online longitudinal assessment will allow for the development of increasingly relevant content that is specific to the scope of specialized radiologist practice patterns, including oncologic imaging.” Radiologists who do not receive adequate results from this process will have to undergo a summative test at a test center of the American Board of Radiology, Wagner said.
INTEGRATION AND COLLABORATION OF SPECIALTIES
Many speakers at the workshop discussed integration and collaboration among the specialties involved in cancer diagnosis and care, including the rationale for greater integration and collaboration, challenges involved, and potential strategies to facilitate better coordination among the diagnostic specialties.
Rationale for Collaboration and Integration
Lawrence Shulman, professor of medicine and deputy director of clinical services at the University of Pennsylvania Abramson Cancer Center, said that greater integration of pathology, radiology, and oncology reports is needed for a more unified interpretation of a patient’s disease and for treatment planning. Stewart and Warner agreed, and added that multidisciplinary conferences or tumor boards can be very helpful in facilitating collaboration. “If we could figure out how to get every patient who has imaging, pathology, and molecular testing into a tumor board situation—where people get together and think about that patient—we’ll get to the right place,” Warner said. Becich added that “pathologists and radiologists have got to get out of their dark reading rooms and get [more] involved in the care pathway.”
Cohen noted that there is growing overlap between the fields of radiology and pathology. “As these molecular reagents [used in imaging] become more sophisticated, the distinction between pathology and radiology is going to narrow,” he said.
Becich suggested prioritizing image sharing as a method to integrate the specialties: “We could have a flawed report, or the interpretation could change, but the image doesn’t change. I’m going to focus on image democratization because we now have enough robust networks, scalable technologies, and deep learning, so putting the image at the center puts truth at the center of our studies and eliminates the noise.”
Schilsky pointed out that some imaging studies are highly sensitive but
lack specificity,30 while some molecular tests, such as those for circulating tumor DNA, have high specificity, but low sensitivity. He asked, “Are we headed toward a time when we can begin to combine technologies to get an optimal diagnostic assessment? If so, where does that integration occur, and how do we prepare the workforce for that integrated diagnostic approach?” Hricak responded that “we hope in the future we will have data analytics that can help with this. Integrated diagnostics is unquestionably in our future. But it will be at least 5 years before it is widely adopted because you have to develop, validate, and disseminate, and this will take us awhile.”
Cohen noted that such integration may not always be possible in some community settings. “If you are in a community hospital with a couple hundred beds where you may have 3 or 4 pathologists and maybe 10 radiologists, that type of work effort may not be feasible given limited health care dollars,” he said. Friedberg added that clinicians in community practice are integrating information from the diagnostic specialties, but may not be trained in the latest tests and technologies.
John Cox, medical director of oncology services at Parkland Health and Hospital System and professor of internal medicine at the University of Texas Southwestern Medical Center, said there is a real need for integrated reports in his community health care system, especially when test results come from separate labs in different formats: “From a community delivery system standpoint, integrating those reports into a common diagnostic portfolio is really key and something our current technologies ought to help us solve,” said Cox.
Challenges to Integration
Several workshop participants discussed challenges to integrating the disciplines involved in cancer diagnosis. Brink and Friedberg noted that it is difficult to foster interactions within an individual specialty, let alone promote greater collaboration among multiple specialties. “We need to start seeing ourselves as having different ways of looking at the same elephant—microscopically and macroscopically with overlap in the middle as the technology advances. But none of us are trained that way,” Friedberg said. He added, “My colleagues who define themselves as microscopists are
___________________
30 Sensitivity is the proportion of people with a disease who are correctly identified from all positive test results for the disease, and specificity is the proportion of people who are correctly identified as not having a disease from all negative test results for the disease.
going to be a little too narrow for the future. They need to be diagnosticians, using all available tools, as the line between radiology and pathology continues to get blurred.”
Cox added that “specialization and how we are paid has put us in many silos.” Becich agreed, and added that increasing subspecialization has prevented the development of comprehensive patient reports. “We can talk about wanting to improve the delivery of care, but really the enemy is us. As we continue to subspecialize and fragment . . . we fail to take it back to the context of the whole patient,” he said.
Several participants noted that data-sharing challenges impede communication. “There are significant challenges related to sharing diagnostic images between and across institutions,” Stewart stressed, despite the progress that has been made in digitizing radiographic images. For example, Stewart said a patient may have an extensive imaging work-up at one institution, but if that patient moves to another institution, obtaining the prior images or reports in order to accurately stage or restage for comparison may be challenging. Some institutions have started importing outside images into their computerized imaging archiving systems, but this is not universal. Some patients may bring in compact discs of prior imaging, but these images may be incomplete, or not viewable or importable, said Stewart. Prior imaging may also be insufficient due to technique limitations, such as a lack of contrast. “Sometimes, exams need to be repeated, which is wasteful, duplicative, and results in increased radiation dose,” Stewart said.
Friedberg noted that anatomic pathology is further behind radiology in terms of digitizing and sharing information, but predicted that “once pathology becomes digital like radiology, the overlap between the two will naturally disappear and AI will be very useful.” Brink agreed that AI is merging pathology and radiology (see also the section on Computational Oncology and Machine Learning).
Elenitoba-Johnson pointed out the need for greater sharing of genetic data, but a lack of EHR interoperability among institutions prevents sharing this information in a usable format. Warner added that in a 2016 ASCO survey,31 less than one-quarter of respondents said their laboratory could deliver genetic profiling data on a patient’s tumor in a format their EHRs could receive. Interoperability has been impeded by a lack of reporting standards for genetic tests, Warner stressed. He noted that if such standards were in place, a clinician could receive results and use third-party applica-
___________________
31 Results from the survey are unpublished.
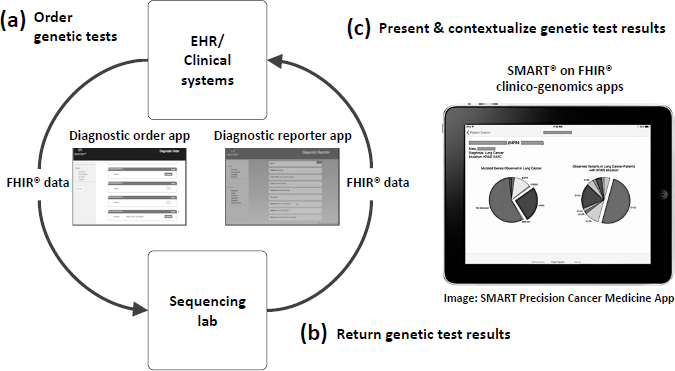
NOTE: App = application; EHR = electronic health record; FHIR = Fast Healthcare Interoperability Resources; SMART = Substitutable Medical Apps, Reusable Technologies.
SOURCES: Warner presentation, February 13, 2018; Reprinted with permission from BioMed Central: Genome Medicine. Integrating cancer genomic data into electronic health records. Warner, J. L., S. K. Jain, and M. A. Levy. © 2016.
tions to present and contextualize genetic test results (Warner et al., 2016) (see Figure 2).
Warner said a recent President’s Cancer Panel report32 called for developing tools and applications that can facilitate oncologists’ use of genomic data, and added that Vanderbilt-Ingram Cancer Center built the SMART Precision Cancer Medicine33 application to incorporate genomic data into its EHRs. Because this application was developed with a publicly available application programming interface and uses the Fast Healthcare Interoperability Resources (FHIR) interoperability standard, it can be adopted by other health care systems. The DIGITizE Action Collaborative34 has also convened experts from academic health centers and EHR vendors Cerner,
___________________
32 See https://prescancerpanel.cancer.gov/report/connectedhealth (accessed May 5, 2018).
33 See http://www.vicc.org/smart-pcm (accessed May 4, 2018).
34 See http://www.nationalacademies.org/hmd/Activities/Research/GenomicBasedResearch/Innovation-Collaboratives/DIGITizE.aspx (accessed July 5, 2018).
EPIC, and Meditech to develop a standard structure for reporting genetic test results, which is being tested by several pilot sites, including Intermountain Healthcare, Duke University, Johns Hopkins University, Mission Health, Partners Healthcare, and St. Jude Children’s Research Hospital. DIGITizE has been taken up by the FHIR Foundation, an implementation body related to Health Level Seven International.35
Stewart pointed out that insurance coverage issues, such as out-of-pocket costs or restrictions on additional testing, may prevent more comprehensive and integrated radiology and/or pathology reports. For example, Cox said self-referral restrictions can impede genetic testing performed by the same institution that performed the initial pathology testing. “When we ask the pathology department to guide us on whether the sequencing should be [conducted] or not, we are met with the barrier that they cannot easily refer to their own lab at UT Southwestern to do the sequencing. As a result, FoundationOne provides those services, but their genetic testing report comes back in a non-integrated way with our systems,” Cox explained.
Misalignment of incentives can also hamper the integration of diagnostic testing results and communication among diagnostic specialists. Stewart noted that cancer centers are more likely to repeat an imaging study rather than try to acquire or discuss imaging results with radiologists from another institution, in part due to reimbursement issues. Hricak agreed, noting that payers will not reimburse radiologists for clinical consultations, hindering collaboration among radiologists, pathologists, and oncologists. Cohen stressed that “to integrate, we need to have alignment of the financial incentives.”
Sause said that interdisciplinary teams are labor- and resource-intensive, but “the way that most of the system is designed, you’re not paid for that collaborative process,” which is a challenge in the current era of increasing subspecialization. He suggested considering changes to payment to foster more interdisciplinary collaboration. Grubbs agreed, and said, “The driving force will be reimbursement. When we’re all responsible for the total cost of care, you’ll find who you want to partner with very quickly to get the job done quickly with high quality that reduces the cost. That will drive us all to do it.” Kline added, “Payment does change the way people organize themselves and does [help] overcome cultural barriers.” Hofmann added that “a small change in payments can make a big difference.”
Becich pointed out that CMS increased reimbursement to pathologists
___________________
35 See http://www.hl7.org (accessed July 23, 2018).
who adopted standards for cytology reporting and quality measurement and within 2 years, every pathology practice and cytology lab conformed to these standards. Becich suggested that integrated pathology and radiology reports could become a quality measure that CMS and other payers require for payment. “We need systems to reimburse for integrated diagnostic reports that are patient-centric and disease-centric,” Becich said. Larson added that Medicare could require hospitals or practices to use integrated diagnostic reports as part of their conditions of participation. “The solution is not going to be piecework and individually focused, but moving toward a whole organization working cohesively together to meet the needs of patients and referring providers,” Larson said. Zutter added that leadership is needed to promote interdisciplinary collaboration in cancer diagnosis. “That’s one of the reasons it has worked at Vanderbilt,” she said.
Amy Abernethy, chief medical officer, scientific officer, and senior vice president of oncology at Flatiron Health, stressed that once reimbursement policies drive integrated diagnostic reports, “the tech industry will build the solutions that will make that happen. Electronic health record companies will build solutions into their systems when it goes along with what clinicians and health systems demand and need.” Becich stressed, “Pathology and radiology can work together [for a] common purpose but they need to partner aggressively.”
Hofmann recommended incorporating patient input when developing better systems for diagnostic integration. “We need to start with the patient and take a patient-centered approach to figuring out what’s wrong in order to figure out how to fix things,” he said. Spears agreed and added, “When you talk about taking a systems approach, make sure to include the patient and make things patient-centered, and get information back to the patient in a way they can understand it. The patient outcome is really what we are going after.”
Alternative Payment Models to Promote Collaboration and High-Quality Care
Hricak noted that major cancer centers can afford to have radiologists integrated in their medical oncology clinics. This set-up enables more collaboration and interaction between radiologists and oncologists, she said, adding, “It’s a luxury that is wonderful for patient care, but [one that] few centers can afford.”
Grubbs suggested that greater collaboration could promote high--
quality care while also resulting in cost savings for health care systems, and recommended financial analyses be conducted to create the business case for better integration of the specialties. He added that this could be a component of current efforts to move from volume- to value-based payment in health care using alternative payment models, which aim to reward health care systems and practices for delivering high-quality, cost-efficient care.
Grubbs reported on the Health Care Payment Learning & Action Network, which the Department of Health and Human Services launched in 2015 to align public and private stakeholders in the transition toward high-quality, value-based payment.36 The Network’s first initiative was the development of a framework of the different types of payment models, which include (Health Care Payment Learning & Action Network, 2017):
- Category 1: Fee-for-service, no link to quality and value
- Category 2: Fee-for-service, link to quality and value
- Category 3: Alternative payment models built on fee-for-service architecture
- Category 4: Population-based payment
Grubbs also discussed the CMS Oncology Care Model (OCM),37 a 5-year voluntary pilot project initiated in 2016 with the goal of improving the quality of care and reducing Medicare spending. Features of OCM include treatment based on clinical practice guidelines, the use of data and feedback for continuous quality improvement, and the documentation and reporting of clinical practice and patient care outcomes. Approximately 190 practices in 31 states have participated in the pilot program, said Grubbs.
Kline said that because OCM takes into account the total cost of care, it has the potential to facilitate collaboration among pathologists, radiologists, and oncologists. The model supports sharing of reports and other multidisciplinary interactions to foster better value and efficiency. For example, an oncologist who has a brief conversation with a radiologist may decide not to order unnecessary and expensive imaging. “You may find that is the path forward,” Kline said. Schilsky agreed that OCM could “foster team building because it provides [the] opportunity for reimbursement for physicians
___________________
36 See https://innovation.cms.gov/initiatives/Health-Care-Payment-Learning-and-Action-Network (accessed July 14, 2018).
37 See https://innovation.cms.gov/initiatives/Oncology-Care (accessed May 4, 2018).
to spend time contributing to those teams. The big challenge is setting the bundled payment at the right level.”
Kline stressed that paying for improved integration “by simply layering it on with another CPT [Current Procedural Terminology] code is not going to happen. Our country can’t afford that,” noting that U.S. health care expenditures equal approximately 18 percent of the gross domestic product. He added that an Institute of Medicine report estimated that approximately 30 percent of health care dollars spent in the United States are wasteful (IOM, 2013a). “If we can reduce that waste by incentivizing high-value care by aligning financial incentives appropriately, we’ll have the money left to pay for some of these computational systems that will improve the quality of care,” Kline said. Brkljačić added that European nations also continue to wrestle with the rising costs of cancer care and stressed that “value-based health care should be translated to all regions in the world.”
Grubbs also reported on ASCO’s efforts to develop an alternative payment model built on oncology clinical pathways, which are detailed protocols for delivering cancer care—including but not limited to anticancer drug regimens—for specific patient populations, including the type, stage, and molecular subtype of disease (Daly et al., 2018). Compared to clinical practice guidelines that list several treatment options, Grubbs said that clinical pathways narrow down these potential options to a single optimal choice, with the goal of facilitating high-quality care (Lawal et al., 2016; Rotter et al., 2012). He added that oncologists are increasingly using clinical pathways in their practice; in 2016, approximately 60 percent of practices reported compliance with clinical pathway programs (ASCO, 2017).
Grubbs said the goals of ASCO’s Patient-Centered Oncology Payment Model include adherence to high-quality, evidence-based clinical pathways; reducing unwarranted variation in oncology care; guiding appropriate survivorship care and monitoring; encouraging participation in clinical trials; and eliminating care disparities and protecting against underutilization (Zon et al., 2017). He noted that the payment model “still preserves physician and patient autonomy, because nobody is expected to be 100 percent compliant with a pathway.” He said the model will also evaluate whether cancer care is consistent with quality standards, using measures from the Quality Oncology Practice Initiative38 and Choosing Wisely®.
___________________
38 See https://practice.asco.org/quality-improvement/quality-programs/quality-oncology-practice-initiative (accessed May 4, 2018).
Real-World Data and Computational Oncology
A number of workshop participants discussed the roles of real-world data, computational oncology, and data sharing and standardization in improving cancer diagnosis and care.39
Real-World Data
Abernethy said large, interconnected datasets can facilitate cancer diagnosis and care. She said Flatiron Health has compiled longitudinal EHR data from more than 43,000 patients with lung cancer at multiple sites in the United States. These data are linked to other data sources, such as cancer registries and administrative claims databases. “Now that we have aggregated data about patients receiving care in the lung cancer setting across the country, we can start to understand histology at scale,” she stressed. “Because as soon as you’re able to understand what exactly happened to that patient across time and pull that back to the diagnostic image or test, it starts to improve both how we evaluate the test and how we understand images with machine learning algorithms. That made a big difference,” she said.
Abernethy noted that in Flatiron Health’s datasets, the interpretation of imaging and pathology results in lymphoma vary substantially over time and among clinicians. She said large aggregated datasets with longitudinal information could help mitigate diagnostic ambiguity in lymphoma by understanding which interpretations are most accurate: “We’re really starting to try and use the longitudinal understanding of the patient to get back to improved diagnosis.”
Abernethy stressed that large datasets are only useful if the data are properly collected, curated, and aggregated. Within a dataset, she noted that it is important to understand the relationships among the diagnostic event, the treatment event, and a patient’s outcome. Abernethy noted that diagnostic events make up combinations of clinical, pathological, radiological, and biomarker data, but these data can be collected at different points in time. During that period of time, a patient’s cancer might progress or interpretations of the data may evolve over time.
___________________
39 This topic will be explored in more detail at the second workshop, The Clinical Application of Computational Methods in Precision Oncology. See http://www.nationalacademies.org/hmd/Activities/Disease/NCPF/2018-OCT-29.aspx (accessed May 4, 2018).
A major challenge in curating large datasets using EHR data is the plethora of unstructured data, Abernethy noted. Flatiron Health primarily uses human abstractors to pull out essential information from patients’ pathology and radiology reports, as well as the notes from oncologists and surgeons. These abstractors then translate this information into a machine-readable form and feed it into a database, using AI to facilitate this process. The accuracy of that curation is important, Abernethy explained. “We need to think about data provenance or traceability back to the source, and all the standards that are behind these datasets. If you want to have confidence in the datasets that you are working with, you need to be able to look under the hood and understand how you got there. You need to document back to the source and maintain full provenance,” she said.
Data completeness and quality also has to be documented, she added, including linking unstructured data from clinician notes to other sources of data, such as reports for genetic testing, PROMs, and administrative claims data. Abernethy suggested applying a consistent approach to ensuring data completeness and quality, and relying on sources of data known for accuracy. “We need gold standards but we don’t have national gold standards for many of the data points in these datasets, and that’s something we hope the National Academies can help to shepherd forward,” Abernethy stressed. She also suggested applying a checklist (Miksad and Abernethy, 2018) to evaluate real-world data quality by considering factors such as
- Clinical depth: Data granularity to enable appropriate interpretation and contextualization of patient information.
- Completeness: Inclusion of both structured and unstructured information to provide a thorough understanding of patient clinical experience.
- Longitudinal follow-up: Ability to review treatment history and track a patient through time.
- Quality monitoring: Systematic processes implemented to ensure data accuracy and quality.
- Timeliness/recency: Timely monitoring of treatment patterns and trends.
- Scalability: Efficient processing of information with data model that evolves with standard of care.
- Generalizability: Representativeness of the data cohorts to the broader patient population.
- Complete provenance: Robust traceability throughout the chain of evidence.
Computational Oncology and Machine Learning
Langlotz emphasized that AI will revolutionize clinical practice, and radiologists and pathologists will need education and training on how to effectively use these tools. Becich agreed, and said AI can help clinicians overcome information overload. He noted that human cognitive capacity is limited to considering approximately 5 to 10 facts per decision. But cancer diagnosis can involve massive amounts of data: “We are swamped with data and that’s why these algorithms applied to large datasets are important for clinicians,” Becich said. Sause added that “we have to work smarter and more efficiently, and AI, interconnectivity, and better computer systems are going to help us create a more dynamic, smarter, and efficient workforce that will allow us to bring experts to the patient.”
Warner pointed out that there were tens of thousands of distinct genomic variants identified in the first public release of data from the American Association for Cancer Research Project GENIE (Genomics Evidence Neoplasia Information Exchange) (AACR Project GENIE Consortium, 2017). Among the 19,000 samples, the top 100 variants accounted for only 10 percent of the distinct variants identified. “To think a clinician or any other human could digest even a fraction of that information is impossible. That’s why we need artificial intelligence,” Warner emphasized. “Artificial intelligence and machine learning will be critical to the practice of precision oncology,” he stressed.
Becich said that AI will not replace pathologists or radiologists. “The number of diagnostic tests being interpreted by pathologists continues to bloom,” he pointed out, and these professionals are needed to guide the development of AI systems and in the interpretation of their findings.
Langlotz provided an overview of different types of AI, including machine learning, neural networks, and deep learning. He said that machine learning offers new opportunities to analyze large datasets. “If you have a large enough dataset that has labeled images as ‘cancer’ or ‘no cancer,’ you can feed it into a machine learning algorithm, and it will automatically learn how to distinguish benign from malignant. . . . It’s the speed, scalability, and scope of these systems that has really changed dramatically,” Langlotz said.
He said neural networks are a more sophisticated form of machine learning, with millions of nodes that enable fine-tuned detection and clas-
sifiers. When those networks are compiled into multiple layers, Langlotz said, deep learning can occur. Although the accuracy of deep learning systems is impressive, he noted that neural networks are a form of “black box” learning—the features that neural networks use to make classifications are unknown. He discussed the ImageNet Large Scale Visual Recognition Challenge,40 a yearly contest that evaluates algorithms for object detection and image classification. He said that in 2015, a Microsoft deep learning system achieved an error rate of less than 5 percent, which exceeds human-level performance (He et al., 2015). “These are very powerful techniques,” Langlotz stressed.
Langlotz and his colleagues are using deep learning systems to analyze imaging data linked to EHRs, genomics, and biobank data through the Medical Image Net repository.41 In this context, the goal of deep learning is to improve clinical decision support tools and provide actionable advice to clinicians. He provided several examples of deep learning in radiology. One study found that a deep-learning neural network model estimating skeletal maturity in pediatric hand radiographs performed with accuracy similar to that of an expert radiologist (Larson et al., 2017). Another deep-learning neural network algorithm was developed to detect pneumonia from a dataset of more than 100,000 chest X-rays; researchers found that the model outperformed radiologists (Rajpurkar et al., 2017).
He said machine learning is also being used to improve image quality (Zaharchuk et al., 2018). This has the potential to reduce the time needed to acquire quality images, and with CT and PET imaging, it could reduce the radiation dose required, Langlotz noted. He added that computer-aided detection and classification, facilitated by machine learning, could help radiologists identify abnormalities when they are evaluating images outside of their specialty.
He noted that none of the labeling techniques used in the development of machine learning algorithms are perfect; however, he said that might not matter for large datasets analyzed by neural networks. Because these systems can process a large number of cases, even weakly labeled images might be equivalent with human-based labeling. “You still need the human label—the reference standard label—to validate the system. But to train the system, you can use these weak labels,” Langlotz pointed out.
However, there are challenges with development and use of deep
___________________
40 See https://aimi.stanford.edu/medical-imagenet (accessed July 17, 2018).
41 See http://langlotzlab.stanford.edu/projects/medical-image-net (accessed May 4, 2018).
learning algorithms, Langlotz noted. “The need for clinical validation is still very important,” he stressed, adding, “These are black boxes. We don’t understand how they’re working so we need to validate them.”
Mia Levy, director of cancer health informatics and strategy at the Vanderbilt-Ingram Cancer Center, stressed the need to ensure patient diversity in the datasets used to develop precision oncology tools: “We need to make sure these publicly available datasets upon which we are training all these algorithms have enough representation of minority populations. We need to make sure we lift everyone up in the process and that we’re not training [algorithms] only against a certain subset of our population,” Levy stressed. Cox agreed and suggested the inclusion of data from safety-net hospitals in training datasets.
Langlotz said that applying machine learning to the medical field will require developing new tools that can process complex, multimodality, and time-based data. He also suggested developing additional methods for automated labeling as well as establishing linkages across various data types, including EHRs, genomics, and imaging. Langlotz noted that deep learning in cancer imaging, integrated with gene expression data, could provide useful input for cancer diagnosis and subsequent care (Korfiatis et al., 2016): “There may be some signal in these images that we’re not detecting with the human eye that may correlate directly with genomic signatures, and that would really change the way we stage cancer.”
In pathology, Cohen noted that a recent study found that some deep-learning algorithms to detect lymph node metastases in women with breast cancer performed better than a panel of 11 pathologists (Bejnordi et al., 2017). Becich added that AI can also highlight areas of interest in a specimen that pathologists should carefully review, so more time is spent evaluating potentially invasive lesions versus those that are more likely to be benign. “This won’t put the pathologist out of business, but will make sure regions of interest are scanned by the pathologist,” Becich stressed.
Becich said the FDA’s recent approval of a whole-slide imaging platform is a watershed moment for computational pathology, which he defined as an approach to diagnosis that combines multiple sources of pathology data and transforms them into clinically actionable knowledge that can facilitate decision support (Louis et al., 2014). The whole-slide imaging platform enables robotic scanning of histopathologic slides for digital distribution and review by pathologists (Hartman et al., 2017) (see Figure 3). Becich added that this platform will enable the compilation of large digital pathology datasets that can be analyzed with AI and facilitate computational
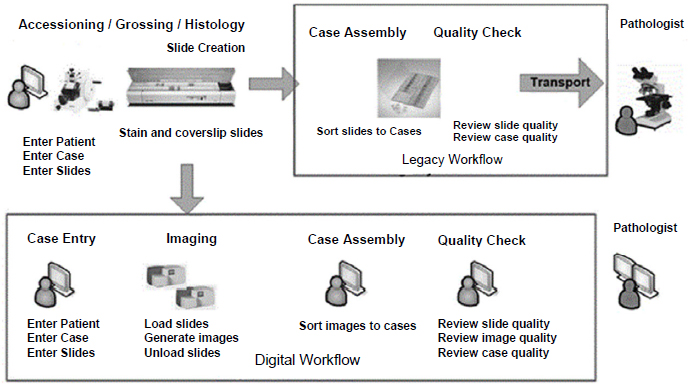
SOURCES: Becich presentation, February 13, 2018; Reprinted by permission from Springer Nature: Journal of Digital Imaging. Enterprise Implementation of digital pathology: Feasibility, challenges, and opportunities, Hartman, D. J., L. Pantanowitz, J. S. McHugh, A. L. Piccoli, M. J. Oleary, and G. R. Lauro. © 2017.
pathology. “It’s a very exciting time in pathology, but it’s even a more exciting time if we get the radiologists and pathologists to work together. We think computational pathology, genomics, and radiology have an awesome partnership that could provide a lot of clinical opportunity to make things better for patients with cancer,” Becich said (see Box 3).
Data Sharing and Standardization
Several workshop participants discussed the importance of data sharing and interoperability to develop and validate machine learning methodologies for cancer diagnosis and care. “No single organization, particularly for low-prevalence conditions, is going to have all the training data needed to develop the kinds of systems that are going to make the maximum impact,” Langlotz said. Becich added, “Data sharing is going to be key in oncology.”
Becich recommended that academic–industry partnerships that develop databases should adhere to FAIR data principles, which stands for
findable, accessible, interoperable, and reusable (Wilkinson et al., 2016).42 He said that bioinformatics specialists and professional societies will be critical for developing large aggregated databases.
Warner noted that for databases to be interoperable they need to share
___________________
42 Examples of FAIR databases that Becich described included Project GENIE, CancerLinQ, Genomic Data Commons, Health Care Systems Research Collaboratory, Oncology Research Information Exchange Network, and National Patient-Centered Clinical Research Network. See http://www.aacr.org/research/research/pages/aacr-project-genie.aspx, https://cancerlinq.org, https://flatiron.com, https://gdc.cancer.gov, http://www.rethinkingclinicaltrials.org, http://oriencancer.org, and http://www.pcornet.org (accessed May 10, 2018).















