4
Nutrigenomics Applications: Dietary Guidance and Food Product Development
Before introducing the first speaker of session 2, moderator Wendy Johnson, Nestlé USA, reflected on a few highlights from the morning’s presentations and discussion that she found particularly interesting. She mentioned Patsy Brannon’s discussion of the circular relationship among the genome, diet, and health outcomes; José Ordovás’s description of evidence suggesting that where people live can influence their genetic makeup; Claudia Morris’s remarks on the elusive defined nutrition requirement that appears to change according to various disease states; discussion of how little is known about how behavior impacts nutrigenomics; and several participants’ thoughts on what can be done commercially with personalized medicine to help people, but also the complexity of making nutrigenomics a practical alternative for consumers. Now in session 2, Johnson continued, the focus would be shifting to “how we really take that information and move it forward in a way that makes a difference in people’s lives.” This chapter summarizes the session 2 presentations and discussion, with highlights provided in Box 4-1.
NUTRIENT REQUIREMENTS AS COMPLEX TRAITS: WHAT CONSUMERS NEED TO KNOW
Patrick Stover, Cornell University, began his presentation by describing an incident that had recently taken place when he missed a flight and was sent an Uber driver to accommodate his changed travel plans. It was about 1:00 AM, but the driver was quite chatty, he recalled. She told him about her family and how she had started a career as a personal training
coach and amassed a large clientele. When she learned that Stover was in the nutrition field, she told him that nutrition was the most important guidance she provided to her clients who were morbidly obese. “I use the blood type diet,” Stover remembered her saying, adding, “that’s the single most important thing that improves their health: the blood type diet.” Stover noted that the blood type diet is based on Peter D’Adamo’s book Eat Right 4 Your Type, and remarked that his experience with the Uber driver
reflects where he thinks a lot of consumers are in terms of their knowledge of nutrition and health.
Stover then showed a picture of the front cover of a 2010 issue of Nature with the headline “Can Science Feed the World?” That is, can science feed the 9 billion people who will be on this planet in 2050? The same issue, Stover continued, contains an article on diabetes. He explained that for him, the question is not whether we can feed that many people, but, “Can we feed them in a way that keeps diabetes off the cover of Nature?” He suggested that another way to frame the question, given today’s unprecedented capacity to formulate the food supply in any way desired, is, “What are our expectations of the food supply? That is, when we think about setting dietary and nutrient guidance and recommendations, what are the outcomes we really want to achieve? And what is achievable?” In fact, Stover observed, the U.S. federal government has asked these questions, as indicated by the request to the National Academies that a committee be assembled to develop a consensus report on developing guidance on dietary reference intakes (DRIs) based on chronic disease endpoints (NASEM, 2017a).
Stover went on to stress that making connections between food and disease prevention poses several challenges. He cited as the first of these that few chronic diseases are affected by a single nutrient or a single pathway. Thinking about the relationship between food and chronic disease, he argued, requires considering systems, or networks of pathways, rather than individual pathways, and how these pathways and nutrients converge and interact. This means, he observed, that thinking about biomarkers requires thinking not about single nutrient biomarkers, but about integrative biomarkers that are the endpoints of systems where pathways and nutrients interact and impact chronic disease. Additionally, he stated, as called for in the above-referenced National Academies report (NASEM, 2017a), it means considering DRIs as ranges, not point estimates. He noted further that, beyond considering integrative biomarkers, thinking about chronic disease endpoints requires considering biomarkers of aging and how they interact with biomarkers of nutrition. As a final challenge, he mentioned that the above report (NASEM, 2017a) recommends use of the Grading of Recommendations Assessment, Development and Evaluation (GRADE) Working Group’s standards of evidence. But, he pointed out, this is a field that is driven primarily by observational data, most of which are not at the GRADE level of evidence needed to set chronic disease endpoints. He added that, given that half of the adult population in the United States is under the care of a physician for some sort of chronic disease, combined with the fact that the DRIs are intended for healthy people and therefore may not apply directly to what is essentially half of the population, the National
Academies convened a separate committee to plan a public workshop to examine nutritional requirements in disease states.1
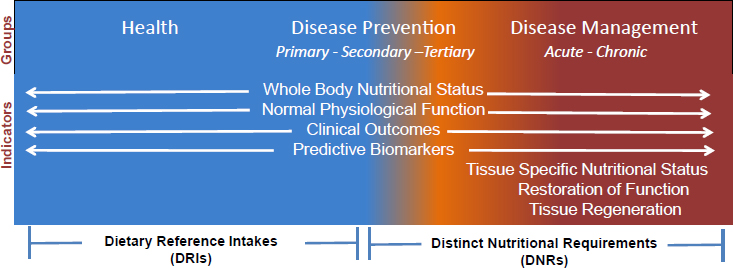
Stover continued by arguing that, while the DRIs are highly focused on the nutritional needs of healthy individuals, nutritionists need to start thinking about the totality of nutritional requirements and how to classify and evaluate human nutrient needs from health, through disease prevention, to disease management. Acknowledging that there are many markers available that can be used in attempting to understand the nutrient needs for health and disease prevention, he asserted that other types of indicators will need to be considered as nutrition scientists work to understand the connection between food and disease management (see Figure 4-1). These include indicators of tissue-specific nutritional status, because chronic disease often exhibits tissue-specific effects that may not relate to whole-body effects; restoration of function, as addressed by Claudia Morris in her presentation on conditionally essential amino acids (as summarized in Chapter 3); and tissue regeneration (i.e., the unique nutritional needs of stem cells as they repair damaged tissue). Stover suggested that in thinking about the connection between food and disease management, one can begin to think about distinct nutritional requirements (DNRs) instead of DRIs. He noted that these requirements would include medical foods, which provide nutrients that may not be accessible from the food supply in the quantity or quality one needs.
Dietary Requirements as Complex Traits
Like previous speakers, Stover described dietary requirements as complex traits. That is, many physiological processes inform the nutrient needs of an individual (i.e., absorption, catabolism, excretion, metabolism, stability, transport, bioactivation, energetic state, nutrient storage), all of which interact with each other as well. He added that nutrient requirements also are affected by a number of modifiers and sensitizers, including disease, but also epigenetics, the food matrix, genetics, nutrient–nutrient interactions, pharmaceuticals, toxins, age and physiological decay, the microbiome, pregnancy, and sex. He noted that although the topic of the workshop was genetics, it does not make sense to consider any one of these modifier or sensitizer variables in the absence of the others, given that nutrient requirements are what he described as “true complex traits.” Of course, he continued, this has been recognized for many years, as indicated by the American Society for Nutrition’s 2013 research agenda, on which one of
___________________
1 For information on this April 2–3, 2018, workshop, see http://www.nationalacademies.org/hmd/Activities/Nutrition/ExaminingSpecialNutritionalRequirementsinDiseaseStatesWorkshop.aspx (accessed April 23, 2018).
SOURCE: Presented by Patrick Stover on December 5, 2017.
the top priorities is to understand variability among individuals in response to diet and food (Olhorst et al., 2013). More recently, he added, the U.S. federal government released the National Nutrition Research Roadmap, 2016–2021, which also focuses not just on the connection between nutrition and disease prevention, but on the importance of understanding individual differences in nutritional status as well (Interagency Committee on Human Nutrition Research, 2016).
Genetics as a Modifier of Nutritional Status
Stover went on to emphasize the importance of remembering, when thinking about the effects of genetics on nutrition, that the purpose of the Human Genome Project was not only to inventory all of the genes in the human genome, as well as variations in these genes, but also to assemble and understand cellular networks, or circuits; variation in these circuits; and how they can be manipulated by inputs, whether these be drugs or nutrients. The impact of genetics on diet is rooted in evolutionary biology, he observed. He showed the cover of a 2002 issue of Scientific American with the headline “Food for Thought.” The article is about dietary change as a driving force in human evolution (Leonard, 2002). Stover explained that the molecular basis of the evolution of a species is the evolution of genes, but not all genes evolve, or change over time, at the same rate. Genes that evolve quickly are those that enable adaptation to a local environment, he noted, adding that upon examination, it appears that one of the most powerful selective pressures in genome evolution is diet. Thus, he argued, today’s human genome—not just its primary sequence, but also genome programming and gene expression—is suited to a particular,
historical nutrient environment. When the genome is exposed to a changed nutrient environment, he said, people experience food intolerances, special dietary requirements, and susceptibility to disease.
Stover explained that there are two major contributors to all of the variation in today’s human genome. First is the introduction of the genome of archaic humans into that of what are now modern humans. More specifically, with the recent sequencing of the Neanderthal genome, it appears that about 500,000 to 1 million years ago, probably through the male germline, Neanderthal DNA was integrated into the human genome. He added that, based on identification of the Denisovan archaic humans, it is now known that Denisovan DNA also became integrated into the modern human genome. Further, he said, it is likely that a third, unknown hominid also contributed DNA to the current human genome. He suggested that, in addition to contributing to understanding of why humans across the planet are so genetically variable, knowing which Neanderthal and Denisovan genes became integrated into the modern human genome and how they affect human traits is of interest to consumers. He cited as an example 23andMe, which offers a genetic test whereby consumers can learn not just about their ancestry but also what percentage of their genome is Neanderthal or Denisovan.
Stover identified as a second major source of human genetic variation mutation events that subsequently extended though a population via either selection or drift. He explained how computers can identify genes or regions in the genome in which what he called a “selective sweep” occurred—that is, where a mutation enabled some sort of selective advantage that displaced all of the preexisting variation. Many of the genes that show evidence of selective sweep are related to nutrition, metabolism, or immunity, he noted, which he interprets as good evidence that nutrition has played an active role in much of the variation that exists today.
Importantly, however, in Stover’s opinion, the relationship between nutrition-related genetic variation and phenotype is not a one-to-one correspondence. The one gene in the human genome that shows the strongest evidence for this type of selection, he observed, is the lactose tolerance gene, with a mutation in the promoter allowing the gene to be expressed through adulthood, and therefore allowing a source of nourishment inaccessible to the rest of the population. However, he noted, when the geographic distribution of this gene is overlaid on a map of the phenotype of lactose intolerance, the two overlap fairly well but not completely (Itan et al., 2010). “No one gene variant completely determines what the phenotypic expression is going to be,” he said.
Stover added that there is also evidence of evolution of nutrition-related copy number variants (CNVs), such as amylase CNVs. Across human populations, he elaborated, individuals have between one and nine copies
of the gene. Those with high copy numbers are from populations with a history of an agrarian lifestyle (for improved digestion of starch), he explained, whereas those with low copy numbers are from populations with a history as hunter-gatherers. Stover pointed to this as another example of an adaptation that has been driven by the food supply.
Stover went on to observe that many other diet-related genes, such as the calcium transporter gene, have similarly displayed genomic signatures of adaptive evolution by selection (Stover, 2007). He finds it interesting that selection for the lactose tolerance gene appears to have occurred before selection for the calcium transporter gene. That is, only after the lactose tolerance gene arose and expanded and enabled milk consumption did the calcium transporter mutation arise and expand.
Stover suggested that while all of this evidence provides a strong biological premise for the link between nutrients and health, one rooted in evolutionary biology, “what we are not so sure about” is the strength, or penetrance, of the effect. Again, he stressed, dietary requirements are complex traits, and genetics is only one of many factors that determine the relationship among food, nutrition, and health. For Stover, this uncertainty raises the question of whether all of this genetic variation really matters in public health. He went on to describe an example in which, in fact, it does.
The MTHFR polymorphism is a fairly common variant in human populations, Stover explained, with about 80 percent of individuals having a C allele, which codes for alanine, and the other 20 percent having a T allele, which codes for valine. The T allele protein is less stable and less active, and individuals carrying that variant have a lower folate status and, all other things being equal, a higher folate requirement and greater risk for birth defects and miscarriage. However, Stover observed, if individuals with the T allele survive to adulthood, their risk of colon cancer is reduced by 70 to 80 percent if they maintain adequate folate status (Ma et al., 1999). He noted that this finding has been replicated in several cohorts. “This is a very powerful effect from a single nucleotide polymorphism,” he said.
The C and T allele frequencies at the MTHFR polymorphism vary across the globe, Stover added. He explained that the polymorphism clearly arose after migration out of Africa, but it does not show a signature of selection, so it either accumulated through drift or entered the human genome from another archaic human.
Stover stated that there is also good evidence from controlled feeding trials that the variant can affect nutrient requirements for folate. When Solis and colleagues (2008) fed adult Mexican men the recommended daily allowance (RDA) for folate, they found that initially, serum folate levels dropped for both CC and TT individuals, but by the end of 12 weeks, the CC individuals had significantly higher levels of serum folate relative to the CC individuals. In addition, homocysteine level, which is a functional
biomarker of folate metabolism, did not change over time among CC individuals but rose markedly among TT individuals. According to Stover, “This is an example of one single polymorphism that definitely affects nutrient requirements.” In fact, he suggested that the RDA is probably not adequate for Mexican American men who harbor the TT genotype. But again, he asked, whether these findings have affected policy. The answer, he said, is yes.
In 2015, the World Health Organization (WHO) published a guideline for optimal serum and red blood cell folate levels to prevent birth defects (WHO, 2015). Stover explained that this action was taken in response to member states approaching WHO and asking how much folate they should be adding to their food supplies. He described the WHO (2015) committee’s work as remarkable because it represented the first time a chronic disease endpoint was being used to help establish a nutrition reference value, and because big data integration was used to reach this conclusion. Regarding the latter point, he explained that there was a paucity of data linking folic acid intake to risk of neural tube defects and that what data did exist were of low quality. However, there were data linking folic acid intake to folate concentration in red blood cells. So through Bayesian modeling, Stover explained, the WHO (2015) committee was able to derive a computed dose-response curve predicting estimated risk of neural tube defects as a function of folate concentration in red blood cells (Crider et al., 2014). He added that, although the curve was computer generated, data from the one empirical study that does exist, a small study in an Irish population, align with the big data curve of Crider et al. (2014) and the WHO (2015) committee. He highlighted this as a case in which evidence of a single polymorphism was used to generate a guideline, in this instance on what the optimal level of folate should be to prevent neural tube defects.
Genetic Testing: What Consumers Will Need to Know
Stover pointed out next that consumers have available to them a range of genetic tests that are providing not only ancestry information but also, increasingly, health information. He raised the question of what consumers really need to know to use this information.
It has been suggested, Stover observed, that individuals be classified into subgroups for diets, which he noted had already been discussed earlier in the workshop. He referenced the recently released National Academies report on redesigning the process for the Dietary Guidelines for Americans (DGA), which emphasizes not only the prevention of chronic disease but also the need to take into account a range of individual factors, including age, gender, and metabolic health (NASEM, 2017b). He identified as a challenge to this approach, however, that although there have been many
studies on various types of diets and how they affect health in human populations, most of those studies are based on observational data, and very few are long-term.
Stover cited a recent long-term study in inbred mice, in which Barrington and colleagues (2018) fed the mice one of four diets—American, Mediterranean, ketogenic/Maasai, or Japanese—for much of their lifetime and compared the diets’ effects on metabolic health. Additionally, they measured certain health-related metabolic and epigenetic markers over time. Stover reported that the study showed that the best diet for maintaining health varied depending on both the health outcome of interest and the genetic background of the mouse. So, for instance, among mice fed the Mediterranean diet, HDL cholesterol levels were at very healthy levels in one strain but not another (see Figure 4-2). In Stover’s opinion, these findings provide a good biological premise for matching diet to genotype. “Of course,” he said, “the challenge is, how are we going to classify people? Because we can’t classify them based on inbred strain, if you will.”
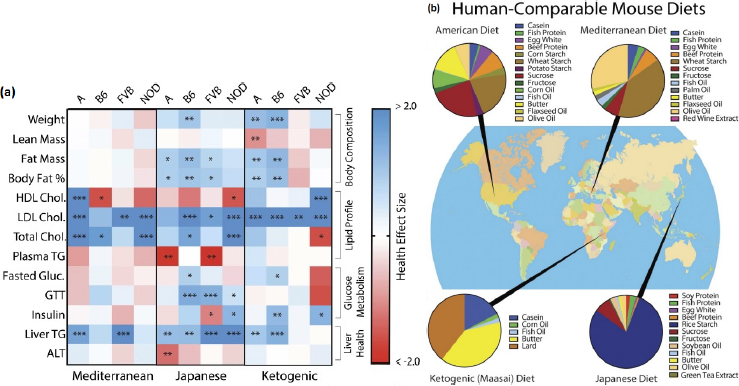
NOTE: A = A/J strain mice; ALT = alanine aminotransferase; B6 = C57BL/6J strain mice; Chol. = cholesterol; FVB = FVB/NJ strain mice; GTT = glucose tolerance test; HDL = high-density lipoprotein; LDL = low-density lipoprotein; NOD = NOD/ShiLtJ strain mice; TG = triglyceride.
SOURCES: Presented by Patrick Stover on December 5, 2017, from Barrington et al., 2018. Reprinted with permission from the Genetics Society of America.
Instead of classifying subgroups of people by diet, Stover continued, it has been suggested that subgroups be classified by nutrients. This approach, he explained, is based on the precision medicine model, in which a prognostic test related to a risk for some chronic disease predicts whether a drug will work or which drug will work, as well as how to dose the drug. According to Stover, however, this approach has been somewhat problematic for nutrition thus far. He cited the example of diabetes, which is such a complex trait affected by so many nutrition-related variables that it that it has made classifying individuals difficult. He asked, “Does it really make sense to split the pie up in so many pieces?”
In fact, in Stover’s opinion, it may not even matter. Instead of classifying subgroups by either diet or nutrient, he elaborated, the trend toward real-time personal readout has created a third possibility. He pointed out that people are collecting a tremendous amount of data on themselves, with apps available to record how many steps they have taken, what their blood pressure is, and so on. Increasingly, he added, high-resolution biomarker data are being collected and will be available to almost everyone. Still, he suggested, the questions are what guidance will be provided to individuals and whether systems/network biology can be applied. He then cited one example of point-of-care measurement of nutrition-related biomarkers, explaining that his colleagues at Cornell University have developed what they call the Nutriphone, a smartphone-based microfluidic device. He described this device as much like a pregnancy test, except that it provides a real-time readout of 8–10 nutrient status markers, functional biomarkers, chronic disease markers, and infectious disease markers (Lee et al., 2016). According to Stover, the price of the device will eventually fall to a few dollars, but again, he raised the question of what consumers will do with the information collected.
In an attempt to understand how best to help consumers with the genetic and other information now emerging, Stover and his group have been working with researchers at the University of Toronto to develop a stochastic model for understanding metabolism networks. They have recreated the entire folate network, running the simulation for hours or days at a time, with different inputs, until the system reaches equilibrium, and they are using the model to conduct sensitivity analyses for changes in gene expression and enzyme levels (Misselbeck et al., 2017). Stover described this as the type of algorithm that will be needed to manage all of the data coming from cell phone apps. He envisions plugging one’s phone into some sort of computational model and then receiving guidance on how to optimize the function of one’s own network.
In closing, Stover mentioned that he was helping to organize the First International Conference on Precision Nutrition and Metabolism in Public Health and Medicine (September 21–26, 2018, in Crete, Greece), with the
goal of bringing together people in the fields of nutrition, engineering, and computer science to address how a systems-level understanding of nutrition can be used to provide better guidance to consumers.
GENE-GUIDED NUTRITION INTERVENTIONS
Steven Zeisel, University of North Carolina at Chapel Hill, began by saying that he would be sharing his thoughts on how to develop a nutrigenomic application in industry based on his work with a number of companies that are doing just that. Additionally, he himself was involved in the early startup phase of Zthera, a gene-guided medical foods company.
“We’ve heard over and over again how metabolically different we are,” Zeisel said, noting that each individual has about 50,000 common, inherited single nucleotide polymorphisms (SNPs) out of the millions that exist. Because people have inherited these SNPs from their ancestors, he explained, the distributions of many of them differ among populations. As an example, he cited the distributions of the GG, GC, and CC genotypes of the PEMTrs12325817 polymorphism. These genotypes vary worldwide, with their proportions differing among populations in Africa, Asia, Europe, and America. Zeisel noted that the C allele is associated with inefficient choline production.
But because so much is already known about metabolism, Zeisel continued, there is no need to be overwhelmed by such complexity. Scientists understand the metabolic pathways, he elaborated; they know that nutrients have to transit these pathways; they know that each of these pathways depends on a number of genes; they know what these genes are; they know how the different pathways interact with each other; and they know that the polymorphisms that affect the function of a gene in a specific pathway will have a metabolic marker, such as buildup of a precursor or less final product. “So we can cut down on the complexity of what we look at,” he said, “and increase the power by having pre hoc hypotheses based on our understanding of metabolism.”
Zeisel pointed out that some polymorphisms that are functionally active will create “roadblocks” in metabolism—for example, by changing the affinity site of the enzyme or by changing the regulator of a gene, thereby making metabolism inefficient. “If we know this,” he said, “we can ask, how do we step around this? . . . How could you develop nutritional solutions or medical foods, if they meet all the other Food and Drug Administration (FDA) criteria, that are specifically designed to bypass each of the roadblocks that you identify as being associated with a health condition related to nutrition?”
Zeisel went on to observe that not all SNPs cause functional change. For example, more than 1,000 polymorphisms for the gene PEMT have
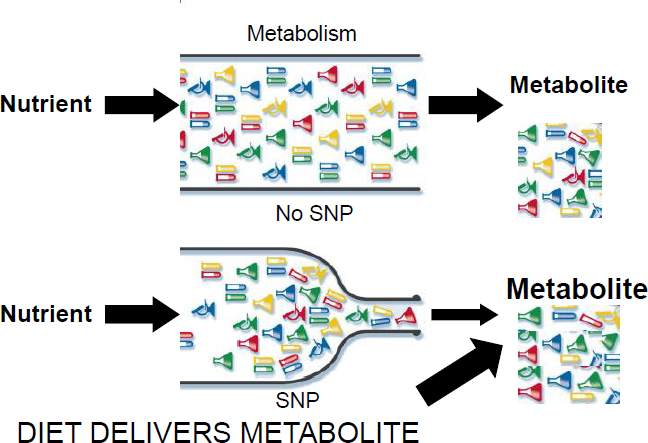
been identified in humans, but only a selected subset—about 10 thus far that have been identified—are functionally important (i.e., because they change, for example, the binding site or a response element). But if one of those SNPs that are functionally active causes a roadblock, Zeisel explained, it creates a bottleneck such that less metabolite is being produced and more precursor is building up, unless diet hides this fact. That is, if someone has a metabolic inefficiency because of one of these SNPs but is eating enough of the relevant nutrient to push through the bottleneck, the SNP will not manifest as functionally important (see Figure 4-3). It is only when one is challenged by having low amounts of the nutrient in one’s diet that an inefficiency becomes important. Zeisel said he would be giving an example of this from his research with choline (see the following section), but first he argued that this is why genome-wide association studies (GWASs) have been “so abysmal” at identifying nutritionally relevant SNPs. By failing to consider dietary intake, he elaborated, GWASs combine people who are consuming large amounts of a nutrient with people who are challenged for the nutrient, thus canceling out any significant effect among those who are challenged.
SOURCE: Presented by Steven Zeisel on December 5, 2017.
Gene-Guided Nutritional Intervention: Choline as an Example
Zeisel began his discussion of choline by remarking that anyone can do with another nutrient what he and his colleagues have done with choline—that is, identify genetic polymorphisms that explain the difference between responders and nonresponders to a nutritional intervention. He then mentioned that in addition to an adequate intake (AI) recommendation set by the Institute of Medicine (IOM, 1998), choline has had approved FDA labeling since 2016. He also explained that choline is important for both the liver and muscle. It can be made in the liver through the enzyme coded by PEMT, and it is contained in such foods as liver and eggs, as well as other high-cholesterol foods. But Zeisel cited National Health and Nutrition Examination Survey (NHANES) data (2009–2014) showing that most Americans are not meeting the recommended AI for choline, which is about half a gram per day for adults, a finding with especially important implications for pregnancy and lactation, as he would explain later. He noted that only small children, who drink a great deal of milk, come close to achieving their recommended AI, making choline what he called a “problem nutrient.”
In one of his first choline studies, Zeisel and his team questioned whether the half a gram per day requirement was really necessary. To explore this question, they removed the nutrient from their study participants’ diets to see if they would develop fatty liver or liver or muscle damage caused by apoptosis. In the process, they noticed that young women needed less choline relative to both men and postmenopausal women (Fischer et al., 2007). After 42 days without choline, only 44 percent of the young women got sick. In contrast, most men (77 percent) and post-menopausal women (80 percent) got sick. Most of the participants who fell ill presented with fatty liver and liver damage, and only about 10 percent presented with muscle damage.
So Zeisel and his collaborators asked the question of what is different about young women. Why is it, they asked, that 56 percent of young women do not need choline? Estrogen appeared to be an obvious answer, Zeisel recalled. Indeed, the researchers found that PEMT is induced by estrogen and is turned on at exactly the concentrations of estrogen that are achieved during pregnancy (Resseguie et al., 2007). But if that is the case, Zeisel’s team asked—that is, if women can turn on PEMT to make their own choline when estrogen is present—why do 44 percent still get sick when deprived of choline? According to Zeisel, it turns out that there is a polymorphism, or set of polymorphisms, in the estrogen response element for PEMT that prevents PEMT from responding to estrogen even if a woman’s estrogen level is high (Resseguie et al., 2011). Women who are homozygous for the C allele at PEMTrs12325817 do not respond to
estrogen at all, he reported, in contrast to women who are homozygous for the G allele, who do respond, and heterozygotes, who respond somewhat. Because women who are CC at this polymorphism cannot increase their production of choline during the high-demand period of pregnancy and lactation, he added, they need to ingest choline. Zeisel explained that it was because of this kind of data—that a polymorphism can help predict which women will need choline in their prenatal diet—that the American Medical Association (AMA) voted for the first time in 2017 to recommend that prenatal vitamins contain choline.
As Zeisel had mentioned earlier in his presentation, the different genotypes at the PEMTrs12325817 polymorphism are distributed differently around the world. In Europe and America, 72 percent of women have one or more alleles of the C allele, and 22 percent are homozygous for it. In contrast, very few women in The Gambia have the C allele. Zeisel explained that the indigenous diet in The Gambia is very low in choline, which he said is an indication that the C allele was selected against. Among the Maasai in Kenya, by contrast, where the diet is higher in choline, the C allele is as common as it is in Europe, where there was enough choline in the diet (e.g., in eggs) so that presumably there was no such selection (Silver et al., 2015).
Regarding the effects of low choline on the fetus, Zeisel shared results from a mouse study showing that when pregnant mice were fed a low-choline diet, their offspring had significantly fewer neural progenitor cells relative to the offspring of control mice (Wang et al., 2016). This phenotype, he explained, has a lifelong effect on memory in mice and probably occurs in humans as well. It has been shown, he reported, that maternal choline intake during the first and second trimesters of pregnancy is correlated with performance at age 7 years on a cognitive test known as the Wide Range Assessment of Memory and Learning, 2nd Edition (WRAML2), with an apparent dose-response (Boeke et al., 2013).
Based on this cumulative evidence, Zeisel envisioned a company forming around this scenario: a gene test (for the PEMT SNP), an obvious intervention (dietary choline), and a common mutation (72 percent of women in the United States having at least one allele for this gene).
Recognizing the Involvement of Multiple SNPs
While single SNP analysis is useful, Zeisel continued, as the field of nutrigenomics evolves, companies will need to start recognizing the complexity of metabolic pathways and the involvement of both multiple pathways and multiple “hits” within pathways. He explained that with choline, for example, other SNPs besides the PEMT SNP can alter sensitivity to low choline and put people at greater risk of becoming depleted and developing liver or muscle problems. In addition to this observation having been
demonstrated in his own work with multiple SNPs (da Costa et al., 2006, 2014; Kohlmeier et al., 2005), Caudill and colleagues (2009) showed that MTHFR alters sensitivity to low choline and increases one’s choline requirement. According to Zeisel, these other SNPs can be found at almost every step of the choline pathway.
Another question that has interested Zeisel is why choline deficiency presents differently, with 90 percent of individuals developing fatty liver and the other 10 percent developing muscle damage. So again, he and his research team examined the responders and nonresponders to see what was different about them and found that people who are choline deficient cannot export fat from the liver as very low-density lipoprotein and thus develop fatty liver (Corbin et al., 2013). He explained that this is because choline is needed to export fat from the liver. That is, the liver makes a “wrapper” out of phosphatidylcholine to export the fat, and without this wrapper, the fat remains in the cytosol. But given that a number of pathways affect how fast the liver can package fat and how much choline is needed, Zeisel and his team genotyped the liver tissues of a population of individuals who had provided liver biopsies, for various reasons. They found a number of SNPs in other pathways (e.g., genes related to choline metabolism, folate metabolism, fatty acid transport, and bile synthesis) that were also associated with fatty liver (Corbin et al., 2013). When they included these genes from the other pathways in their prediction model, they were able to predict susceptibility to liver disease with 95 percent accuracy, compared with 70 percent when they included only the choline polymorphisms. Additionally, however, they found that the predictive power of these polymorphisms was what Zeisel termed “totally useless” in lean people, because lean people are not making a large amount of fat in their liver and can afford to be inefficient at exporting it. Thus it is only individuals with a high body mass index (BMI) for whom export of fat from the liver is important because their liver produces much more fat from the excess calories they consume, and for whom these polymorphisms predict the development of fatty liver.
As a result of this research, Zeisel’s team now has a gene test for 19 SNPs in women and 21 in men that predict susceptibility to developing fatty liver with 90 percent accuracy among people gaining weight. According to Zeisel, a nutritional intervention that bypassed the choline-related SNPs probably would be about 70 percent effective, while an intervention that bypassed all of the roadblocks probably would be about 90 percent effective. In his opinion, given the difficulty of delivering all of these metabolites with a normal diet, the best intervention likely will be a medical food. The idea, he reiterated, is that by giving people whatever metabolite they are unable to make because of a metabolic roadblock, the problem is solved theoretically. And in fact, he asserted, in the case of choline it works: when
people who are choline deficient and who have developed fatty liver are given choline, their fatty liver resolves.
Zeisel then turned to the 10 percent of people who are choline deficient and have developed muscle defects. It turned out, he reported, that everyone in his team’s study who developed muscle damage (rhabdomyolysis2) had SNPs that resulted in a problem in the transport of choline into the muscle cells and in phosphorylating choline once it had entered the muscle cells. He explained that, as with glucose, choline metabolism entails phosphorylating the choline to give it a charge so that it cannot leak out of the cell. In addition to the choline polymorphisms he had discussed, the MTHFD1 polymorphism also appears to predict who is going to develop rhabdomyolysis, as measured by leakage of creatine kinase from muscle cells (da Costa et al., 2014). More specifically, he continued, during exercise, people break down their muscles more if they have these polymorphisms and are eating a diet lower in choline. So again, he predicted the opportunity for another medical food as a solution to a blocked metabolic pathway, or pathways—in this case the SNPs responsible for muscle breakdown during exercise.
SNPs and Sperm Dysfunction: Another Example of Gene-Guided Intervention
Zeisel went on to explain that when a study is conducted in humans and polymorphisms are identified that appear to be important, as was the case with choline deficiency, those polymorphisms can be knocked out in mice and other effects examined. So he and his team knocked out the choline dehydrogenase gene, Chdh, in mice because it was one of the genes that affected the choline requirement in humans, and they found that the mice developed what he characterized as horrible-looking mitochondria in their sperm (Johnson et al., 2010). The sperm were infertile, unable to swim, and unable to make adenosine triphosphate (ATP). The researchers then found that they could restore sperm function in the mice by giving them the metabolite (betaine) that was being blocked from production. Although Chdh is a nuclear gene, Zeisel noted, the protein resides in the mitochondria.
Zeisel’s team next examined this same polymorphism in men (Johnson et al., 2012). It is a common polymorphism, he observed, existing in about 5–9 percent of men, depending on lineage. The researchers found that men who are homozygous for the T allele make little ATP in their sperm, and heterozygotes can make only half as much ATP as GG homozygotes. Although the sperm in the men with the T allele “look terrible,” as they did in mice, Zeisel and his team have yet to test whether they are less fertile.
___________________
2 Rhabdomyolysis is a condition in which damaged skeletal muscle breaks down.
He suspects that they are, as their sperm do not swim well. “So again,” he said, “you can imagine if you were trying to think of a company, you could genotype men who are unable to have a baby, predict poor sperm function using ATP in sperm as a biomarker, and conduct a clinical trial to see if their low sperm ATP is reversible by delivering the metabolite that bypasses the genetic roadblock.” And again, he emphasized his underlying thought process: “Take what you know about metabolism and use that to predict what nutrigenetic test and treatment should work.”
Future Developments: Medical Foods as a Starting Point for Developing Nutrigenomic Products
To move forward, Zeisel called for better methods for working with complex metabolic pathways involving multiple SNPs. He remarked that he had been focusing on choline because that is his area of expertise. But he argued that the same approach could be used to study vitamin D, for example, to design interventions that would bypass specific inefficiencies in metabolism that contribute to the problems people have with vitamin D.
Additionally, Zeisel called for a better way to include diet information in GWASs. He remarked that, as part of many precision medicine initiatives, researchers are going to be measuring genes and metabolome, but, he said, “they are not thinking of collecting nutritional data.” Yet without those data, he asserted, it will be impossible to know who is being challenged by low or high intake. If people with a polymorphism are not being challenged, he added, they will look the same as people without the polymorphism.
Finally, Zeisel encouraged the workshop participants to think about medical foods, as defined by FDA, as a potentially good starting point for developing nutrigenomic products. He referred to José Ordovás’s description of what Zeisel called the “prototype medical food”—a food for rare mutations that cause aminoacidopathies, such as phenylketonuria. He argued that the same strategy could be used for metabolic roadblocks, except that instead of bypassing a rare mutation, the medical food would be bypassing a common polymorphism. In his opinion, an intervention that specifically bypasses a multitude of blocked pathways would not be easily deliverable in a normal diet; indeed, it would not even necessarily be easy to calculate, he opined. Therefore, he said, “it is the perfect fit to the FDA’s current definition of what a medical food should be.”
DISCUSSION
Following Zeisel’s presentation, he and Stover participated in an open discussion with the audience, summarized here.
Questions About the Potential Role of Medical Foods in Nutrigenomics
Much of the discussion revolved around the potential role of medical foods in nutrigenomics, beginning with a question from an audience member about Zeisel’s rationale for working toward developing choline as a medical food when it could be a dietary supplement. Zeisel agreed that if choline alone were the issue, a dietary supplement would be a good nutritional solution, but he argued that when multiple pathways are involved, a medical food makes more sense. The advantage of a medical food, he asserted, is that a physician or other health professional would be overseeing the interpretation of the gene test for its need. Additionally, he observed, physicians understand prescription medicine, and a gene test tells them what to prescribe. The test, he elaborated, validates that there is a problem and that they are really treating something, providing both physician and consumer with more confidence. He added that, because developing this type of nutritional intervention will require research and investment, it would be beneficial to make it something (i.e., a medical food) that brings slightly higher returns to account for that extra cost.
Another audience member, noting that “Food Product Development” was in the title of this session, questioned how nutrigenomics tools will impact what kinds of food products become available, how widely used these products will be, and what the regulations around them will be. She pointed to gluten-free foods as an example of a class of food products that has become very mainstream and is related to a medical condition that she presumed could be predicted with nutrigenomics tools.
“I would say we are at a frontier,” Zeisel replied. He predicted that someday, it will be obvious how to develop foods that are not commodities, that is, foods that have a little extra value because they are designed specifically to deal with a health problem. They may or may not be based on genetics, he added. “We’re going to have a lot of snake oil,” he acknowledged, “but we have a lot of snake oil diets with regular foods.” He emphasized that he liked the idea of medical foods as a starting point because FDA has already set some ground rules in this area. The agency has not fully defined the problem, he remarked, but with what it has defined, there is at least some oversight. At the same time, he characterized the development of a medical food as “kludgy,” requiring a much greater financial investment and being more difficult relative to developing a dietary supplement. Yet, he said, using the term “medical foods” is a way to differentiate these products from those developed by what he called “mom and pop operations” that sell products that do not work. Once it can be shown that a nutrition intervention works as a medical food, he suggested, one can work backward and consider how to make it an over-the-counter intervention and distribute it to the public at large.
Douglas Wallace remarked that one of the goals of nutrigenomics is to minimize chronic disease. “The pharmaceutical industry would love to develop drugs that have the same effect,” he said. He asked how these conflicting perspectives on how to deal with chronic disease are going to be managed when an estimated 50 to 60 percent of the population is being prescribed pharmaceuticals to minimize a chronic disease.
Stover replied that FDA is very clear about the effect of chronic disease treatment on nutrition; that is, if a pharmaceutical changes one’s requirement for a nutrient, the change must become part of the formulation for the drug. He suggested that perhaps this rationale needs to be revisited given how widespread use of these drugs is today, but that is how it currently stands. With respect to which is going to play the bigger role in lowering rates of chronic disease—food or drugs—Stover expressed the view that both need to move forward, but, he said, “I think a lot of the evidence is that these are diet-driven, and in the long run, the diet-driven approaches are going to be the best approaches in terms of lowering rates of chronic disease without side effects and without the added health care cost.” At the same time, he added, those are decisions made by politicians, not scientists.
Tim Morck agreed with Zeisel that medical food appears to be an appropriate category for nutrigenomics products. However, he observed, not everyone is familiar with what he described as “all the narrowness” that this characterization entails from a regulatory standpoint. Choline is a nutrient, he said, “so at least it gets that part of the box checked.” But he argued that many people are trying to make medical foods out of things that are not actually nutrients. He identified as a second challenge with the medical food category that although medical foods require medical supervision, which he agrees is an appropriate approach, it is important to emphasize that these products are for nutritional management. One would not be able to say, he elaborated, that they are being used to treat a disease because that characterization is allowed only for drugs; instead, one would have to say that they are being used to address the nutritional imbalance a patient has by virtue of a genetic condition, or in some cases, a disease impact. He argued that this then calls into question how to talk about nutritional balance, or normalization—in other words, the restoration of normal metabolism—rather than about a pathological condition. Keeping this in mind, he said, medical foods are the only categorization that would allow for a nutritional treatment for patients with disease, as dietary supplements can only help support normal function.
Zeisel clarified that he was not suggesting a medical food is the only way to start implementing nutrigenomics; rather, he was suggesting that it might be the best way to get the field started. It is credible, he argued, because it has science behind it. Moreover, he suggested, given the complexity of nutrigenomics, developing a medical food rather than focusing
on dietary advice is a way to simplify and focus on a small problem with a well-understood outcome. He predicted that eventually, products will be developed that address multiple problems, and the idea that nutritional intervention works will begin to permeate medicine. At that point, physicians will be able to tell their patients, “Go out and eat more eggs.” (During his presentation, he had mentioned eggs as a dietary source of choline.) But at present, in his opinion, most physicians would not be comfortable telling their patients, “Go out and eat more eggs”; they would be more comfortable with conducting a genetic test and then prescribing a nutritional intervention on the basis of its results.
The audience member observed that, given the direction this field appears to be going, it eventually will require the involvement of both clinical nutrition and clinical genetics, as well as medical supervision. Yet, she asserted, even adults who have inborn errors of metabolism caused by single gene defects are not well handled right now in what she described as “the great hospitals of America.” She acknowledged that perhaps children with these disorders are being well handled, but argued that their adult counterparts are not being integrated into internal medicine, adding that she was unaware of the situation with surgery. She asked, “How is this all going to be handled, this brave new world?”
Medical foods fit the precision medicine model, Zeisel replied. He described precision medicine as a powerful movement at present, with the support of both federal researchers and many large medical schools, and as a subject being taught to every medical student today. “I think everybody will be practicing that way,” he said, so “this will just look the same to them.”
Stover suggested that the formulated medical foods being used to treat loss of function in individuals with inborn errors of metabolism also serve as a model. In the case of inborn errors of metabolism, he noted, the loss of function is due to a single-gene defect, and many chronic diseases are similarly due to a loss of function, not in a single gene, but across multiple genes, and other factors, such as inflammation. Thus, he argued, just as medical foods are used to restore function in individuals with single-gene inborn errors of metabolism, they could also be used to restore function in people with chronic disease.
Foods: From Wellness to Therapeutics
Naomi Fukagawa called attention to what she called a “spectrum” of classifications for food, from wellness to therapeutics, and asked how categories of food along this spectrum will be differentiated from each other and from supplements.
Stover clarified that some chronic disease can be initiated prior to
conception. It is a trajectory over time, he said, and nutrition can change or modify that trajectory. He suggested that complete prevention may not be possible in all cases, and the idea of the food supply and wellness does not always align well with the idea that chronic disease manifests over a lifetime as the result of multiple factors, including genetics and others, in addition to diet. For Stover, the question to ask is when the requirements of an individual fall outside the realm of what can be achieved through a natural, food-based diet because of genetic or other age-related factors. For example, he observed, depending on one’s genetic makeup, one may not be able to get enough folate from the food supply without fortification or supplementation to prevent the risk of neural tube defects.
Zeisel suggested that another way to think about Fukagawa’s question is that without a challenge, a person may be phenotypically healthy, but when challenged, that person’s genetic or other limitations are revealed, and they get sick. For example, he noted, there is no need to worry about fatty liver in someone who is not consuming excess calories or is not making a large amount of lipid in the liver. In that case, he said, transport of fat out of the liver does not matter; it becomes important only when a person is challenged. Thus, he observed, one could anticipate the fatty liver phenotype appearing as a person moves toward becoming overweight, which is when the need to transport fat out of the liver increases. So the question for Zeisel is, What is the challenge that is going to reveal someone’s metabolic inefficiency? The limitation of this approach, he added, is that it misses things that were never conceived as being likely. The other approach, which will reveal things not anticipated to be present, is to measure everything and see what is correlated with health outcomes. But this approach, Zeisel argued, is limited by the fact that the results will be associational and not based on an understanding of the person’s biochemistry and metabolism.
Nutrigenomics in the Context of Preexisting Conditions
An audience member asked about the future of genomics, metabolomics, and proteomics given that preexisting conditions may cause a problem with insurance for today’s children. Stover responded that insurance coverage is one of the greatest challenges with medical foods—for example, for children with inflammatory bowel disease who are dependent on a liquid diet for their survival. It is very difficult, he observed, for those families to get insurance to pay for these products. He stated that political interest in this issue is currently growing, and a bill now pending, the Medical Nutrition Equity Act of 2017, would address the problem. The question is, he suggested, What will be the standard of evidence that justifies this huge expansion in reimbursement, that demonstrates these products actually work and are meaningful for
managing disease? “That is something that this community is going to have to wrestle with,” he said.
In Zeisel’s opinion, if someone already has a problem, such as fatty liver or rhabdomyolysis, his or her physician is already looking for the optimal treatment at the lowest cost. He argued that testing is standard for physicians, and if a test comes back positive, the physician feels confident prescribing a treatment. If a test shows that someone has a specific metabolic effect, for example, and there is a medical food designed to treat that effect, Zeisel would argue that prescribing that medical food as an intervention poses no greater risk than would be the case if there were no such option.
Zeisel suggested that eventually, babies will be genotyped at birth. Thus, for example, if a person were prescribed warfarin later in life, that person would not need to be genetically tested because his or her genotype would be on file. Zeisel cautioned, however, that legislation will be necessary to ensure that this type of screening does not place people at a disadvantage. He mentioned a study in which he was involved with the National Aeronautics and Space Administration (NASA), in which 16 of 32 astronauts who spent more than 16 months in space were found to have permanently lost visual acuity by the time they returned. It is not clear why, he said, but the best clue is that they have higher homocysteine levels, not out of the normal range, but still higher than those of the other astronauts who did not lose visual acuity. They have some polymorphisms as well, he added. Once NASA knows the genotype associated with lost vision, he observed, it will not want to send those astronauts to Mars. So again, he added, there is a risk to genetic testing, and there must be either legislation or a treatment. He mentioned a similar situation with the U.S. Army Special Forces, among whom a significant percentage have developed rhabdomyolysis and had to be sent home from the field. Again, he noted, the army would like to avoid rhabdomyolysis in those people, so soldiers are not going to want to be genotyped. On the other hand, he said, if there is a solution related to the genotype that cures the problem such that muscle breakdown does not develop, it will be worth being genotyped.
Implications for Dietary Guidelines
Wendy Johnson posed the question of what understanding of genetic variation and how the environment impacts gene expression means for dietary guidance. Will dietary guidelines still exist in “this new world,” she asked, or are they to be abandoned altogether and replaced with something more progressive?
For Stover, the question is whether the focus will be on clinical nutrition in a patient or in a population. “I think the answer is both,” he said.
He referred to the description in his presentation of the WHO recommendation for fortifying the food supply to prevent neural tube defects, and how the WHO committee included in its model the effects of the MTHFR polymorphism on folate status and the prevalence of the polymorphism in the population. He explained that the question will always be the degree to which any sort of modifier or sensitizer (e.g., the MTHFR polymorphism) affects the requirement for a given nutrient. If there is an identifiable group that falls outside of the normal population distribution, he observed, then either the value for the nutrient will need to be changed to accommodate that minority group, or the group will need to be considered a separate subgroup with separate nutritional requirements. Then the question becomes, he suggested, how many subgroups there can be. “But there is no way I think we are going to be able to walk away from having population-based nutritional requirements,” he asserted, “because that’s the basis of the food supply. The question is, how do we deal with people who fall outside the distribution?”
In Zeisel’s view, a polymorphism found in 72 percent of U.S. women (i.e., those with at least one PEMT C allele), represents a large enough identifiable group to be accommodated. On the other hand, he suggested, a polymorphism that occurs in only 1 percent of the population may be too infrequent to accommodate. As far as what accommodation entails, he said it could involve a genetic test indicating whether someone needs, for example, more of a particular nutrient, or it could involve extending the upper limit of the nutrient to meet that higher nutritional requirement. He cautioned that the latter approach would be pushing toward toxicity.
With respect to the number of identifiable groups, Zeisel suggested that the next step for the dietary guidelines may be to base recommendations on haplotypes. He described a haplotype as a “lumping of genes that tend to travel together with an ancestry,” and remarked that there are many fewer of them than SNPs.
Stover urged the workshop participants to keep in mind, especially when thinking about chronic disease endpoints for either dietary or nutrient guidelines, that the risk factors for chronic disease include genetics, but they also include age and diet and the interactions among genetics, age, and diet. “The aging genome has just as much effect on a chronic disease, and maybe even on nutritional requirements, as genetics,” he asserted.
Consolidation and Collaboration
Nathan Price remarked that both Stover and Zeisel had addressed the notion of “genetics under challenge,” which he subscribes to as well. He noted that for GWASs, a 1 percent effect is considered large, yet speakers in this session had presented examples of much larger effects under challenge.
He asked about the degree to which this information has been catalogued or consolidated such that other people can learn from it. Additionally, he asked how the field can move beyond a few examples to begin studying genetics under challenge on a large scale.
Stover replied that many databases are available to aid in developing the architecture of a metabolic network, such as those for gene expression, SNPs, the proteome, and the transcriptome, any of which can be used to set a range for or circumscribe the magnitude of an effect at any one node. Yet, he added, although the information is available, one must have some familiarity with the field to understand it. Once the dynamic range of an effect is known, he observed, one can perform a sensitivity analysis to determine how different inputs of nutrients affect the range of responsiveness. But, he added, there has been no coordinated effort to examine and determine, node by node, the degree of stochasticity of expression that is associated with each node, something he believes is clearly needed.
Zeisel called for greater collaboration between experts who know how to measure the genome or metabolome but do not really understand metabolism and experts in nutrition and other fields who understand metabolism and know the inputs, outputs, and challenges along a pathway. He stressed that most GWASs are focused only on statistical association, without accounting for the fact that both challenged and unchallenged individuals are involved.
























