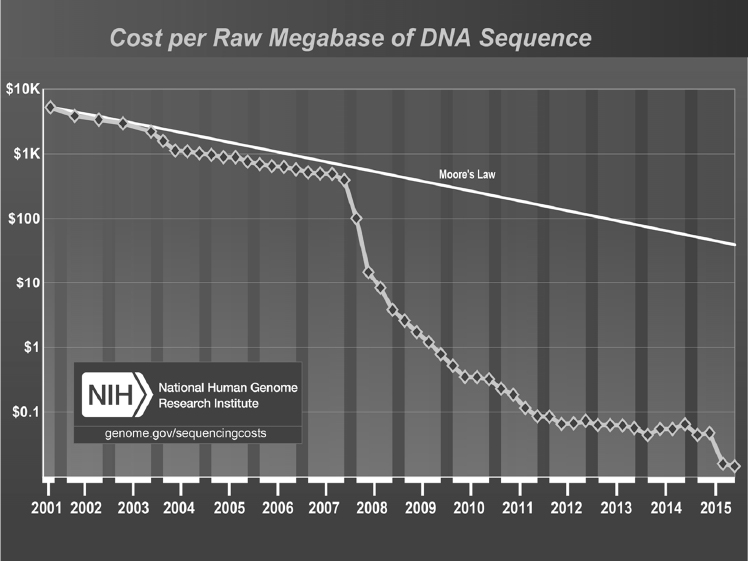
The development of next-generation sequencing technologies, said Sam Reed, has triggered a dramatic decline in the cost of sequencing, one that since 2008 has occurred at a pace faster than that of the semiconductor world (see Figure 3-6). As a result, next-generation sequencing is making it possible to detect an extremely broad range of pathogens, strains, and antibiotic resistance in a single test and to catch “unexpected” or rare organisms. It also makes possible assays that are less sensitive to genetic drift and shift and that might have a reasonable chance of detecting something someone designed using synthetic biology, issues that several participants raised in earlier discussions at the workshop. One advantage of a sequencing-based assay, said Reed, is that even if it detects something that was not anticipated, it still generates sequence data for further analysis. “You still have a chance to interpret those data and act, based on that information, even if it is an organism you did not plan for in years past,” he explained.
Sequencing data have an advantage over other broad-based methods such as mass spectroscopy because the data are always in a format that institutions can exchange and archive for later analysis. “The significance of that is that sequence may be something that weeks or months later can be used in unforeseen ways and can have new meaning,” said Reed.
There are, however, some significant limitations to next-generation sequencing. Turnaround time from collecting a sample to generating sequence data typically requires days or even weeks, in large part because of the extensive sample preparation needed before sequencing can begin. Reed noted that while the instruments themselves have been refined and shrunk, the processes leading up to feeding a sample into those machines still need a great deal of work. The complexity of the process, he said, is such that it requires specialist operators and bioinformaticians to generate reliable and meaningful data, and standardizing and hardening the process has proven to be challenging, with even the few FDA-approved sequencing-based assays essentially being run as experiments. Currently, he added, there is a trade-off between turnaround time and cost. “The limitations are extensive, and when most sequencing companies talk about what they are doing to move closer to diagnostics, I think those are baby steps and they still have a mile to go,” said Reed.
Current platforms include those developed by Illumina, ThermoFisher Ion Torrent, Oxford Nanopore, and his company, DNA Electronics. The Illumina technology, currently the dominant market player, uses fluorescence-based imaging. It has gone through incredible scaling, said Reed, and is a powerful technology. In general, the various versions of Illumina’s technology operate in its original paradigm as a research tool. ThermoFisher Ion Torrent licensed its technology from DNA Electronics, and after trying to go head-to-head with Illumina the company decided to move into targeted diagnostic applications in the ecology and human genetics settings rather than in infectious diseases. Reed called Oxford Nanopore’s technology exciting, with a great deal of potential, but also plagued by error rates in its current form. For all of these technologies, what is needed is to develop them to the point that they are robust from end to end and fast.
With regard to the two main approaches to next-generation sequencing—targeted amplicon sequencing and unbiased shotgun sequencing—Reed explained that shotgun sequencing has significant challenges when it comes to infectious disease applications because the target produces only a tiny percentage of the total sequence data generated from a raw sample. While the sample preparation is easier with shotgun sequencing, the sequencing process is expensive and the data analysis is lengthy and difficult and not really appropriate for BioWatch’s purposes. Reed predicted that shotgun sequencing could gain traction in unusual clinical cases for which no other technologies are providing an answer, and indeed, there have been several reports in the literature where it has been used in that way.
For most applications, then, targeted or amplicon-based sequencing will be the preferred technology. “Targeted or amplicon-based sequencing can be a near-universal method,” said Reed. “You can go after conserved regions that maximize
the chances you will get sequencing reads off of whatever is present in your sample.” He cautioned not to believe anyone claiming to have a universal method that works for every target. Nonetheless, he said, the cost-benefit trade-off of using an amplicon-based method is that there is minimal loss and tremendous benefit.
DNA Electronics’ goal is to refine amplicon-based sequencing so that it produces results from as little as one target per milliliter, directly from raw specimens, in 3 to 4 hours, including sample preparation and analysis. The resulting system also needs to be user friendly, robust enough that a general-purpose user can operate it, and provide broad coverage of organisms, strains, and antibiotic resistance. “Our vision is to move sequencing from a high-skill, high-capital-expenditure, centralized process to something that essentially anyone can use,” said Reed. Realizing that vision, he explained, requires automating the entire process from initial specimen preparation through sequencing. He added that raw sequence data by themselves are not clinically relevant. What is relevant is using those data to identify an organism or a resistance determinant, and that requires a curated database.
In terms of clinical applications, what motivates Reed and his colleagues is the fact that time is often of essence when it comes to infectious diseases. The survival rate of patients with sepsis, for example, falls 7.6 percent for every hour delay in administering an appropriate antibiotic (Kumar et al., 2006). In his opinion, the only way to achieve a clinically meaningful time to result is to do away with the existing workflows associated with sequencing, particularly the dependence on culturing, the requirement for microbiologists with sequencing training, and specialists who can analyze the sequencing data.
The basic principle of DNA Electronics’ technology is that hydrogen ions are released as part of the nucleotide incorporation reaction during PCR or sequencing, and that when these hydrogen ions bind to the sensing layer on the surface of a standard complementary metal oxide (CMOS) semiconductor chip, they create an electric field that modulates the current flowing through a transistor sitting underneath the sensing layer. Thus, by monitoring the current through this transistor, the chip detects and monitors the biochemical event, a process replicated in thousands or millions of wells across the chip. By taking advantage of the billions of dollars that have been invested in the semiconductor industry and its economy of scale, Reed’s company has incorporated PCR, clonal amplification, and other up-front steps onto the chips as well. “Essentially, that comes for free because we are leveraging the economies of scale that already exist in that industry,” said Reed.
Recognizing early on that DNA Electronics’ product needed to include sample preparation in its system, it acquired NanoMR and its proprietary antibody and bead-based technology that can capture pathogens directly from samples even at concentrations of one colony-forming unit per milliliter. Equally important, said Reed, this technology can be incorporated in a user-friendly, closed fluidic cartridge, one that is not prone to contamination and that can also incorporate semiconductor sequencing functionality.
In addition to designing this integrated pathogen capture/sequencing cartridge, Reed and his colleagues developed a unique sequencing workflow that is well suited to take advantage of cartridge-based operation, offers rapid turnaround and user-friendly operation by nonspecialists, and is applicable in a wide range of clinical environments. In closing, Reed noted that DNA Electronics received funding from the Biomedical Advanced Research and Development Authority (BARDA) in September 2016 to develop this technology for use in detecting influenza virus and for identifying antibiotic resistance.
Discussion
David Cullin began the discussion by asking the panelists to comment on where they felt their technologies fit into the BioWatch picture. Bryden said that mass spectrometry is potentially an outstanding trigger and would be good for toxin detection, and Probst thought the portability of his company’s technology would make it a good fit for confirmation and for use by first responders. Reed said that next-generation sequencing would be of value in terms of the extra information it can provide with regard to developing extremely broad panels for organism identification, strain identification, and finding more challenging resistance markers.
An unidentified participant asked Reed about the length of the reads his technology generates and the error rates for homopolymer regions. Reed replied that, while the technology can generate sequence reads of hundreds of bases, many applications will only require 50 to 75 base reads, which would optimize turnaround time. With regard to error rates for the lengths of homopolymer regions, while important in the whole genome sequencing world, this technology is not meant to be used for general sequencing purposes and so any clinical assay the company developed would not look at those regions and would not attempt to distinguish between two organisms based on the homopolymer characteristics.
Donna Boston from BARDA asked the panelists to estimate when their technologies will be coming to market and how they plan to get the market to adopt their products. Bryden replied that while mass spectrometry is already in clinical use in the microbiology setting, it is not fast and it is limited to things that are easily culturable. For his company’s technology, he predicted it would probably be 2 years before it was proven for more rapid diagnostic applications. Probst said the first market for his company’s technology is likely to be in the agricultural space, such as food processing and detection of genetically modified organisms, when the portable version of FireflyDX is launched in 2018. The company is also preparing an FDA submission and expects to launch an FDA-approved version in 2019. Reed said his company’s first application is a few years away and will likely be in sepsis, followed by other high-value applications in the infectious disease space.
An unidentified participant asked if multiple instruments would be needed to analyze samples from multiple people. Reed replied that the instrument his
company is developing will have the capability to accept 24 cartridges and still fit within the confines of limited bench space.
Concluding the day’s presentations, Adel Mahmoud, planning committee chair, noted that the horizon of science presented over the course of the first day of the workshop has applicability not only in protecting the nation against biological agents, but on the wide spectrum of health in the United States. He then adjourned the workshop for the day.
INDOOR SURVEILLANCE REQUIREMENTS
The first session of day two of the workshop featured three presentations on the challenges of deploying BioWatch in an indoor environment. James Liljegren, research scientist at Argonne National Laboratory (ANL), discussed modeling; Chuck Burris, deputy chief of the Department of Security at the Metropolitan Transportation Authority/New York City Transit, then spoke about using BioWatch in a transit environment; and Suzet McKinney, executive director of the Illinois Medical District Commissions, addressed the public health implications of an indoor release of a biological agent. Colin Stimmler moderated an open discussion following the three presentations.
Modeling
One characteristic of an indoor environment is that there is no one indoor environment, said James Liljegren. A sports arena, for example, comprises a single large open space with simple airflow and a largely static population. A convention center represents a more complex system, with multiple large exhibit halls, meeting facilities, and corridors, each with their own air-handling systems, and a more variable population moving between these spaces. A transit system represents the extreme end of complexity, with its network of tunnels, stations, connected transit centers, very complex airflows, and a highly variable and transient population.
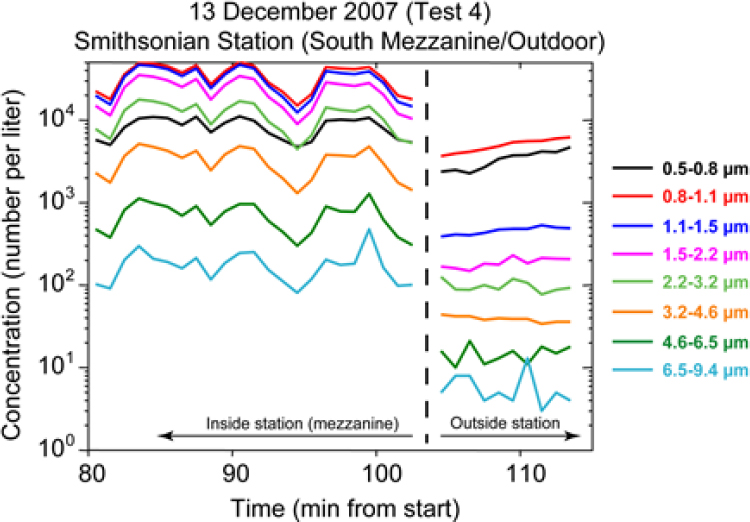
A major source of ambient particles in a typical transit system is “wheel dust,” the iron oxide that forms on rails and is rubbed off by wheels in the trains. Liljegren, who spent most of his presentation on transit systems, noted that the concentration of all but sub-micron-sized particles is orders of magnitude greater inside a subway system than in the outside air (see Figure 3-7). In addition, fluorescent shed skin cells and clothing fibers, each of which may inhibit the PCR process, are also present at lower concentrations outdoors (see Figure 3-8).
There are two important mechanisms that can drive the dispersion of biological agents in an underground transit system. Trains move most of the air in a subway system, pushing air through train tunnels in what is known as the piston effect. Tunnel and station geometry, train speed and length, and the spacing of vents, entrances, and portal areas determine how the piston effect moves particles in a transit system. The train cars themselves can carry particles that stick to the cars, that the
ventilation system pulls into the cars, and that people carry into the cars. In this way, train cars can transport materials across bridges above ground. Natural airflows, driven by winds and temperature differences and mechanical ventilation, serve as the principal drivers of particle transport in other indoor environments.
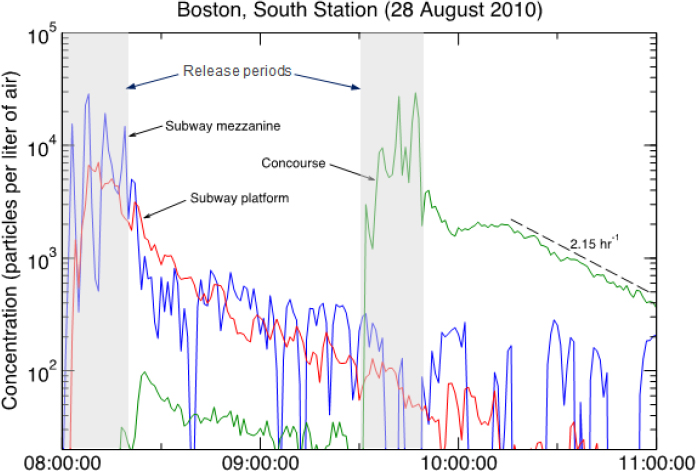
As an example of how particles move in a transit system, Liljegren described an experiment in which he and his colleagues released tracer particles on the mezzanine of an intermodal subway station. Within minutes, the tracer particles dispersed to the subway platform and over the next 90 minutes, out of the station on trains passing through (see Figure 3-9). Very little material, he added, got into the multistory concourse above the mezzanine. A subsequent release in the multistory concourse demonstrated that the filters in the building’s air-handling system removed 90 percent of the particles per hour, which means that the bulk of the material would be removed from that facility within 2 to 3 hours. Even with removal of one order of magnitude an hour, Liljegren explained, “the material is pretty persistent indoors, whether it is in the subway or whether it is in a building,”
As David Brown discussed the previous day, the ANL Below Ground Model (BGM) is the standard modeling tool for transit systems, and field studies in several transit systems have validated the model. The BGM predicts transport and fate of a chemical or biological agent in an underground subway system and predicts the amount of material released to the aboveground environment via vents, station exists, and portals. Output from the BGM also includes projections for subway passenger movement and reaction, if any, to an incident, as well as passenger exposure to and transport of material.
As Brown also noted, the ANL team has coupled the BGM to the Los Alamos National Laboratory Quick Urban and Industrial Complex (QUIC) model, and the combined model predicts that Bacillus anthracis released in a model subway system would spread and persist throughout the subway system, while also being ejected in multiple plumes into the aboveground environment to be dispersed by wind. The result would be a citywide impact from a single-point release; a 1-hour exposure to even the lowest level of particles would lead to a lethal infection for 10 percent of the exposed population (see Figure 3-10). Fortunately, other than transit system employees, few passengers would be in the system for an hour, said Liljegren, but even at a 10-minute exposure, a substantial number of people would receive a lethal dose of bacteria.
Turning to the subject of fomite transport, Liljegren explained that subway passengers become fomites for biological agent particles, and these particles can resuspend and deposit elsewhere in the subway system and beyond. In addition, passengers can track particles on their shoes to points distant from where they are exposed to the biological agent. BGM modeling shows that fomite transport and tracking both disperse particles, though tracking enables particles to spread farther than would be expected by fomite transport alone. Similarly, the dose of particles people would encounter remains elevated beyond what would be expected without human transport of particles. As a consequence, said Liljegren, the potential for many low-level detections increases, the probability of having a single-detection scenario becomes less likely, and the ability to reconstruct where the release occurred becomes more difficult.
How fomite transport affects the eventual distribution of material depends on the time of day at which a release occurs. Modeling, said Liljegren, shows that when a release occurs in the morning, when commuters are traveling into the city, the bulk of the material deposits in the central business district, whereas release during the evening rush hour disperses material over a much larger area, with the highest concentrations occurring at terminal points in the transit system, including at airports and in neighboring towns.
In addition to dispersion modeling, Liljegren and his colleagues have modeled the consequences of a release to provide postevent support with regard to source identification, predicting which areas would be contaminated or clean, and how many people are likely to become ill. Source identification includes estimating particle size, which is important for determining how respirable the material is. Liljegren and his colleagues have developed an artificial neural network capable of serving as a consequence model, but the challenge is to relate the model, which is based on mass, to laboratory PCR results. Addressing this challenge to improve this model will require more quantitative detection methods with well-characterized uncertainty and variability.
In summary, Liljegren said the challenges in dealing with the effects of an indoor release are fourfold and are associated with
- How the rapid rise of indoor aerosol amounts and constituents may affect detection;
- The time during which material aerosolized indoors can spread rapidly through a venue or transit system;
- The confined space that enables airborne concentrations to persist for several hours and increase both exposures and consequences; and
- The mobile population that can enhance the spread of a material within a venue or transit system and beyond.
The detection requirements for dealing with those challenges include
- Rapid, reliable, and actionable detection to enable a low-regret initial reaction and an informed, robust response;
- Widespread, optimally sited detectors to provide situational awareness of the evolution and extent of an event and protection of the largest population possible; and
- Computational modeling to inform the number and location of detectors for an optimally effective system, the efficacy of response strategies, and postevent responses.
To tie this together, said Liljegren, test beds in actual venues are needed to foster the development and document the efficacy of detection methods and model predictions. He concluded his comments with a quote from Richard Feynman, who in the report on the space shuttle Challenger disaster said, “For a successful technology, reality must take precedence over public relations, for Nature cannot be fooled” (Presidential Commission, 1986).
Transit Security
The goal of mass transit is to provide safe, on-time, reliable, and clean transportation services for the public and a safe and healthy work environment for transit employees, said Chuck Burrus. To illustrate the complexity of his job fulfilling the “safe” part of that mission, he explained that the Metropolitan Transportation Authority (MTA), which serves 12 counties in downstate New York and two counties in southern Connecticut, comprises six subsidiaries: the Long Island Railroad, Metro-North Railroad, MTA Bus, Capital Construction, Bridges and Tunnels, and New York City Transit, which by itself is the largest subway and bus system in the United States. The MTA and its 48,000 employees move some seven million passengers on any given weekday, and 5.5 million of those use the subway system for at least part of their journey. The MTA operates 24 hours per day and manages 24 subway lines, more than 6,000 railcars, 4,000 buses, and
472 stations, including hubs with Amtrak and New Jersey Transit. It also is responsible for more than 600 mainline track miles, 43 bridges, 14 underwater tunnels or tubes, 13 railcar maintenance facilities, and 28 bus depots and maintenance facilities.
When Burrus briefs the MTA executive staff on monitoring, the most important question he gets is, “Why monitor?” to which he always answers that the threat of biological or chemical attack is real. “If you recognize that the threat is real, then a sophisticated and effective enough detection system can serve as a deterrence,” said Burrus, “and when it comes to public health, prevention is the name of the game. If we can deter an attack by having an effective detection system, we have done our job, and if we cannot deter, then we at least have a system in place that allows us to lessen the effect of an attack.” Lessening the effect of an attack, he said, means limiting the extent of the customer and employee exposures and the extent of the contamination of the infrastructure.
New York City, said Burrus, is a target-rich environment, and both the city in general and New York City Transit in particular have a history of being targeted (see Figure 3-11). As a result, the city has received several hazard mitigation grants from the Federal Emergency Management Agency. He noted that the results of one exercise simulating a biological attack on New York City Transit was so sobering that he had an easy time convincing the MTA executive staff that it should pursue a monitoring system. Another analysis showed that the economic cost of investing in a detection system paled in comparison to the cost to get the transit system running again once it is contaminated.
Burrus said that he and his security colleagues in New York City are more concerned about a biological, chemical, or radiological attack than a suicide bombing or an improvised explosive device (IED) because it takes very little material to produce a devastating effect. “Unlike with an IED or something that is obvious when the attack takes place, what makes the biological weapon much more effective is the fact that we do not know we have been exposed until people become symptomatic, and by that point in time, there is not necessarily a lot that the medical community can do for them,” he explained.
He noted that the U.S. government has spent some $90 billion since 2001 to prepare for a biological attack, of which approximately $12 million went to New York City Transit to develop the first subway-vetted Detect-to-Protect prototype that was eventually validated by researchers at the Massachusetts Institute of Technology. His department has worked closely with Liljegren and the ANL team to conduct a range of pathogen dispersion studies in the city’s subway system to help with modeling and portable sampling unit siting. Should deterrence fail, he added, a biodetection system capable of detecting a release in real time would enable transit management to limit customer and employee exposure and limit contamination of the larger transit system.
Given the MTA’s size, key questions include where to monitor, how often to monitor, and which organisms a monitoring system should target. Burrus said that there are other organisms besides those in BioWatch’s repertoire that could be problematic, such as hemorrhagic viruses, multidrug-resistant tuberculosis, and genetically modified influenza virus. The problem is that adding monitoring capability costs money, and funding is not limitless. For him, another issue relates to who in his organization decides the answers to those questions. While he can offer advice, the executive staff has to make the ultimate decision, which usually comes down to the amount of funding available. Another decision that has to be made regards who is responsible for managing the biodetection system. “Should it be us? A combination of us, law enforcement, and public health? If you cannot get that resolved, should something happen, there is the making of a great deal of confusion and finger pointing,” said Burrus.
Deciding what technology to select requires determining the attributes and requirements of the system. In an ideal world, the system would be inexpensive to install and operate, comprehensive with regard to threat agents, quantitative, able to detect in real time with no false positives or negatives, and be functional in and tolerant of a subway environment that is not technology friendly. The problem, said Burrus, is deciding whether to wait for the perfect system or deploy something that is currently available, practical for the operating environment and the available budget, and adaptable as technology improvements are developed. In his opinion, he would prefer a combination of a trigger technology based on a sensor array, to minimize someone spoofing a single detector, and a confirmation technology.
With regard to who collects and who analyzes samples, Burrus said his preference is to leave the analysis up to staff at the New York City Department of Health and Mental Hygiene. They developed the concept of operations that spells out who initiates a response, if any, who responds to a response and at what level
and with what personal protection equipment, and what the type and extent of the response should be. The concept of operations also includes what, if anything, the transit system does with and says to its customers, employees, vendors, and the media in the event of an alarm or trigger.
Public Health Implications of an Indoor Release
One indoor environment that the previous speakers had not mentioned, said Suzet McKinney, is the interconnection between buildings. In larger cities such as Chicago, tunnels and walkways connect a number of buildings. In addition, Chicago has quite a few buildings that share heating, ventilation, and air conditioning systems and other mechanical systems. “Those interconnected buildings and systems pose an even greater level of complexity when we are talking about the potential for an indoor release,” she said. The bottom line is that cities present complex indoor environments serving a high population density. In addition, major cities serve as transportation hubs with regional, national, and international connections. All of these factors, said McKinney, mean there are many mechanisms by which a point release of a biological agent can become a much larger public health threat.
Regarding the response to a public health emergency such as release of a biological agent, every single level of government has a role in both planning and responding. “At the end of the day, though, all events are local, and the initial response is going to start at the local level,” said McKinney. At some point, she added, state and federal officials will become involved, but public health is responsible for the critical initial response to a threat.
According to the CDC’s Public Health Preparedness Capabilities document,3 “Public health threats are always present. Whether caused by natural, accidental, or intentional means, these threats can lead to the onset of public health incidents. Being prepared to prevent, respond to, and rapidly recover from public health threats is critical for protecting and securing our nation’s public health.” That document also states, “Public health has made great progress since 2001, but continues to face multiple challenges, including an ever-evolving list of public health threats. Regardless of the threat, an effective public health response begins with an effective public health system with robust systems in place to conduct routine public health activities.” McKinney’s point in providing those quotes was to convey the idea that, while she and her public health colleagues worry about a biological release detected by BioWatch, there are other threats that do not go away during a BioWatch response.
Some of the responsibilities of public health after the detection of an indoor release include
___________________
3 See Executive Summary, p. 2 of https://www.cdc.gov/phpr/readiness/00_docs/DSLR_capabilities_July.pdf (accessed October 27, 2017).
- Taking the lead role in joint operations,
- Advising elected officials and facility owners and operators,
- Conducting the epidemiologic investigation in coordination with law enforcement’s forensic investigation to assess the hazard and determine the ongoing risk to the public,
- Conducting surveillance for ill and potentially exposed individuals and continuing to test samples in the public health laboratory, and
- Consulting with environmental health specialists.
In the aftermath of a biological attack, public health also maintains responsibility for making treatment recommendations to hospitals and medical providers, distributing and dispensing medical countermeasures and other medical supplies, protecting the health and safety of first responders, and issuing emergency public information and warnings.
Over 2 days in May 2009, Chicago conducted a BioWatch exercise that shut down one of the subway stations adjacent to city hall and had sampling teams in personal protective gear conducting Phase I environmental sampling of the “contaminated” subway platform. This exercise used modeling data and other data points to aid with the decision-making process, and even included conducting a simulated press conference. However, the subsequent laboratory analysis of the Phase I samples had to be rescheduled until July because the city laboratory was in the middle of its H1N1 influenza response exercise. “Rather than canceling the exercise, we decided to really test our system, stress our resources, and see exactly how much we could get done with the resources we had on hand with seemingly two very different emergencies happening at the same time,” said McKinney.
That exercise, she said, taught the city a great deal and helped her and her colleagues understand what some of the most critical challenges would be for decision makers after the detection of a release in an indoor environment. “Clearly, state and local officials will be conducting decision making in the context of unique jurisdictional and political constraints, and every city would operate differently in the event of one of these detections,” said McKinney. Decision makers have to balance multiple interests, including the threat to public health, resource constraints, and even the current political environment regarding whether or not to close large buildings that house major corporations or airports. “Those decisions have large business and economic implications that need to be considered, both in the immediate term as well as in the longer term when you begin to look at issues around remediation, re-occupancy, and recovery,” said McKinney. Decision makers also have to consider the safety of first responders who have to go into a contaminated indoor environment.
One thing she learned from this exercise was that there are many issues that still need addressing, including
- Whether a facility should be closed and who makes that decision,
- What the rights of facility owners and operators are and do those rights supersede public interest,
- Who has the authority to make decisions about remediation and when a facility can be occupied again, and
- How to balance the public health and law enforcement investigations to avoid interference with one another.
Public health messaging is another ongoing concern, particularly with respect to who leads and who drives messaging. How much information is needed to make decisions is another open question, as is whether Phase I and II sampling is sufficient to make informed decisions. Given the possibility there will be what McKinney called high-regret decisions, she questioned how much support public health will receive from the public and the federal government.
Moving forward, McKinney stated BioWatch will continue to be a critical component of the U.S. biodefense effort, one that requires increased coordination among local, state, and federal officials to frame the local response in the context of an event of national significance. “From my public health perspective, there is still a need for understanding the vast array of federal agency responsibilities, clarification on what federal assets can be brought to bear in one of these response efforts, and how quickly can those assets be brought to bear,” said McKinney in concluding her remarks. She would also like more clarity on acceptable practices for sampling, testing, and characterizing a threat, particularly with respect to remediation and reoccupancy.
Discussion
To start the discussion, Colin Stimmler asked the panelists whether they believe a 10-year horizon is sufficient time for BioWatch to deploy a detect-to-mitigate system or even a detect-to-prevent system. Burrus replied there are technologies available today that have been through trials and challenges in different subway systems and could be deployed as an array-based trigger system. Those technologies would give him the ability to react in real time to suspend the movement of trains and minimize the number of people exposed to an attack. “Years from now, you may improve upon that, but I do not see why we cannot do what we are able to do right now,” he said. In the future, the number of target agents could be expanded and an autonomous system deployed, he added, but for now he would not let the pursuit of the next great technology slow down implementation of technology that is readily available today.
Liljegren, noting that time is of the essence, agreed with Burrus regarding the idea of having a trigger system that indicated something was afoot before receiving laboratory analysis is complete. “It would be nice to have something that
would allow you to implement these lower-regret reactions before you even knew that it was a pathogen,” said Liljegren. He noted that colleagues at ANL developed a network of sensors for chemical detection that has been operational in the Washington, DC, Metrorail system for many years. “If the sensor technology was vetted and if the system was integrated, I think that it might be a possibility in the next 5 to 10 years to have something like that operational,” said Liljegren.
McKinney said she agreed with both Burrus and Liljegren, but added that it will be critically important for public health, first responders, and decision makers to accept such a system and have confidence in the ability of that technology. “Whether we use something that is currently on the market now or something new is developed, that technology and its use need to be planned in conjunction with the very folks that would be the end users,” she said. The user community, she added, would include public health and facility owners and operators.
An unidentified participant asked if stopping or slowing a train was a low-regret decision and what others might be. Burrus said that stopping or slowing trains in a limited part of the system would be a low-regret decision. Liljegren said shutting down air handlers to prevent circulation of material would be another low-regret decision, or confining the potential agent to a zone that the air handlers could remove would be another one. Modeling, he added, can help develop and test such strategies.
Another unidentified participant asked the panelists what they would do to mitigate the current lack of a real-time detection system. McKinney said more training exercises and public announcements to educate the public about biological weapons and possible responses to an attack would be a help as far as speeding the decision-making process. Burrus agreed, particularly with the need to educate the public. “The public should not be hearing about a BioWatch Actionable Result for the first time when we are having a crisis,” he said. In his opinion, educating the public up front about preparedness activities would go a long way toward the public not panicking more than might be expected. The pushback he got from law enforcement about advertising that such a system exists is that bad actors might take that as an invitation to challenge the system to see how good it is. His response is to say, “If nobody knows about it, how is it going to be a deterrent?”
One step the MTA advisory committee took, Burrus said, was to write a script detailing what would happen and who would be in charge of what activities in the event of declaring a BioWatch Actionable Result. They shared that script with the BioWatch program’s stakeholder panel, the New York City Department of Health and Mental Hygiene, and law enforcement to get everyone on the same page.
Toby Merlin asked if data exist to support the type of trigger system the panelists suggested deploying. If not, he proposed deploying such a trigger system throughout a system and collecting data for 2 years about the background to build confidence that an aerosol release would create a signal above background. The short answer, said Burrus, is yes. He said he has been playing with triggers for years, and acknowledged that it took time to ensure that the system learned about background levels and how they fluctuate. “That data has been available to us for long enough for me to make the recommendation to senior management that this
is what we can do now,” said Burrus. He added that the sensor network is connected to closed circuit television, which provides the ability to immediately look where the trigger was tripped to see if there is any unusual activity in that area.
Derek Carver from the DoD’s JUPITR program noted that this program has deployed a sensor array at Busan in Korea and has been collecting operational data for approximately 9 months. He characterized the array as “fairly successful” and suggested it would be worth examining that system for use with BioWatch.
Thomas Slezak asked the panelists how early they would like to be engaged in the process of selecting and developing the new technologies that will be coming out over the next 5 to 10 years. McKinney replied that, from her perspective, yesterday would have been the right time based on her experience with field testing the Generation 3 BioWatch system. “When the field test was completed, I felt as though I had enough experience with the system and had had enough coordination and involvement with the team from Homeland Security that I could at least provide information and advice to elected officials in my jurisdiction about how well protected we were with that technology,” she said. “My advice to those who have the responsibility for determining what the new technology might be would be to involve public health as soon as you can.”
Referring to her experience with the Generation 3 system test, McKinney said that there were only a few people from public health involved, which created problems when trying to go to other cities to discuss the proposed system. Burrus agreed that the earlier public health is brought into the development and testing process, the better. Liljegren added that this is why local test beds are so important, and he credited New York City with being in the vanguard of hosting such test beds for both biological and chemical sensors.
Mark Buttner commented that the panelists made a compelling case for moving the BioWatch program indoors and about ways to enhance technology to hasten a response. One theme he noted in the presentations was the need to educate decision makers, and he asked if the panelists had ideas on the best approach for doing that. McKinney said education needs to start as soon as possible and be as comprehensive as possible. She credits BioWatch for bringing together the entire response community in Chicago when it conducted the 2009 exercise, saying that was the catalyst that enabled the rest of the city’s public safety community to understand how critical the public health world would be in the event of an attack. She recommended holding such exercises as often as possible because each exercise brings new questions, new insights, and sometimes new solutions.
Burrus added that he advocates making sure MTA’s employees and the public have at least a basic understanding of what MTA is doing to prepare for an attack and what the potential threats are. One pushback he got for disseminating response plans to MTA’s employees was that it might enable a nefarious actor to plan a second release. “There is this balance about how much of the procedures can you share even internally and just where do you draw the line, and that is a discussion we are still having today,” said Burrus. He noted that the public has been receptive to seeing tests being run in the New York City subway system.

































