2
Guiding Principles for Identifying Species and Subspecies
Determining the taxonomic status of the Mexican gray wolf and the red wolf requires an understanding of the contemporary meanings of “species” and “subspecies” and access to relevant genomic, ecological, and morphological data. The goal of this chapter is to provide background in all of these topics, with a strong focus on those considerations directly relevant to wolf taxonomy. The analytic strategies and standards for assessing the phylogenetic and taxonomic implications of those data are equally important to understand, and they are addressed in Chapter 3.
The chapter begins with an overview of species and subspecies concepts that are especially relevant to canids in North America. The discussion emphasizes how these concepts can be applied in light of the modern understanding that gene flow between species has played a major role in the evolutionary history of many well-accepted mammalian species, including the canids of North America. The chapter then describes a framework for integrating data of multiple types to address core questions in the designation of taxonomic groups. It ends with a brief introduction to some analytic strategies that are useful for understanding genomic data, and it directs the reader to Chapter 3 for a more detailed discussion of those strategies.
WHAT IS A “SPECIES”? WHAT IS A “SUBSPECIES”?
Attempts to classify species are based on the longstanding view that species are real biological entities. Ideas about the best way to define species have changed over time due, in part, to the increasing amounts of data and to the availability of new types of data. Aristotle’s classification scheme focused on morphological similarities and differences, with species—a term that translates from Latin as “kinds”—assumed to be entities static from their inception and distinct from one another. Carl Linnaeus, who established the hierarchical naming system that is still used to classify organisms, was among the first to note hybridization as a possible mechanism for the initiation of new species. In the mid-1800s, Charles Darwin proposed that species arise through gradual change
over time, embracing the ideas that traits can evolve and that new species can form through the divergence of existing ones. This led to the notion that the evolutionary relationships among organisms can be depicted using a branching structure that is today known as a phylogenetic tree.
Darwin’s view of evolutionary relationships among organisms remains the guiding principle for modern taxonomy. Indeed, modern species concepts have universally moved away from the type-specimen approach of early taxonomists, which used an individual specimen to establish the defining characteristics of a taxon, to focus instead on population-level features (Coyne and Orr, 2004; Howard and Berlocher, 1998; Mayr, 1963), especially the range of individual phenotypes. Modern concepts pay particular attention to the delineation of independently evolving lineages (Hey, 2001; Mace, 2004).
Ecological, morphological, and behavioral data can all be of use in identifying lineages. But challenges can arise when data of different types suggest different conclusions. For example, organisms that have similar ecological roles are more likely to encounter and mate with one another, yet maintain distinct genetic groups. Organisms that are closely related tend to have similar morphology and behavior. However, ecological and phenotypic similarities do not necessarily indicate a close genetic relationship because the traits in question could have been strongly influenced by similar environmental factors.
Modern genetic data and analytical tools can help to address these challenges. DNA sequence data complement ecological, morphological, and behavioral data by providing information on the genetic makeup of individuals and populations and making it possible to draw inferences about their genetic relatedness and history. Key insights on population history can emerge through an analysis of genomic data from ancient samples collected using paleontological or archaeological approaches combined with genomic data from modern populations that inhabit the same locations. Assembling data from both modern and ancient individuals can be challenging, however. For North American canids, for example, while multiple whole-genome sequences are available from a large set of modern individuals, data from older samples are comparatively sparse.
The challenge for modern taxonomy is to use available data of diverse types to assess the taxonomic status of various groups of organisms under modern species concepts. Although these concepts differ in the emphasis they place on various factors, they all seek to identify groups of organisms whose reproductive compatibility sustains genetic continuity, and they can all be evaluated using a combination of ecological, phenotypic, and genomic data, as described in detail later in the chapter. This section focuses on three widely used species concepts that are specifically relevant to the systematics of canids.
The Biological Species Concept
Ernst Mayr (1942) first defined the biological species concept, which sees species as groups of potentially or actually interbreeding individuals that are reproductively isolated from other such groups. The vast majority of biologists now agree on the value of this basic approach, but they also appreciate the complexities involved in this approach in light of emerging evidence that hybridization is a key part of the history of many taxonomically recognized species.
Reproductive isolation can occur in two different ways. First, pre-mating isolation refers to a situation in which individuals from the separate populations never mate with one another due, for example, to geographic barriers or to ecological or behavioral differences. Such isolation will set two populations on independent evolutionary trajectories. Second, post-mating isolation refers to the inability of individuals from two different species to produce fertile descendants from a mating. This inability can be due to a number of reasons, including that the eggs are not fertilized, that any fertilized egg is not viable, or that descendants are viable but sterile. In such a situation, contemporary biologists would agree that the two populations potentially belong to different species. It is not
always practical to test for the presence of these post-mating isolating mechanisms since assessing hybrid viability and fertility requires direct observation in the wild or in a controlled setting.
Even if two populations have some levels of pre- and post-mating isolation, scientists have found that gene flow can, on occasion, occur between those populations. In other words, even when two populations have some degree of isolation, hybridization occasionally occurs. This indicates that absolute reproductive isolation is likely an overly stringent criterion for designating species. In any attempt to classify North American canids, which are both evolutionarily young and geographically overlapping (due in part to human-induced habitat alterations), it is important to keep in mind that hybridization is a common feature in the history of many well-accepted species—and thus may appear between separate canid species as well. Hybridization and its relevance to modern species concepts will be discussed in more detail later in this chapter.
The Phylogenetic Species Concept
The phylogenetic species concept is largely compatible with the biological species concept, but it has an added emphasis on shared ancestry—that is, individuals are inferred to belong to the same species if they are descended from a common ancestor and share lineage-specific mutations. As a natural consequence of the relentless processes of mutation and genetic drift (i.e., the random changes in the frequency of alleles due to chance), reproductively isolated species diverge at the DNA level over time, with or without changes in their morphology or behavior. The divergence of groups is a natural consequence of reduced gene flow between them, and is sometimes hastened by natural selection favoring different mutations in divergent lineages. At the same time, natural selection can also preserve similar phenotypes in divergent lineages, even though the underlying DNA sequences are changing. This can lead to cryptic species that are morphologically similar but so divergent genetically that they are reproductively incompatible and thus have been set on different evolutionary trajectories.
The Chronospecies Concept
Changes can accumulate across generations of a given lineage, making modern members of a given species highly distinct from their predecessors, despite genetic continuity among generations (Stanley, 1978). Thus, taxonomists have the concept of chronospecies, which is a group of organisms at one point in time that evolved as a single lineage into a later group of organisms that had changed enough over time to be considered a separate species. The determination of a chronospecies is generally based on time-series documentation from morphological data alone. However, a chronospecies can be challenging to define because historical samples are often limited, and it is challenging or impossible to assess how strongly organismal phenotypes are shaped by contemporary environmental factors. As noted below, DNA extracted from paleontological specimens can offer unprecedented opportunities to assess genetic cohesion between past and present generations. In the case of North American canids, while DNA from historical individuals is not always available, some key specimens and data have been obtained from the skeletal remains of a few individuals.
In summary, new species are constantly arising, and existing ones are constantly changing (Hey, 2001; Mace, 2004). While no species concept is ideal, the principles underlying the biological species concept, the phylogenetic species concept, and the chronospecies concept—some level of reproductive isolation mediated by genetic and ecological factors, and phylogenetic continuity mediated by the processes of mutation, drift, selection, and inheritance—provide a compelling, comprehensive platform for identifying species. By making observations at the genomic, ecological, and behavioral levels, it is possible to transcend the limitations of the individual species concepts and
provide a strong framework for evaluating the taxonomic status of groups of North American canids within the context of all three modern concepts. This framework is outlined later in this chapter.
The Subspecies Designation
Some classification schemes also recognize taxonomic groups below the species level. The earliest definitions of subspecies distinguished sets of populations whose members share pattern, color, or morphological attributes not found in other, geographically separated populations of the same species. Most modern concepts (Haig et al., 2006), including one suggested by Darwin, rely on the notion of a partial restriction of gene flow. Virtually all modern definitions of subspecies follow the spirit of these original definitions, with the general view being that subspecies are groups of actually or potentially interbreeding populations that are phylogenetically distinguishable from, but reproductively compatible with, other such groups (Mayr, 1953, 1963; Mayr and Ashlock, 1991).
“Species” as Defined Under the Endangered Species Act
The U.S. Endangered Species Act (ESA) defines a species—for the purposes of the act—as not only a species, but also “any subspecies of fish or wildlife or plants, and any distinct population segment of any species or vertebrate fish or wildlife which interbreeds when mature.” In other words, if the survival of a subspecies that occupies a distinct geographic area is in jeopardy, then the ESA allows that subspecies to be treated as a “species” for the purposes of restoration and recovery. The ESA thus provides the opportunity for legal protection of subspecies as well as “distinct population segments” (DPSs) of vertebrates as if they were recognized taxonomic species. DPSs have been defined under official policy (U.S. FWS and NMFS, 1996) on the basis of two characteristics: (1) the discreteness of the population segment in relation to the remainder of the species to which it belongs; and (2) the significance of the population segment to the species to which it belongs. Thus, the ESA uses the word “species” in a broader sense than the usual biology-based definitions discussed above and throughout the remainder of this chapter, providing for stability of conservation policy even in the face of ever changing taxonomic designations and shifts in the species concept itself.
UNDERSTANDING “SPECIES” IN LIGHT OF HYBRIDIZATION
As noted above, all contemporary species concepts are based on the idea of different species being independently evolving lineages. This concept is relatively easy to apply if gene flow is nonexistent or at least extremely rare among groups of organisms that are considered to belong to distinct species. Increasingly, though, genomic data have revealed that the cross-species exchange of genetic material through hybridization is a core feature of the evolutionary history of many widely accepted species (Arnold, 2006; Grant and Grant, 1992; Mallet, 2005), raising challenges for the strict application of existing species concepts. This section has two primary objectives: (1) to provide an overview of the hybridization process and examples of its outcomes and (2) to describe how species concepts are understood and applied in light of emerging data on hybridization.
Several terms are used to describe the processes and outcomes of gene flow across taxonomic groups. In the contemporary literature, “hybridization,” “introgression,” and “admixture” are sometimes used interchangeably. In this report, hybridization refers to the mating of two individuals from different species to yield offspring that have ancestry from both parents (Abbott et al., 2013) (Figure 2-1).
Interspecific mating does not always result in sustained hybridization. In some cases, when individuals from different species mate they are unable to produce offspring, or their offspring are
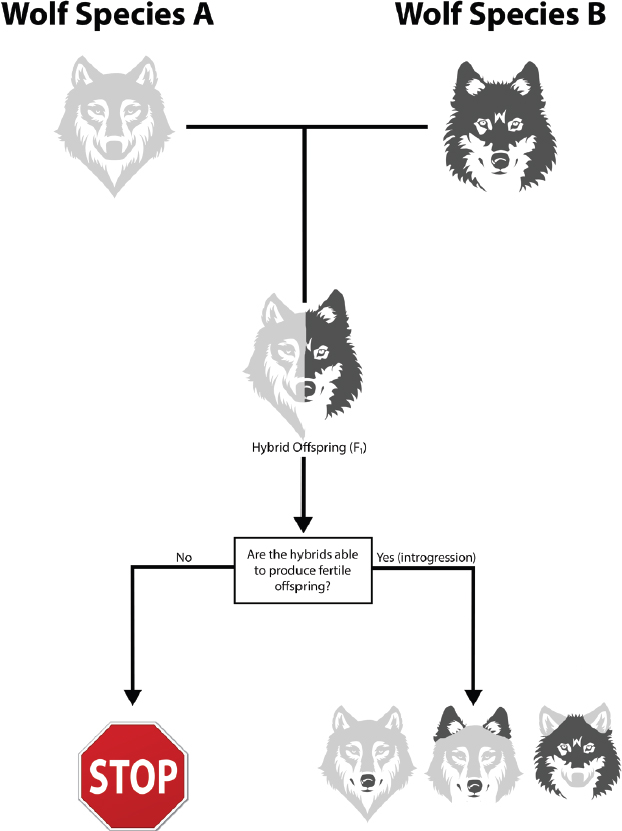
infertile. In other cases, the offspring of a first generation (F1) hybrid and a member of the parental species or other hybrids are infertile. Reduced hybrid viability or reduced hybrid fertility imposes a strict barrier to gene flow between the two hybridizing species. It can also drive the evolution of pre-mating isolation because of the higher reproductive success of individuals that mate exclusively with members of their own species.
If hybrid individuals are able to produce fertile offspring, this can lead to the development of an admixed population. The admixed individuals can have a wide range of genetic contributions from each of the two “parental” species. A number of long-term outcomes are possible. The survival and mating of hybrid individuals with one of the parental species can result in introgression, the movement of specific regions of the genome of one parental species into the homologous chromosomes of some individuals of the other species. If these chromosome regions contain differences between species (i.e., different alleles), the presence of the new regions may or may not affect the fitness of the individuals who possess them. These differences will remain in the population if they provide advantage or are not detrimental to the individuals who carry them. Chromosome regions that carry alleles that are extremely detrimental (e.g., if the hybrids that have those alleles die or cannot reproduce) will be removed by natural selection. Chromosomal regions conferring alleles that only moderately reduce fitness can linger for a long time or even become fixed, especially in small populations, where natural selection is weak as compared to genetic drift.
Hybridization produces specific molecular patterns. The DNA in the nucleus of each cell is divided into a number of separate chromosomes. In mammals, including wolves, each cell contains two copies of each chromosome. One chromosome within each of these pairs is inherited from the mother, the other from the father. A hybrid individual, then, would have one chromosome in each pair from parental species 1, the other from parental species 2. During the production of eggs and sperm, the two chromosomes within a pair trade segments through genetic recombination, producing new chromosome copies composed of new combinations of the DNA from the original chromosomes. If hybrids are fertile, their offspring therefore inherit individual chromosomes that contain segments from both parental species. With each successive generation, each segment is broken into smaller and smaller pieces, producing increasingly complex mosaics of DNA from the two parental species.
Admixture
Admixture, the formation of novel genetic combinations through hybridization of genetically distinct groups, is now recognized as a core component of the history of many taxonomically recognized species. For example, genomic analyses indicate that modern Europeans and Asians inherited a fraction of their genome from Neanderthals and Denisovans. On average, 2–3 percent of the genomes of humans originating from areas outside sub-Saharan Africa consists of DNA that introgressed from Neanderthals (Green et al., 2010), while Melanesians and aboriginal Australians trace 3–4 percent of their DNA to introgression with Denisovans (Reich et al., 2010). These findings, coupled with information on the distribution of the sizes of the introgressed regions that remained in the genome of modern humans, have led to the inference that the hybridization of modern humans with Neanderthals and Denisovans occurred some tens of thousands of years ago (Green et al., 2010).
Indeed, hybridization is known to have occurred in all major lineages of non-human primates (Tung and Barreiro, 2017; Zinner et al., 2011), including mangabeys (Rungucebus) (Zinner et al., 2009b), baboons (Papio) (Zinner et al., 2009a), guenons (Guschanski et al., 2013), macaques (Fan et al., 2014; Guschanski et al., 2013; Tosi et al., 2003), langurs (Ting et al., 2008), howler monkeys (Cortés-Ortiz et al., 2007; Mourthe et al., 2018), marmosets (Malukiewicz et al., 2015), and lemurs (Wyner et al., 2002).
Introgression is possible even across highly diverged lineages. Perhaps the most extreme example of introgression in mammals involves pigs (Sus scrofa), which show evidence of gene flow from an extinct species outside the Sus genus that had diverged from pigs an estimated 8.5 million years ago (Ai et al., 2015). The divergent fragment of introgressed DNA harbors genetic variation associated with climate adaptation and may be an example of adaptive introgression among species that have been diverging for millions of years.
In summary, genome sequencing has revealed evidence that introgression between pheno-typically and genotypically divergent species has occurred in many mammalian lineages. As such, complete genetic separation and a complete absence of admixture appears no longer to be a strict criterion for defining species.
Hybridization
Hybridization is possible when populations that have been geographically isolated for some time regain some level of contact. Contact can arise through either natural processes, such as a change in the course of rivers, or through anthropogenic effects, such as the intentional or unintentional transport of animals. For example, the pet trade in Brazil has led some species of marmosets (genus Callithrix) to be introduced into urban areas that have been historically inhabited only by other marmoset species (Malukiewicz et al., 2015). These introductions have resulted in hybridization among several species of marmosets that were adapted to different ecological and social environments.
The Implications of Hybridization
Hybridization can have diverse implications for the conservation (Box 2-1) and evolutionary trajectories of the species involved. Each of the outcomes described below is potentially relevant to the evolutionary history of canids in North America.
Continued Strict Genetic Separation of Species
Species may remain genetically distinct following hybridization. This outcome is most likely if the two species involved harbor genetic incompatibilities that reduce the fitness or fertility of hybrid individuals. When the expression of such incompatibilities is extreme, it can result in the
complete inviability or sterility of hybrids. Under such a scenario, the alleles from one species never enter the gene pool of the other. Even in the absence of complete sterility, though, hybrids can be unfit in ways that tend to limit introgression. For example, admixed individuals two or three generations subsequent to the initial hybridization event sometimes suffer reduced fitness (Lynch and Walsh, 1998). In such cases, features may evolve that reduce or prevent the occurrence of interspecific matings. These features might be genetic, behavioral, ecological, or morphological; one example would be the call of a frog that allows an individual to identify potential mates in that species (Butlin, 1987; Butlin and Smadja, 2018). Such features can limit introgression and, over the course of many generations, potentially eliminate introgressed genes, leaving negligible evidence of hybridization in the genomes of the species involved.
Partial Genetic Separation of Species with Introgression of Some Genetic Elements
Species may remain distinct following hybridization but still retain the capacity for occasional exchange of genetic material. In such cases individual genomes are likely to be mosaics, with individuals of one species having segments of DNA that are derived from another (e.g., Ellegren et al., 2012). The genetic elements exchanged may in some cases be adaptive, and the exchange of genetic material may allow the recipient species to adapt quickly to new environmental conditions through shared genetic variation.
Indeed, there is evidence for this adaptive introgression in several different species (e.g., Whitney et al., 2006), including modern humans. For example, genomic regions that introgressed from Neanderthals and Denisovans into modern human populations include genes associated with immune defense functions (Quach et al., 2016) and with human survival in high-altitude environments (Huerta-Sánchez et al., 2014). Similar examples abound in other lineages. Wu et al. (2018) inferred introgression among yak (Bos grunniens), domesticated cattle (Bos taurus), the wisent (Bison bonasus), the gayal (Bos frontalis), and the banteng (Bos javanicus). The introgressed regions contain genes that regulate red blood cell production and high-altitude adaptation in both the yak and Tibetan cattle (Wu et al., 2018). These examples underscore the capacity of hybridization to introduce beneficial variation without undermining the genomic distinctness of the species involved.
Formation of Stable Hybrid Zones
It is possible for stable hybrid zones to form when species with ecological adaptations to different environments inhabit neighboring or overlapping ranges or when two formerly geographically separated species meet as a result of a range expansion of one or both of them. In the first case, ecological selection may counteract the effect of hybridization and maintain distinct species. Even so, varying degrees of introgression can occur in the area where the species meet. If the selection is sufficiently strong, separate species can be maintained despite gene flow. However, at the genomic level the species will eventually become genetically similar—that is, will come to have nearly identical collections of genes—except for the regions harboring genes associated with local adaptations (see, e.g., Barton and Hewitt, 1981). Such species might be functionally distinct, but they end up being genetically almost identical in some genomic regions. When species experience secondary contact, stable hybrid zones (tension zones, cf. Barton and Hewitt, 1981) can also be maintained by a balance between selection against hybrids and the dispersal of parental species into the area of contact. In such cases, neutral introgression is expected, and adaptive introgression is possible, but genomic regions that are associated with reproductive isolation will help reduce the production of hybrids, thereby maintaining species distinctness even in the face of gene flow.
Formation of Hybrid Species, Sometimes with Loss of Parental Species
If hybridization is common and there are no strong ecological selection or genetic incompatibilities that prevent introgression, an entirely new hybrid species can emerge (e.g., Mallet, 2007). This type of speciation by hybridization may result in a third species that replaces the two parental species or that coexists with them. Many plant species (e.g., wild sunflowers, cf. Ungerer et al., 1998) and some animal species (e.g., Heliconius butterflies, see Mavárez et al., 2006; Ungerer et al., 1998; virgin chub, DeMarais et al., 1992; and Caribbean bats, DeMarais et al., 1992; Larsen et al., 2010) have originated from hybridization.
ESTABLISHING GUIDELINES FOR DETERMINING TAXONOMIC STATUS
The increasing evidence of gene flow among taxonomic groups conflicts with earlier views that strict reproductive isolation is a defining feature of taxonomically valid species. As noted above, all modern species concepts focus on whether a given group of organisms constitutes a distinct, independently evolving lineage that merits recognition as a taxonomically valid species. Combining several approaches is often essential. Morphological and paleontological analyses can be informative in assessing the continuity of organismal phenotypes over time and geographical areas, but they are often limited by the availability of fossils, which may be few in number, in poor condition, or from a limited geographic range. Genetic and genomic data can provide an enormous number of quantifiable characters, but they are not always available for specimens of diverse ages. Behavioral traits and ecological roles have long been recognized as important in the designation of taxa, especially below the species level (Crandall et al., 2000; Haig et al., 2006), as they explicitly address differences in adaptive characters, but they are not readily useful for assessing continuity between historical and contemporary populations. Support for the validity of a taxonomic designation of species and subspecies relies upon the strength of morphological, genetic, and ecological evidence, individually and collectively.
A FRAMEWORK FOR ESTABLISHING TAXONOMIC DESIGNATIONS
As noted above, although modern species concepts do have some differences, they are united by the goal of identifying groups of organisms whose reproductive compatibility sustains genetic continuity. The levels of genetic differentiation do not need to be the sole—or even the primary—evidence considered. It is tempting to prioritize molecular data, as they are readily quantifiable and can be used to define phylogenetic and past population relationships. However, exclusively focusing on molecular characters risks over-splitting taxa or, conversely, failing to recognize as distinct those that have diverged recently and now occupy different ecological niches but have not yet had time to accrue substantial genetic differentiation (Coates et al., 2018; Haig et al., 2006).
Fossils, when available, can provide useful data on morphology of ancestral populations. Notably though, the current fossil record represents less than 5 percent of the known living species (Prothero, 2007). The formation and preservation of fossils is highly dependent on local conditions including moisture, rapid burial, and vegetation cover. Fossils from organisms that inhabit open environments therefore far outnumber those from organisms that live in tropical forests, as organic remains degrade quickly in the forest environment (Behrensmeyer, 1978; Tappen, 1994). Therefore, while fossil evidence can be useful for assessing the morphologic distinctness of a given lineage, absence from the fossil record may reflect conditions unfavorable for fossil formation, rather than the true absence of a given lineage from a given area at a given time.
Combining genomic data with morphological and ecological data can provide a more complete picture of the taxonomy and evolutionary history of species and subspecies.
Strong support for the validity of a taxonomic designation of a species may include the following:
- Morphological, paleontological, or fossil evidence that the taxon under consideration is evolving independently or morphological evidence that specimens possess a phenotype that is distinct from other defined species.
- Evidence of genetic or genomic distinctiveness, based on data from several independently segregating genetic loci, with evidence that extant subpopulations are connected by gene flow. If hybridization with other defined species is also detected, it must be clear that introgression does not substantially affect the discreteness of the taxon under consideration.
- Ecological or behavioral data indicating adaptive differences and reproductive incompatibilities separating the taxon under consideration from other closely related species. Relevant differences may be ecological, behavioral, or physiological. Genomic data, if available, may reveal genetic variation underlying these differences but are not required to confirm the importance of ecological or behavioral differences.
Strong support for the validity of a taxonomic designation of subspecies may include the following:
- Morphological, paleontological, or fossil evidence of a geographically and historically isolated lineage within the species to which it belongs. Morphological evidence may be especially useful for identifying distinct, locally adapted phenotypes that evolved during isolation from other lineages of the same species.
- Genetic or genomic evidence of distinctness based on data from multiple independently inherited genetic loci, with no evidence of reproductive isolation from other populations of the same species in regions of range overlap. Phylogeographic analyses may be especially useful in assessing the distinctness of the lineage under consideration from other populations belonging to the same lineage.
- Ecological, behavioral, or physiological characters that provide evidence of adaptive differences between the lineage and other groups belonging to the same species. Available genetic or genomic data may reveal the presence of alleles, or of differences in allele frequencies, at loci that underlie these differences.
These criteria provide a framework for assessing diverse data types useful for evaluating the taxonomic status of the Mexican gray wolf (Chapter 4) and the red wolf (Chapter 5). By way of background, Chapter 3 first provides an overview of strategies for the analysis of relevant genetic and genomic data.
REFERENCES
Abbott, R., D. Albach, S. Ansell, J. W. Arntzen, S. J. E. Baird, N. Bierne, J. Boughman, A. Brelsford, C. A. Buerkle, R. Buggs, R. K. Butlin, U. Dieckmann, F. Eroukhmanoff, A. Grill, S. H. Cahan, J. S. Hermansen, G. Hewitt, A. G. Hudson, C. Jiggins, J. Jones, B. Keller, T. Marczewski, J. Mallet, P. Martinez-Rodriguez, M. Möst, S. Mullen, R. Nichols, A. W. Nolte, C. Parisod, K. Pfennig, A. M. Rice, M. G. Ritchie, B. Seifert, C. M. Smadja, R. Stelkens, J. M. Szymura, R. Väinölä, J. B. W. Wolf, and D. Zinner. 2013. Hybridization and speciation. Journal of Evolutionary Biology 26(2):229-246.
Ai, H., X. Fang, B. Yang, Z. Huang, H. Chen, L. Mao, F. Zhang, L. Zhang, L. Cui, W. He, J. Yang, X. Yao, L. Zhou, L. Han, J. Li, S. Sun, X. Xie, B. Lai, Y. Su, Y. Lu, H. Yang, T. Huang, W. Deng, R. Nielsen, J. Ren, and L. Huang. 2015. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nature Genetics 47(3):217-225.
Arnold, M. L. 2006. Evolution through Genetic Exchange. Oxford, UK: Oxford University Press.
Barton, N. H., and G. M. Hewitt. 1981. A chromosomal cline in the grasshopper Podisma pedestris. Evolution 35(5):1008-1018.
Behrensmeyer, A. K. 1978. Taphonomic and ecologic information from bone weathering. Paleobiology 4:150-162.
Butlin, R. 1987. Speciation by reinforcement. Trends in Ecology & Evolution 2(1):8-13.
Butlin, R. K., and C. M. Smadja. 2018. Coupling, reinforcement, and speciation. The American Naturalist 191(2):155-172.
Coates, D. J., M. Byrne, and C. Moritz. 2018. Genetic diversity and conservation units: Dealing with the species–population continuum in the age of genomics. Frontiers in Ecology and Evolution 6:165.
Cortés-Ortiz, L., T. F. Duda, D. Canales-Espinosa, F. García-Orduna, E. Rodríguez-Luna, E. Bermingham. 2007. Hybridization in large-bodied New World primates. Genetics 176:2421-2425.
Coyne, J. A., and H. A. Orr. 2004. Speciation. New York: Oxford University Press.
Crandall, K. A., O. R. P. Bininda-Emonds, G. M. Mace, and R. K. Wayne. 2000. Considering evolutionary processes in conservation biology. Trends in Ecology & Evolution 15(7):290-295.
DeMarais, B. D., T. E. Dowling, M. E. Douglas, W. L. Minckley, and P. C. Marsh. 1992. Origin of Gila seminuda (Teleostei: Cyprinidae) through introgressive hybridization: Implications for evolution and conservation. Proceedings of the National Academy of Sciences 89(7):2747-2751.
Ellegren, H., L. Smeds, R. Burri, P. I. Olason, N. Backström, T. Kawakami, A. Künstner, H. Mäkinen, K. Nadachowska-Brzyska, A. Qvarnström, S. Uebbing, and J. B. Wolf. 2012. The genomic landscape of species divergence in Ficedula flycatchers. Nature 491(7426):756-760.
Fan, Z., G. Zhao, P. Li, N. Osada, J. Xing, Y. Yi, L. Du, P. Silva, H. Wang, R. Sakate, X. Zhang, H. Xu, B. Yue, and J. Li. 2014. Whole-genome sequencing of tibetan macaque (Macaca thibetana) provides new insight into the macaque evolutionary history. Molecular Biology and Evolution 31:1475-1489.
Grant, P. R., and B. R. Grant. 1992. Hybridization of bird species. Science 256(5054):193-197.
Green, R. E., J. Krause, A. W. Briggs, T. Maricic, U. Stenzel, M. Kircher, N. Patterson, H. Li, W. Zhai, M. H.-Y. Fritz, N. F. Hansen, E. Y. Durand, A.-S. Malaspinas, J. D. Jensen, T. Marques-Bonet, C. Alkan, K. Prüfer, M. Meyer, Hernán A. Burbano, J. M. Good, R. Schultz, A. Aximu-Petri, A. Butthof, B. Höber, B. Höffner, M. Siegemund, A. Weihmann, C. Nusbaum, E. S. Lander, C. Russ, N. Novod, J. Affourtit, M. Egholm, C. Verna, P. Rudan, D. Brajkovic, Ž. Kucan, I. Gušic, V. B. Doronichev, L. V. Golovanova, C. Lalueza-Fox, M. de la Rasilla, J. Fortea, A. Rosas, R. W. Schmitz, P. L. F. Johnson, E. E. Eichler, D. Falush, E. Birney, J. C. Mullikin, M. Slatkin, R. Nielsen, J. Kelso, M. Lachmann, D. Reich, and S. Pääbo. 2010. A draft sequence of the Neandertal genome. Science 328(5979):710-722.
Guschanski, K., J. Krause, S. Sawyer, L. M. Valente, S. Bailey, K. Finstermeier, R. Sabin, E. Gilissen, G. Sonet, Z. T. Nagy, G. Lenglet, F. Mayer, and V. Savolainen. 2013. Next-generation museomics disentangles one of the largest primate radiations. Systematic Biology 62(4):539-554.
Haig, S. M., E. A. Beever, S. M. Chambers, H. M. Draheim, B. D. Dugger, S. Dunham, E. Elliott-Smith, J. B. Fontaine, D. C. Kesler, B. J. Knaus, I. F. Lopes, P. Loschl, T. D. Mullins, and L. M. Sheffield. 2006. Taxonomic considerations in listing subspecies under the U.S. Endangered Species Act. Conservation Biology 20(6):1584-1594.
Hey, J. 2001. Genes, Categories, and Species: The Evolutionary and Ccognitive Cause of the Species Problem. New York: Oxford University Press.
Howard, D. J., and S. H. Berlocher. 1998. Endless Forms: Species and Speciation. New York: Oxford University Press.
Huerta-Sánchez, E., X. Jin, Asan, Z. Bianba, B. M. Peter, N. Vinckenbosch, Y. Liang, X. Yi, M. He, M. Somel, P. Ni, B. Wang, X. Ou, J. Huasang, Z. Luosang, X. Cuo, K. Li, G. Gao, Y. Yin, W. Wang, X. Zhang, X. Xu, H. Yang, Y. Li, J. Wang, J. Wang, and R. Nielsen. 2014. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512(7513):194-197.
Larsen, P. A., M. R. Marchán-Rivadeneira, and R. J. Baker. 2010. Natural hybridization generates mammalian lineage with species characteristics. Proceedings of the National Academy of Sciences 107(25):11447-11452.
Lynch, M., and B. Walsh. 1998. Genetics and Analysis of Quantitative Traits. New York: Oxford University Press.
Mace, G. M. 2004. The role of taxonomy in species conservation. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 359(1444):711-719.
Mallet, J. 2005. Hybridization as an invasion of the genome. Trends in Ecology & Evolution 20(5):229-237.
Mallet, J. 2007. Hybrid speciation. Nature 446(7133):279-283.
Malukiewicz, J., V. Boere, L. F. Fuzessy, A. D. Grativol, I. de Oliveira e Silva, L. C. M. Pereira, C. R. Ruiz-Miranda, Y. M. Valença, and A. C. Stone. 2015. Natural and anthropogenic hybridization in two species of eastern Brazilian marmosets (Callithrix jacchus and C. penicillata). PLOS One 10(6):e0127268.
Mavárez, J., C. A. Salazar, E. Bermingham, C. Salcedo, C. D. Jiggins, and M. Linares. 2006. Speciation by hybridization in Heliconius butterflies. Nature 441(7095):868-871.
Mayr, E. 1942. Systematics and the Origin of Species, from the Viewpoint of a Zoologist. Cambridge, MA: Harvard University Press.
Mayr, E. 1953. Methods and Principles of Systematic Zoology. Cambridge, MA: Harvard University Press.
Mayr, E. 1963. Animal Species and Evolution. Cambridge, MA: Harvard University Press.
Mayr, E., and P. D. Ashlock. 1991. Principles of Systematic Zoology, 2nd ed. New York: McGraw-Hill.
Mourthe, I., R. A. Trindade, M. Aguiar, T. C. Trigo, J. C. Bicca-Marques, and S. L. Bonatto. 2018. Hybridization between Neotropical primates with contrasting sexual dichromatism. International Journal of Primatology. https://doi.org/10.1007/s10764-017-0011-9.285.
Muhlfeld, C. C., R. P. Kovach, R. Al-Chokhachy, S. J. Amish, J. Kershner, R. F. Leary, W. H. Lowe, G. Luikart, P. Matson, B. Shepard, P. Westley, D. Whited, A. Whiteley, and F. W. Allendorf. 2017. Legacy introductions and climatic variation explain spatiotemporal patterns of invasive hybridization in a native trout. Global Change Biology 23:4663-4674.
Prothero, D. B. 2007. Evolution: What the Fossils Say and Why It Matters. New York: Columbia University Press.
Quach, H., M. Rotival, J. Pothlichet, Y.-H. E. Loh, M. Dannemann, N. Zidane, G. Laval, E. Patin, C. Harmant, M. Lopez, M. Deschamps, N. Naffakh, D. Duffy, A. Coen, G. Leroux-Roels, F. Clément, A. Boland, J. F. Deleuze, J. Kelso, M. L. Albert, and L. Quintana-Murci. 2016. Genetic adaptation and Neandertal admixture shaped the immune system of human populations. Cell 167(3):643-656.
Reich, D., R. E. Green, M. Kircher, J. Krause, N. Patterson, E. Y. Durand, B. Viola, A. W. Briggs, U. Stenzel, P. L. F. Johnson, T. Maricic, J. M. Good, T. Marques-Bonet, C. Alkan, Q. Fu, S. Mallick, H. Li, M. Meyer, E. E. Eichler, M. Stoneking, M. Richards, S. Talamo, M. V. Shunkov, A. P. Derevianko, J.-J. Hublin, J. Kelso, M. Slatkin, and S. Pääbo. 2010. Genetic history of an archaic hominin group from Denisova cave in Siberia. Nature 468(7327):1053-1060.
Stanley, S. M. 1978. Chronospecies’ longevities, the origin of genera, and the punctuational model of evolution. Paleobiology 4(01):26-40.
Tappen, M. 1994. Bone weathering in the tropical rain forest. Journal of Archaeological Science 21:667-673.
Ting, N., A. J. Tosi, Y. Li, Y.-P. Zhang, and T. R. Disotell. 2008. Phylogenetic incongruence between nuclear and mitochondrial markers in the Asian colobines and the evolution of the langurs and leaf monkeys. Molecular Phylogenetics and Evolution 46(2):466-474.
Tosi, A. J., J. C. Morales, and D. J. Melnick. 2003. Paternal, maternal, and biparental molecular markers provide unique windows onto the evolutionary history of macaque monkeys. Evolution 57(6):1419-1435.
Tung, J., and L. B. Barreiro. 2017. The contribution of admixture to primate evolution. Current Opinion in Genetics & Development 47:61-68.
Ungerer, M. C., S. J. E. Baird, J. Pan, and L. H. Rieseberg. 1998. Rapid hybrid speciation in wild sunflowers. Proceedings of the National Academy of Sciences 95(20):11757-11762.
U.S. FWS (U.S. Fish and Wildlife Service) and NMFS (National Marine Fisheries Service). 1996. Policy regarding the recognition of distinct vertebrate population segments under the Endangered Species Act. Federal Register [February 7, 1996] 61(26):4721-4725.
Whitney, K. D., R. A. Randell, and L. H. Rieseberg. 2006. Adaptive introgression of herbivore resistance traits in the weedy sunflower Helianthus annuus. The American Naturalist 167(6):794-807.
Wu, D.-D., X. -D. Ding, S. Wang, J. M. Wójcik, Y. Zhang, M. Tokarska, Y. Li, M.-S. Wang, O. Faruque, R. Nielsen, Q. Zhang, and Y.-P. Zhang. 2018. Pervasive introgression facilitated domestication and adaptation in the Bos species complex. Nature Ecology & Evolution 2(7):1139-1145.
Wyner, Y. M., S. E. Johnson, R. M. Stumpf, and R. Desalle. 2002. Genetic assessment of a white-collared x redfronted lemur hybrid zone at Andringitra, Madagascar. American Journal of Primatology 67:51-66.
Zinner, D., M. L. Arnold, and C. Roos. 2009a. Is the new primate genus Rungwecebus a baboon? PLOS One 4(3):e4859.
Zinner, D., L. F. Groeneveld, C. Keller, and C. Roos. 2009b. Mitochondrial phylogeography of baboons (Papio spp.): Indication for introgressive hybridization? BMC Evolutionary Biology 9:83.
Zinner, D., M. L. Arnold, and C. Roos. 2011. The strange blood: Natural hybridization in primates. Evolutionary Anthropology 20(3):96-103.












