5
Is the Red Wolf a Valid Taxonomic Species?
The red wolf is a currently recognized species that historically inhabited much of the eastern United States. During the 20th century, populations were driven to very low numbers by predator eradication programs and habitat loss and were largely replaced by coyotes spreading eastwards from their original range in the western United States. A few remaining specimens from Texas and Louisiana with apparent red wolf morphology were captured before the red wolf was declared to be extinct in the wild and were used to establish a breeding program. The descendants of this breeding program were reintroduced in North Carolina and are now a managed population in the wild. There has been substantial controversy regarding the species status of the red wolf. In particular, the individuals that were used to found the breeding program were captured from a region where there had already been substantial admixture between eastward-expanding coyotes and local gray wolves and red wolves. Some of the earliest genetic studies of red wolves found that historical specimens had mitochondrial genes that were similar to coyotes, suggesting the possibility of admixture. As a result, substantial research has focused on examining whether and to what extent wolves and coyotes in the eastern United States are genetically distinct. The controversy currently stands on two main issues: (1) the degree to which red wolves historically were a separate species or could be better categorized as either coyotes or gray wolves, and (2) the degree to which the extant (captive and managed) population descends from the original red wolf population or instead is the result of admixture between coyotes and gray wolves.
A BRIEF TAXONOMIC HISTORY OF THE RED WOLF
Audubon and Bachman (1851) described the red wolf (Canis lupus rufus) as a subspecies of gray wolf having long legs and slender proportions and inhabiting a south-central Texas range. In 1937 Goldman elevated the taxon to species status (C. rufus) with three subspecies, distinguishing individual red wolves from gray wolves based on cranial and dental characters. During his study, C. r. gregoryi was found in the lower Mississippi River basin of southeastern Missouri, Arkansas, southeastern Oklahoma, eastern Texas, and Louisiana. Goldman described two additional subspecies, both of which were likely extinct: C. r. rufus, which had been found in Texas, and
C. r. floridanus, which had been found in Florida. Later analyses by Lawrence and Bossert (1967) recognized the red wolf only as a subspecies of gray wolf, but Nowak (1979, 2002) supported the designation of the red wolf as a distinct species. However, the designation of the red wolf as a distinct species remains controversial.
In order to assess whether the red wolf is a valid taxonomic species, there are three questions that need to be answered: (1) Is there evidence that the historical population of red wolves was a distinct lineage?, (2) Is there evidence for the distinctiveness of contemporary red wolf populations from gray wolves and coyotes?, and (3) Is there evidence for continuity between the historical red wolf population and the present managed populations?
IS THERE EVIDENCE THAT THE HISTORICAL POPULATION OF RED WOLVES WAS A DISTINCT LINEAGE?
Historical Distribution of Wolves and Coyotes
The earliest of the modern canids arose in North America. It was a medium-sized animal (C. davisi or a species in this lineage) that crossed Beringia into Eurasia late in the Miocene (approximately 8 million years ago), leaving only a likely ancestor of the coyote (C. lepophagus) and an ancestral gray fox (genus Urocyon) in North America. In Asia, canids underwent a major radiation of wolves, jackals, and hunting dogs (Martin, 1989). Near the beginning of the Pleistocene, the descendants of the Asian canids dispersed back to North America, resulting in the major lineages of the gray wolf (C. lupus), the dire wolf (C. dirus), and the red wolf (C. rufus) (Martin, 1989). Analyses of morphological and biogeographical data by Nowak (1983, 1995) and of genetic data by Vilà et al. (1999) suggest that there were as many as three episodes of gray wolf dispersal into North America, likely in response to habitat expansions and contractions during glacial cycles. The earliest North American fossils of C. lupus are from animals who lived approximately 1 million years ago in the Yukon (Tedford et al., 2009). Following the dispersal back into North America, C. lupus colonized habitats from the Arctic to Mexico, and as many as 24 subspecies have been described (Hall, 1981; Hall and Kelson, 1959). Using canonical discriminant analysis, Nowak (1995) reduced the number of C. lupus subspecies to five and recognized the presence of C. rufus, with a smaller skull and relatively narrower dimensions, in eastern North America. The earliest fossils interpreted to come from C. rufus are dated at 10,000 years ago and were found in Florida.
While C. lupus arose in Eurasia, fossil evidence indicates that the presumed ancestor (C. leophagus) of the coyote (C. latrans) arose in North America. The coyote appeared in the fossil record of North America approximately 1 million years ago and quickly expanded across the continent (Tedford et al., 2009). Nowak found evidence of C. latrans in eastern North America during the late Rancholabrean (approximately 11,000 years ago), but fossil evidence of this species disappeared from the east between 10,000 years ago and the early 1900s (Nowak, 2002; Parker, 1995).
Hody and Kays (2018) examined 347 records from FAUNMAP (Graham and Lundelius, 2010) from the Holocene (10,000 years ago until present) and 12,319 records from VertNet,1 which contains data on animals collected since the 1800s. They found that C. latrans specimens that date to up to 10,000 years ago were found west of the Mississippi River and through the arid western regions of the United States. Although two specimens were recorded for eastern North America (one in New Brunswick, Canada, and another in Florida), the authors argue that these specimens are misidentified remains of other canids (domestic dogs) rather than evidence of coyotes in the eastern United States.
Beginning in the 1900s, coyotes expanded their range steadily toward the north, east, and south into non-forested areas (Hody and Kays, 2018). This range expansion was likely aided by severe
___________________
1 See http://vertnet.org/.
declines of large predators to the east (wolves and cougars) and to the south (cougars and jaguars) as a result of predator eradication programs as well as by the fragmentation and conversion of previously forested habitats due to the expansion of agriculture (Moore and Parker, 1992).
Finding: Fossil evidence suggests that at least five subspecies of Canis lupus were present in North America after 1 million years ago. The earliest fossils attributed to Canis rufus were found in Florida and dated at 10,000 years ago.
Finding: Fossil evidence indicates that Canis latrans arose in North America and spread across the continent but that it disappeared from the eastern North America approximately 10,000 years ago and returned in the 1900s.
Paleontology and Morphology
Morphological data concerning historical or ancient populations of red wolves are based on a very limited set of wolf fossils. As described in Chapter 2, the probability that an organism will become part of the fossil record depends strongly on local conditions such as moisture and vegetation cover (Tappen, 1994). Organic remains degrade quickly in moist, forested habitats such as those of southeastern North America. Thus, most canid fossils from eastern North America that date from before 1800 consist of fragments of skulls or teeth that mineralized prior to the decay of the organism. In addition, the fossil remains of wolves are difficult to analyze because many canids overlap in cranial characters (Nowak, 2002). Although Tedford et al. (2009) performed extensive analyses of fossil canids, their study did not include red wolves. There is no known fossil evidence of a small wolf in eastern North America for nearly a million years before the late Rancholabrean (~11,000 years ago) (Nowak, 2002). It is unclear whether the lack of an early fossil record is due to the absence of the taxon or to the scarcity of the canid fossil record in the area.
The most extensive morphological studies of wolves in eastern North America have been done by Lawrence and Bossert (1967) and by Nowak (1979, 1992, 1995, 2002). The sample size for red wolves analyzed by Lawrence and Bossert (1967) is not provided. Lawrence and Bossert (1967) examined all available red wolf specimens collected before 1920 from Louisiana, Alabama, and Florida, a period in which coyotes were absent from the southeastern United States (Nowak, 2002), and evaluated nine cranial and six tooth characters that they had previously found to differentiate among wolves, coyotes, and domestic dogs (C. familiaris). They noted that overall size was an important discriminating factor among species, and they divided each of their measures by the greatest length of skull to account for this. Their results indicated that red wolves grouped closely with gray wolves and eastern timber wolves (C. lupus lycaon), and they concluded that their sample contained only two species: coyotes and gray wolves. While the cranial variation within specimens they considered to be gray wolves was unusually large for a canid species, they concluded that red wolves are no more distinct from C. lupus than a subspecies.
Only nine complete skulls were available for the period dating from the end of the Pleistocene to 1917, and none was from coyotes. Of these nine, Nowak based his work on the six adult male wolf specimens. Three of the nine specimens were not included in Nowak’s analyses, as one of those was from an immature individual and two were from females, which are smaller and more variable in size than males. Because coyotes were absent from southeastern North America during this period (Nowak, 2002), these skulls represent populations from the range of the red wolf prior to modern contact with coyotes.
Nowak argued that there are many measures of skull size and that corrections using only one of these measures may produce biased results. In an analysis of wild canids in Arkansas, Gipson et al. (1974) found that dividing by total length did not correct for the factor of width, which was an
equally important factor for distinguishing canid species. Furthermore, Nowak argued that using such a correction risked losing important information because body size is an important biological factor in distinguishing wild species of Canis; thus, in most of his analyses Nowak separated specimens by age (analyzing only adults) and sex (analyzing only males) and used raw measurements.
Nowak measured 10 skull characteristics and used them in a canonical discriminant analysis, which weighs them by their ability to distinguish designated groups. The first canonical variable weighs the best combination of characters and assigns a value to each specimen, the second selects the next best set of uncorrelated characters and assigns a value, and so forth. A plot of canonical variables 1 and 2 for six groups of C. lupus from western North America and eastern wolves prior to modern contact with coyotes shows that the western wolves group together despite being from different geographical areas and that there is no overlap between them and eastern red wolves (Figure 5-1).
Nowak (1979) also examined these specimens within an analysis of the same skull characters from specimens of gray wolves, coyotes, and domestic dogs (Nowak, 1979; Figures 8-13). Using the raw data in canonical analyses, Nowak initially showed that the three groups separated neatly along the first two axes, with the first axis separating coyotes and wolves, with dogs appearing intermediate, and the second axis separating dogs from the other groups (Nowak, 1979; Figure 8). Nowak then plotted the positions of specimens diagnosed as hybrids between coyotes and dogs (including captive hybrids) and between coyotes and wolves (Nowak, 1979; Figure 10). The coyote–wolf hybrids were aligned with the two parental groups on axis 2, but were clearly intermediate along axis 1. When Nowak placed the pre-1920 specimens of red wolves on these graphs (Nowak, 1979; Figure 11), they occupied the same position as coyote–wolf hybrids, the position
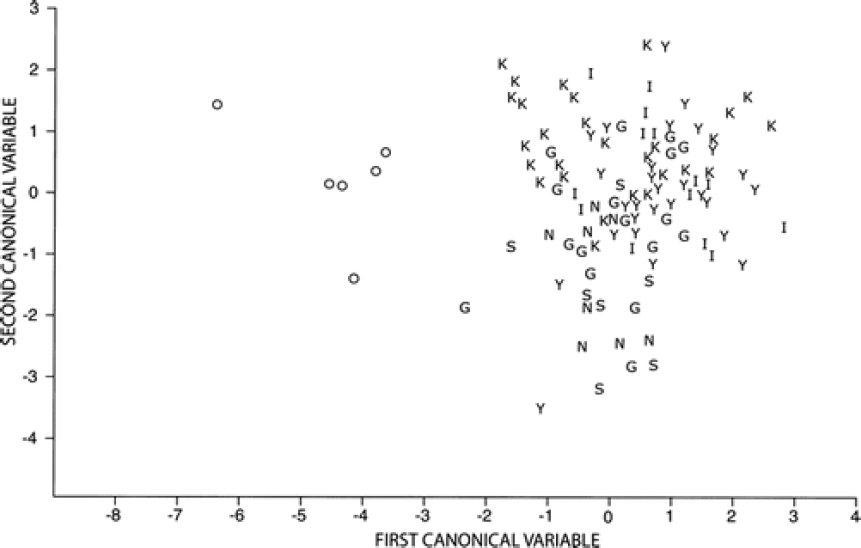
along axis 1 intermediate between coyotes and wolves. However, given that coyotes were absent from the southeastern United States before 1920, these red wolf specimens could not represent first- or early-generation hybrids between wolves and coyotes. Whether they represent a long-standing hybrid lineage or a longstanding species intermediate in size and morphology between coyotes and wolves cannot be determined from this analysis alone.
Nowak (1979, pp. 29–30) noted that although the proportions of pre-1920 skulls of C. rufus from Louisiana may seem to be intermediate to C. lupus and C. latrans, they differ in two important characters. First, when compared to C. latrans, the frontal shield of C. rufus is narrower. Second, when compared to C. lupus, the post-orbital constriction of C. rufus is narrower. Nowak suggested that the slender proportions may have resulted in the ecological niche of red wolves being more similar to that of coyotes (see Behavioral Ecology section below). Finally, Atkins and Dillon (1971) compared the morphology of the cerebellum of six Canis species (C. lupus, n = 13; C. rufus, n = 5; C. familiaris, n = 24; C. latrans, n = 54; C. aureus, n = 1; and C. mesomelas, n = 1) and found that C. rufus differed from all the other species and appeared to be more primitive in multiple characteristics. Although this study was carried out after C. latrans had started to appear in the east, with the potential implication of hybridization affecting the C. rufus population, C. rufus specimens shared multiple features with C. lupus and bore little resemblance to C. latrans. In fact, they more closely resembled foxes, suggesting a retention of traits that may have characterized the ancestral species of Canis.
The ideal approach to the analyses of skull morphology would be geometric morphometrics, either two-dimensional landmark-based methods (Slice, 2007) or three-dimensional landmark-free methods (Pomidor et al., 2016). However, the usefulness of these methods for historical analyses is limited by the small number of historical red wolf samples. These multivariate methods create a high-dimensional multivariate data set and, for hypothesis testing purposes such as robustly distinguishing taxa, require sample sizes much larger than are presently available.
Finding: Based on the limited set of specimens available for analysis, prior to contact with modern coyotes, populations of Canis rufus could be morphologically distinguished from Canis lupus using canonical discriminant analysis. Although conclusions from studies based on skull morphology differ as to whether Canis rufus represented a subspecies of Canis lupus or a distinct species, an analysis of the anatomy of the cerebellum supports the recognition of Canis rufus as historically a distinct species.
Genetics
Recent genetic evidence has established admixture as a common feature of canid evolution and has shown that a bifurcating tree does not provide an adequate model of canid diversification (Gopalakrishnan et al., 2018). As such, one should expect that canid species in North America were not fully reproductively isolated from each other. Furthermore, the radiation of North American canid species may have been a relatively recent event (vonHoldt et al., 2016). Consequently, the three extant nominal canid species in North America are all genetically very similar. For example, based on whole-genome sequence data, the pairwise FST value between gray wolves and coyotes is 0.153, between gray wolves and red wolves is 0.177, and between coyotes and red wolves is 0.108 (vonHoldt et al., 2016). To put this into context, when analyzing a range of different organisms including plants, insects, and mammals, Hey and Pinho (2012) found that when FST was larger than 0.35, populations tended to be characterized as different species and that below this value, they were not. The close genetic similarity among canids in general is a challenge for those assigning species and subspecies status. Each species is non-monophyletic in many genomic positions and shows extensive allele sharing because of past gene flow or recent divergence. Nonetheless, gray
wolves and coyotes are clearly distinct species with pre-zygotic isolation mechanisms (Bohling et al., 2016; Smith et al., 2003), even if the isolation mechanisms are not complete, as well as different ecological specializations, and unique heritable morphology and behavior. The genetic evidence concerning red wolves should be interpreted in this context.
Finding: North American canid species are genetically very similar to each other and have substantial amounts of shared genetic variation.
At the moment, there are no nuclear genomic data available from red wolves dating from before the anthropogenically driven expansion of the coyote into the natural distribution range of the red wolf. However, there have been a limited number of studies of mitochondrial DNA (mtDNA) from museum specimens. Wilson et al. (2003) sequenced mtDNA from two museum specimens of wolves collected in Maine and New York State in, respectively, the 1880s and the 1890s. While they found that generally there was reciprocal monophyly of mtDNA between gray wolves and coyotes, the New York specimen grouped within the coyote clade, and the Maine specimen appeared as the nearest outgroup to coyotes in a phylogenetic analysis. Wilson et al. (2003) interpreted this as supporting the presence of an eastern wolf with an evolutionary history independent of that of the gray wolf. Rutledge et al. (2010a) sequenced mtDNA from a number of 16th century canids and found that the two DNA sequences from the eastern United States grouped with coyote sequences in a phylogenetic analysis, although morphometric comparisons indicated that they were similar to eastern wolves and not to coyotes. Brzeski et al. (2016) studied three ancient specimens from the eastern United States that were 350–1,900 year old and of markedly larger size than modern coyotes. They found that the mtDNA haplotypes of these specimens clustered within the general coyote clade. All three studies are compatible with the hypothesis that the historical red wolf in the eastern United States was more closely related to modern coyotes than to gray wolves, which is in line with what has been found with the extant managed red wolf population (see later sections). Specimens with a wolf-like appearance sampled in the southern United States between 1915 and 1943 (in a region where admixture between red wolves, gray wolves, and coyotes has been proposed) have a mixture of coyote-like and red wolf mtDNA haplotypes, with a preponderance of coyote-like haplotypes except for two specimens from Missouri and one from Oklahoma (Roy et al., 1996), although these analyses were based on only 236 base pairs and may have limited resolution.
Finding: The mtDNA haplotypes from historical wolf-like canids (previous to the recent sympatry with coyotes) in the eastern United States cluster within the coyote clade.
IS THERE EVIDENCE FOR DISTINCTIVENESS OF CONTEMPORARY RED WOLVES FROM GRAY WOLVES AND COYOTES?
Morphology
The current population of red wolves was founded by 14 individuals captured in Texas and Louisiana that were artificially selected based on preconceived notions of what the red wolf phenotype was like, and it has experienced both demographic bottlenecks and hybridization with coyotes. These circumstances have led some to question whether the extant population represents a lineage unique to southeastern North America. To address this question, Hinton and Chamberlain (2014) examined red wolves, coyotes, and hybrids from the Albemarle Peninsula in northeastern North Carolina using 12 morphological traits. They assigned individuals to each category based on 17 microsatellite loci using methods that had been previously developed to specifically discriminate among the three categories. Their study analyzed 528 red wolves, 264 coyotes, and 259 hybrids to
evaluate the use of morphometric measures in identifying these taxa. They then used a polytomous logistical regression analysis with a subset of samples (171 red wolves, 134 coyotes, 47 hybrids) to evaluate the reliability of morphometrics in identifying individuals in each category. They found that while all characteristics were correlated, four measures that characterized overall size were especially useful for distinguishing among them (Table 5-1).
Their polytomous logistical regression model correctly classified 99 percent of coyotes, 98 percent of red wolves, and 13 percent of hybrids. Misclassified hybrids were identified as coyotes 61 percent of the time versus red wolves 35 percent of the time. Thus, the Hinton and Chamberlain (2014) study indicates that, despite hybridization between red wolves and coyotes that took place either at historical or current time scales, these two taxa have maintained morphological differences. These differences likely contribute to ecological differences among populations, especially with respect to their territory sizes and prey selection, and they support the notion that the red wolf is a distinct canid in southeastern North America.
Finding: The contemporary population of red wolves in North Carolina is morphologically distinguishable from sympatric coyotes and red wolf–coyote hybrids.
Genetics
Substantial genetic and genomic work has been done on the original captive and managed red wolf population in North Carolina. The data that have been analyzed include mtDNA sequences (Brzewski et al., 2016; Hailer and Leonard, 2008; Rutledge et al., 2010b; Vilà et al., 1999; Wayne and Jenks, 1001; Wilson et al., 2003), autosomal microsatellites (Bohling et al., 2013), Y chromosome microsatellites (Hailer and Leonard, 2008), single nucleotide polymorphism (SNP) chip array data (vonHoldt et al., 2011), and whole-genome sequencing data (vonHoldt et al., 2016). Here we will briefly review some of the main findings as they pertain to the species status of the red wolf and the issue of distinctiveness.
Analyses of Admixture
Results of most analyses of admixture suggest that the red wolf population is admixed with genetic components of gray wolves, coyotes, and perhaps other canids. The most direct evidence for admixture is the D (ABBA-BABA) test (Green et al., 2010), which was applied by vonHoldt et al. (2016) to genomic sequencing data. This analysis showed that neither the phylogenetic hypothesis of a sister taxa relationship between red wolf and coyote (i.e., a tree of the form [(red wolf, coyote),
TABLE 5-1 Means (± SE) for Morphological Characters of Red Wolves, Coyotes, and Their Hybrids in Northeastern North Carolina, 1987–2011
| Character | Red wolf | Coyote | Hybrid | |||
|---|---|---|---|---|---|---|
| n | Avg ± SE | n | Avg ± SE | n | Avg ± SE | |
| Hind-foot length | 460 | 22.3 ± 0.06 | 256 | 18.7 ± 0.06 | 153 | 20.4 ± 0.11 |
| Body mass | 509 | 23.2 ±0.23 | 240 | 13.4 ± 0.12 | 147 | 17.0 ±0.34 |
| Head width | 182 | 11.9 ± 0.08 | 146 | 10.4 ± 0.05 | 51 | 11.1 ± 0.11 |
| Tail length | 456 | 36.4 ± 0.15 | 241 | 33.9 ± 0.20 | 151 | 35.7 ± 0.25 |
NOTE: SE denotes standard error, which is a measure of the accuracy of the estimated mean, +/- 2SEs being approximately equal to 95% confidence limits.
SOURCE: Hinton and Chamberlain, 2014.
gray wolf]) nor the phylogenetic hypothesis of a sister taxa relationship between red wolf and gray wolf (i.e., a tree of the form [(red wolf, gray wolf), coyote]) is supported by the data. Sinding et al. (2018) also showed that if red wolves were modeled as an admixture between reference coyote and gray wolf populations, the best match was a 70/30 admixture, respectively, of coyote and gray wolf DNA. These findings can be explained by introgression into red wolves by either coyotes or gray wolves or both, or by a separate introgression of DNA from different divergent species into both gray wolves and coyotes. The latter hypothesis is less likely, although given the historical presence of other canid species in North America such as the now extinct dire wolf (Canis dirus), it is not impossible that this happened.
Analyses such as those done by STRUCTURE (see Chapter 3; Pritchard et al., 2000) and inferences of admixture segments (ancestry painting, as described in Chapter 3; Tang et al., 2006) are also compatible with the hypothesis of admixture (vonHoldt et al., 2011). However, a limitation of these analyses is that there are no genomic reference panels for the original wolves in the southern segments of eastern North America. In STRUCTURE analyses, the extant red wolf population is identified as an admixture of gray wolves, as the minority component, and coyotes, as the majority component, when K (the number of admixture components) is low (vonHoldt et al., 2011). This may be because the red wolves truly are admixed, or may be because of sample size issues, so that for low values of K, a higher likelihood is obtained by allowing other components rather than a unique red wolf component (see, e.g., Lawson et al., 2018 for a discussion of some of these issues). When K is large enough to split up the general coyote clade into more than one component, the first component that appears is the red wolf (vonHoldt et al., 2011). It is important to note that this cannot be interpreted as evidence for an old unique coyote ancestry different from gray wolves and coyotes because the emergence of this component could be a consequence of inbreeding and genetic relatedness in the extant red wolf population. These analyses do not provide a quantitative assessment of the proportion of extant red wolf genetic ancestry that is shared with extant gray wolves and coyotes through interbreeding in recent history.
The ancestry painting analyses in vonHoldt et al. (2011) includes only gray wolf, coyote, and dog ancestry. The identification of red wolves as admixed between coyote and gray wolves in these analyses is, therefore, a mathematical consequence of the statistical modeling. It would be useful to carry out more analyses regarding the genetic affinity of inferred ancestry segments to determine whether the mathematical abstraction of the red wolf being a composite of other extant North American canids can be validated by a genetic similarity between the inferred ancestry segments and DNA from the species to which they are assigned. In particular, if the inferred ancestry fragments from red wolves closely match potential donor gray wolf and coyote populations from regions where they might have introgressed during the coyote eastward expansion, this would provide support for the recent admixture hypothesis.
Finding: The red wolf population shows evidence of past genetic contributions from populations related to gray wolves, coyotes, or both.
Genetic Affinity to Other Canid Species
Several different analyses of genetic data, including FST estimates from various data sets (vonHoldt et al., 2016), phylogenetic inference of mtDNA (Wilson et al., 2003) and whole-genome sequences (Sinding et al., 2018), and STRUCTURE analyses of SNP chip data (vonHoldt et al., 2011), suggest that red wolves are more closely related to coyotes than they are to gray wolves. For example, FST between red wolves and western coyotes is smaller than that between red wolves and gray wolves, despite the fact that western coyotes generally appear to be the population with the least genetic drift and, therefore, generally will have lower FST values relative to divergence times
(vonHoldt et al., 2016). Also, when using fewer than five ancestry components in a STRUCTURE analysis to model the structure of canids and dogs in North America, coyotes and red wolves form a single majority ancestry component (vonHoldt et al., 2011). Furthermore, mtDNAs from red wolves generally tend to group with coyotes (e.g., Brzeski et al., 2016; Wilson et al., 2003). Finally, D (ABBA-BABA) statistics based on genome sequencing data identify more derived alleles that are shared between coyotes and red wolves and not shared with western gray wolves than derived alleles that are shared between gray wolves and red wolves and not shared with coyotes (vonHoldt et al., 2016).
Finding: The red wolf is genetically more closely related to coyotes than to western gray wolves.
Time Scale of Admixture or Divergence
The timing of a possible admixture between red wolves and other species is of primary relevance to the species status of the red wolf. The presence of admixture does not in itself challenge the species status of the red wolf. As discussed in Chapter 2, species are on occasion formed by hybridization between two existing species. Furthermore, introgression between different species, as revealed in genomic analyses, increasingly seems to be the rule rather than the exception. However, if the current red wolf population has been formed as an anthropogenic mixture between gray wolves and coyotes postdating the European colonization (i.e., after ~1500 AD) of the Americas, this could affect considerations regarding species status (see Chapter 2, Box 2-1).
The most direct analyses of the time scale of the admixture are provided by vonHoldt et al. (2011, 2016). In 2011 vonHoldt et al. used ancestry painting to estimate the timing of admixture. This method is based on estimates of the mean length of admixture tracts (the contiguous blocks of the genome inherited from a population or a species). Admixture tracts are broken up by recombination so that short tracts indicate old admixture times and long tracts point toward more recent admixture times (see, e.g., Liang and Nielsen, 2014). The estimates obtained in vonHoldt et al. (2011) are 140 and 180 generations for, respectively, the coyote and the gray wolf. The authors then used a generation time of 2–3 years to conclude that the admixture between coyotes and red wolves happened 287–430 years ago. However, recent direct estimates based on observations of gray wolves in Minnesota suggest a generation time of 4.3–4.7 years (Mech et al., 2016), which is also largely compatible with the estimates in vonHoldt et al. (2008) of 4.16 years. Using the Mech et al. (2016) generation time estimate for the coyote and the gray wolf admixtures into red wolves would indicate that the coyote and gray wolf admixtures took place, respectively, approximately 600 and 800 years ago. While these dates are earlier than the recent anthropogenic expansion of coyotes, these somewhat recent dates could still be interpreted as support for a recent origin of red wolves as an admixed coyote/gray wolf population. However, as previously discussed, this analysis is challenged by the lack of appropriate original reference panels for red wolves and by the implicit assumption that the ancestry of red wolves can be modeled as a combination of genomic segments from dogs, western coyotes, and western gray wolves. It is unclear exactly how to interpret the lengths of the inferred ancestry tracts if the reference panels are not representative of the ancestry of the specimen analyzed. Also, it is worth noting that the method used to estimate admixture, SABER (Tang et al., 2006), identifies the timing of a single pulse of admixture. However, the apparent admixture of red wolves and other canids may possibly be a composite of recent coyote admixture and much older shared ancestry with the coyote population. It is possible that, in reality, the admixture time inferred reflects a mixture of old red wolf ancestry dating back thousands of years, and very recent admixture with coyotes or gray wolves, the combined effects of which would appear as admixture 600–800 years ago. A more detailed analysis of the distribution of tract lengths might in the future help determine this issue.
A more relevant modeling effort is provided in vonHoldt et al. (2016) using the program G-PhoCS (Gronau et al., 2016) applied to genomic sequence data (Figure 5-2). The method implemented in G-PhoCS can be used to calculate posterior probabilities for migration rates and divergence times using genomic data, assuming multiple independent non-recombining loci, in other words, assuming that the genome can be divided into segments with unique independent coalescence trees with no recombination within segments. Using this program, vonHoldt et al. (2016) identified that the best fitting model was for red wolves having evolved as a sister group to coyotes. They also estimated the divergence time to be 55,000–117,000 years ago. This estimate should be compared to an estimate of divergence between gray wolves and coyotes generated by vonHoldt et al. that was somewhat larger than 160,000–170,000 years (see Figure S5 in vonHoldt et al., 2016). These divergence time estimates are all based on the old estimates of 3 years between generations. Using the Mech et al. (2016) generation time estimate, the coyote–red wolf divergence would instead be approximately 82,000–175,000 years. Furthermore, the best fitting model includes substantial amounts of subsequent gene flow from coyotes and gray wolves into red wolves. The best fitting G-PhoCS model suggests that red wolves make up an old divergent subgroup in the lineage that gave rise to coyotes, with a divergence time roughly half that of coyotes and western gray wolves, and that this subgroup has received substantial gene flow from both coyotes and gray wolves since the time of divergence (see Figure 5-2 for estimated of levels of gene flow).
The divergence time for coyotes and gray wolves is estimated to be substantially more recent in this study than has been suggested by most analyses of the fossil record. This discrepancy could be caused by several different factors, including a misinterpretation of the fossil record or an erroneous calibration of the molecular clock. The discrepancy in divergence time cannot be resolved in this report, but the considerations herein are based on the relative divergence between various canids. The
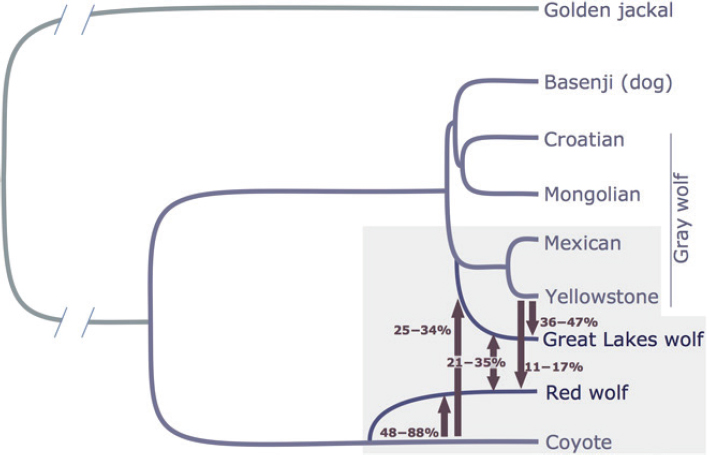
absolute values of the time estimates are less relevant for the committee’s conclusions, and a possible miscalibration of the molecular clock does not affect the committee’s discussion. There are multiple reasons why parametric analyses, such as the ones carried out using G-PhoCS could be cause for concern. It is often difficult to ascertain convergence and it may be unclear how perturbations of the model (such as varying population size and varying migration rates) will affect the estimates. Nonetheless, the G-PhoCS results are the only attempts in the published literature to estimate a history of the red wolf population in terms of parameters such as divergence times and migration rates. Additional future analyses using more data and other methods will likely provide improved estimates. At the time of the writing of this report, the G-PhoCS analyses remain the most direct estimate of the history of the extant red wolves. Future analyses focusing on obtaining more precise estimates of the divergence time using more data would be a significant help in examining species questions about the red wolf.
Mitochondrial DNA represents only a single non-recombining DNA segment, but the analyses of mtDNA are roughly compatible with the G-PhoCS analyses. As previously mentioned, mtDNAs from historical specimens group within the coyote mtDNA clade. Early analyses suggested that red wolves form an outgroup to other coyotes (Wilson et al., 2003). However, later mtDNA analyses showed that historical wolves in the eastern United States group within a larger coyote clade (Rutledge et al., 2010a). This observation agrees with the findings of the G-PhoCS analyses, suggesting that red wolves harbor both recent admixture and a more divergent genetic component related to coyotes. However, it is worth noting that DNA from historical specimens of red wolves does not group together with mtDNA from extant red wolves, an observation that can be explained by recent or old coyote admixture or ancestral lineage sorting. No attempts have been made in the published literature to use the mtDNA sequences to formally test these different scenarios.
Finally, when modeling red wolves as an admixture between gray wolves and coyotes, Sinding et al. (2018) found that there was substantial genetic drift in the estimated admixture trees, not only on the red wolf lineage, but also on both the gray wolf and coyote lineages after the split from red wolves, suggesting that the admixture is not very recent. The proportion of genetic drift in coyotes since the split with red wolves is approximately 30 percent (36/122) of the total drift that has happened in the coyote lineage since the divergence with gray wolves. This is qualitatively compatible with the findings of the G-PhoCS analyses as the shared drift between coyotes and red wolves (70 percent) would result both from the shared ancestral lineage after splitting from gray wolves and from the subsequent gene flow between the coyote and red wolf lineages.
Other methods, including those analyzing the molecular divergence in inferred ancestry tracts, could have been used to determine whether red wolves show evidence of substantial recent admixture and, generally, to determine the time scale of admixture. Such analyses would be greatly facilitated by the availability of genomic reference data from ancient red wolf genomes.
Finding: The timing of the admixture between red wolves and other canids is still unresolved, but red wolves have divergent genetic ancestry that predates European colonization.
Distinct Genomic Ancestry
A relevant question is the degree to which the current red wolf population harbors genetic material of potential adaptive value that is not found in gray wolves or coyotes. Two analyses, in particular, are relevant for this question. vonHoldt et al. (2016) identifies SNPs in the red wolf population not found in a reference set of Eurasian wolves and coyotes. The number of new SNPs (private alleles) that were identified tends to be larger, after accounting for sampling effects, than for other North American canids except for two coyotes. However, vonHoldt et al. (2016) argue that, nonetheless, the number is so small that it provides evidence that red wolves are recent gray wolf–coyote hybrid populations. This assertion is challenged by Hohenlohe et al. (2017) who criticize the
vonHoldt et al. (2016) study because it lacks an appropriate null model. In response, vonHoldt et al. (2017) performed a leave-one-out analysis to assess whether red wolves have more private alleles than would be expected from a gray wolf–coyote admixture; they concluded that they do not. Such analyses rely on strong assumptions regarding demographic history, as bottlenecks will reduce the number of private alleles in the population undergoing the bottleneck. In particular, inbreeding in the captive red wolf population is likely to lead to reduced allelic variation, thereby reducing the number of private alleles expected in small samples. The degree to which red wolves have a different number of private alleles than expected from a gray wolf–coyote admixture is still an open question. This question could likely be resolved by using simulations that take the recent breeding structure (i.e., bottleneck and inbreeding) of red wolves into account. Without such simulations (or analytical calculations) it is not possible to determine whether the number of private alleles is larger or smaller than expected from a recent gray wolf–coyote admixture.
Another line of evidence comes from two recent studies: one identifying a previously overlooked population of canids on Galveston Island with apparent red wolf-like features (Heppenheimer et al., 2018) and another demonstrating the existence of substantial red wolf genetic ancestry in wild canids of southwestern Louisiana (Murphy et al., 2018). The Heppenheimer et al. (2018) study demonstrates an excess of allele sharing between the managed red wolf population and the Galveston canids, particularly of alleles of low frequency. This observation, in itself, demonstrates the presence of genetic ancestry in the red wolf that is not present in gray wolves and coyotes, although this conclusion might be affected by the limited size of the reference panels used (29 coyotes and 10 gray wolves). Furthermore, the degree to which this allele sharing is larger than expected from an admixture of currently extinct population isolates of gray wolves and coyotes cannot be determined from the current analyses. Murphy et al. (2018) found evidence of red wolf mitochondrial or nuclear DNA ancestry in 55 percent of the samples they collected in southwestern Louisiana within the historical red wolf range, and detected 75–100 percent ancestry in one individual. While both of these analyses show that red wolves harbor genetic variability that would not be expected in a simple mixture of chosen reference gray wolf and coyote populations, they still do not show a connection between this genetic variability and the historical red wolf populations in the eastern United States.
Finding: The red wolf has some degree of genetic ancestry not found in reference populations of western gray wolves or coyotes.
Behavioral Ecology
In terms of social behavior and reproduction, red wolves are very similar to gray wolves and quite distinct from coyotes. Red wolves live in social groups or packs that are composed of extended families that include a breeding pair and some offspring, usually from previous years, but that could also include 2-year-old individuals (Phillips and Henry, 1992). This social structure contrasts with that of coyotes, which usually have breeding pairs or small family groups and “pups that often disperse before their second summer” (Phillips and Henry, 1992). Riley and McBride (1972) considered red wolves to be more sociable than coyotes, although less social than gray wolves.
Red wolves have one breeding season during the year, with matings occurring in January and February and births in March and April (Riley and McBride, 1972), which is similar to what is reported for coyotes (Carlson and Gese, 2008). Red wolves typically become sexually mature by their second year, which contrasts with coyotes, where breeding by yearlings is common (Phillips et al., 2003). Both sexes disperse by 2 years of age, around the onset of sexual maturity (Phillips et al., 2003).
In the extant managed population, red wolves live in sympatry with coyotes, and on some occasions interspecific breeding pairs are formed. However, Hinton et al. (2017) reported that when
conspecific mates are available, both species exhibit assortative mating, and red wolves effectively displace coyotes. This suggests that there is some level of reproductive isolation between red wolves and coyotes. Evidence of reproductive isolation is also supported by Bohling et al. (2016). In this genetic study, 6 to 17 (average 12.5) microsatellite loci were used to analyze fecal samples from 311 individuals on the Albemarle peninsula in North Carolina. The sampling area included the red wolf experimental protected areas (RWEPAs) where red wolves and coyotes co-exist. The researchers compared the number of “hybrids” (individuals with a q value between 0.126 and 0.824) in the area to expectations based on simulations of hybridization under three scenarios (random breeding between coyotes and red wolves, weak positive assortative mating, and both assortative mating and territorial challenges by red wolves against coyotes). Despite the presence of the two species throughout the RWEPA only 9 (5 percent) out of 180 sampled individuals were hybrids. This was contrasting to the expectations based on the three hybridization scenarios, suggesting that while hybridization in this area occurs, it is less frequent than expected given the proportion of individuals of the parental species in the area. Bohling et al. (2016) mention that selection against hybrids or low hybrid fitness is unlikely based on previous studies showing a lack of outbreeding depression in red wolves and other canids, and they argue ecological segregation is improbable given the ecological adaptability of coyotes. However, the observed bimodal distribution of genotypes in this area strongly suggests the presence of isolating mechanisms limiting intermixing, perhaps associated with assortative mating (Jiggins and Mallet, 2000), which are likely combined with some effect from human management in this system.
Additionally, Murphy et al. (2018) found high levels of red wolf ancestry in the canid population of southwestern Louisiana, where the last wild population of red wolves resided before the intensive removal efforts in the area during the 1970s, which were thought to have depleted the area of red wolves. The presence of individuals with approximately 43 to 78 percent (or more, depending on estimates) red wolf ancestry is highly surprising given the extremely low population of wolves that must have remained in the area and the extensive interbreeding with coyotes over approximately 12 generations. These findings also support the notion of the presence of isolating mechanisms, albeit incomplete, between red wolves and coyotes.
Finding: Red wolves have a social organization and reproductive behavior that is more similar to gray wolves than to coyotes, and when mates are available they exhibit assortative mating.
IS THERE EVIDENCE FOR CONTINUITY BETWEEN THE HISTORICAL RED WOLF POPULATION AND THE PRESENT MANAGED POPULATIONS?
Paleontology and Morphology
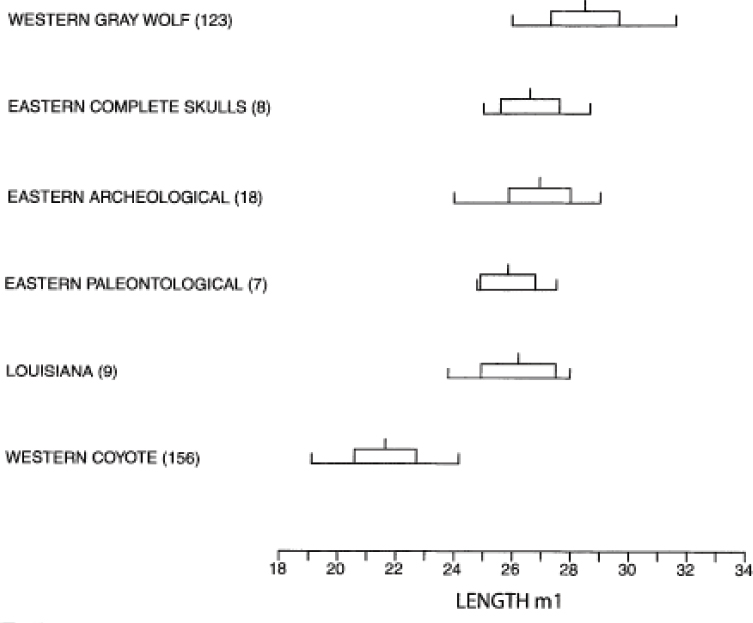
Based on dental analyses of paleontological (around 10,000 years ago), archaeological (2,000–200 years ago), and modern specimens from the 19th and 20th century before the modern expansion of coyotes into the range of red wolves, Nowak (2002) concluded that the red wolf had morphological continuity from about 10,000 years ago to the present (Nowak, 2002) (Figure 5-3). The continuity in morphology of the length of the lower carnassial tooth (m1) among wolves west of the Mississippi and across this time period greatly overlapped with that present in gray wolves (and is similar to what was found by Lawrence and Bossert, 1967).
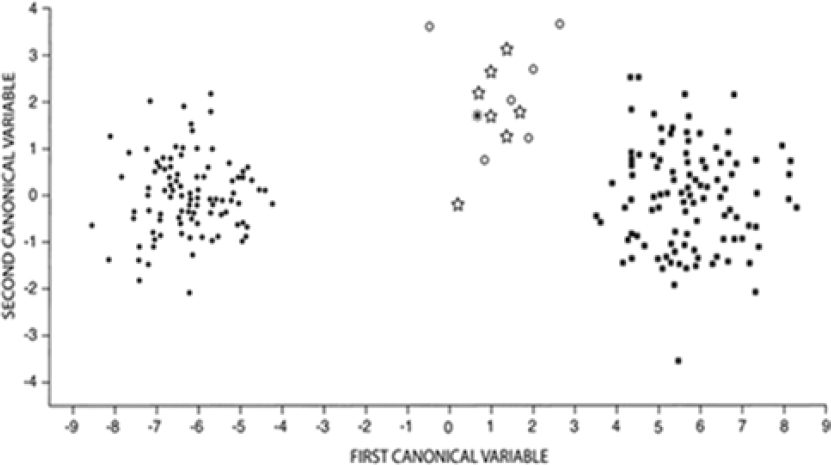
Nowak (2002) compared individuals from the historical population of red wolves to individuals from northeastern Louisiana, which represent the region from which the current managed population was founded. He compared the six specimens of red wolf from east of the Mississippi River that dated from between the time of the Pleistocene up to 1918 with seven red wolves from northeastern Louisiana that were collected during 1898–1905 as well as a single red wolf from Oklahoma. In this
analysis, red wolves from across the range of the species grouped together to the exclusion of both C. lupus and C. latrans (Nowak, 2002). These results suggest continuity of C. rufus populations across time and space as well as a lack of overlap with C. lupus or C. latrans (Figure 5-4).
Finding: Morphological analyses suggest cohesiveness among red wolf specimens from the end of the Pleistocene to the early 1900s, but it remains unclear whether this continuity is shared with the extant captive and managed populations.
Genetics
Because wolves are extinct in the southwestern United States, the only sure way to determine whether the managed red wolf population is a remnant of the original canid species in southwestern United States is to use ancient DNA from museum specimens. Unfortunately, at the moment there are no genome-wide data available from red wolves from before the anthropogenic expansion of the coyote range. However, as previously discussed, both the historical red wolf and the captive or managed population have predominantly coyote-like mtDNA haplotypes.
Because of the absence of genomic data from ancient samples, no firm inferences can be made regarding the genetic continuity between the modern managed red wolf population and the ancient wolves in the middle and southern United States. However, mtDNA analyses suggest that the historical wolf in the eastern United States was not a gray wolf, but instead that it was more closely related to coyotes, much like the modern managed red wolf population (Brzeski et al., 2016; Rutledge et al., 2010a; Wilson et al., 2003). The mtDNA in the managed red wolf population is not identical—or closely related—to the mtDNA lineages from the historical red wolf, but that might be expected under models of lineage sorting or subsequent coyote introgression. The committee notes that there (perhaps surprisingly) have been no model-based approaches providing bounds on the possible divergence times between coyotes, the managed red wolf population, and the historical red wolf population based on mtDNA sequences.
The signal of a deep ancestral divergence between the captive and managed red wolves and coyotes that has been observed in the G-PhoCS analyses (vonHoldt et al., 2016) is compatible with the idea that this population retains ancestry from a now extinct canid population. Additionally, the existence of distinct alleles shared with the Galveston Island population (Heppenheimer et al., 2018) is compatible with the hypotheses of the captive/managed population tracing its ancestry, at least partially, to a wolf population distinct from modern gray wolves and coyotes.
Finding: Genetic continuity between the managed red wolf population and the historical wolf in the eastern United States cannot be firmly established without genomic data from ancient specimens. However, the patterns of genetic variability are compatible with the hypothesis that the red wolf shares a fraction of its genetic history with a canid distinct from modern reference coyotes and gray wolves.
Behavioral Ecology
Only a handful of studies were conducted on the ecology and behavior of the red wolves before the local extinction of the remnant wild population, so an understanding of the natural behavior of this original population is limited. The available data are limited to social and reproductive behavior, home range sizes, and prey consumed.
Social Behavior and Reproduction
Riley and McBride (1972) reported the typical red wolf social structure (i.e., packs of extended families including a breeding pair and some yearlings) prior to the red wolf’s extinction in the wild, and they mentioned that it was common to find three or more individuals traveling as a group throughout the year. The social behavior and reproductive behavior of the natural population are consistent with the behaviors reported in studies of the restored populations. After individuals were reintroduced in the mid-1980s, studies found a social structure that was similar to the one reported for the original wild population, with most individuals forming packs of extended families (Phillips and Henry, 1992). In both the captive and the restored populations, the average litter size is three to four pups (Phillips et al., 2003; Riley and McBride, 1972).
Before the natural population was extirpated, red wolves reared their young in dens dug in the slopes or crests of low natural sand mounds or in human-modified environments such as drain pipes and culverts or the banks of irrigation and drainage ditches (Riley and McBride, 1972). However, red wolves of the restored population place their dens both above ground as nests and also underground (Phillips et al., 2003), possibly as a consequence of ecological differences between now and the earlier time. Both males and females rear the pups, and offspring from the previous year sometimes do (Phillips et al., 2003) and sometimes do not (Riley and McBride, 1972) participate in rearing their siblings.
Finding: The reported social behaviors of the natural and restored populations are very similar.
Habitat and Home Range
Historically, red wolves occupied most of the United States east of the prairies and south of the Great Lakes (Waples et al., 2018). Their original distribution range is highly concordant with the distribution of the eastern temperate forests (North America’s ecoregion 8), and it is hypothesized that the small red wolf was adapted to this historically heavily forested region (Waples et al., 2018). While the red wolf was originally the only canid present in the eastern United States, the anthropogenic alteration of the eastern forests and the reduction of the wolf population caused by wolf control programs allowed the expansion of the coyote into this area in the second half of the 20th century (Hinton et al., 2017; Kilgo et al., 2010; Levy, 2012). Currently, the restored population of red wolves is sympatric with coyotes. This has important implications for the ecology and behavior of the red wolves, particularly as coyotes now represent a competitor for space and resources. Nonetheless, some ecological features remain consistent between the original and managed red wolf populations. For example, before the 1970s the estimated home range of an average red wolf was 65–129.5 km2 (Riley and McBride, 1972). Similarly, Phillips et al. (2003) reported an average of 90.6 ± 18 km2 for individuals and 124 ± 54 km2 for packs in the restored population. These values are larger than those estimated for southeastern coyotes (30.5 km2, range 3.7–137.0 km2, from Schrecengost et al., 2009; 22.7 km2, confidence interval = 9.7–35.8 km2, from Hickman et al., 2015). More recently, Hinton et al. (2017) reported that in the managed population in eastern North
Carolina, red wolves maintain larger home ranges than coyotes, with most red wolves having home ranges over 47 km2, while the coyotes do not keep home ranges above 47 km2.
Finding: The original distribution of red wolves seems to have been tied to the distribution of the eastern temperate forests. The red wolves require larger home ranges to obtain their prey than coyotes. This requirement of larger home ranges is consistent between the original natural population and the extant managed population in North Carolina.
Food Habits
It has been hypothesized that the red wolf was originally adapted to hunt for white-tailed deer (Odocoileus virginianus; Waples et al., 2018). However, overharvesting and habitat loss and fragmentation had driven white-tailed deer populations across the southeastern United States to near extinction by the early 1900s. Based on scat analyses and from direct observations, Riley and McBride (1972) found that the predominant prey species of the last wild populations of red wolves were nutria (Myocastor coypus), swamp rabbit (Sylvilagus aquaticus), and cottontail rabbit (Sylvilagus floridanus); the investigators also reported red wolves preying to a lesser extent on rice rat (Oryzomys palustris), cotton rat (Sigmodon hispidus), and muskrat (Ondatra zibethicus).
In contrast, the diet of the restored population relies on white-tailed deer, raccoon (Procyon lotor), and rabbits (Phillips et al., 2003). However, there is variation in food habits among packs in the restored population that is related to variation in the abundance and distribution of prey, which in turn is due to differences in the habitat used by each pack (agricultural fields or pine-hardwood swamps).
Two recent studies of the diets of restored wolves and coyotes on the Albemarle peninsula produced similar results overall but differed in whether they found wolves and coyotes to have different diets. Hinton et al. (2017) analyzed 1,485 scats collected over 24 months from 13 identified wolf pairs, 17 identified coyote pairs, and 10 identified congeneric pairs. They found that red wolves and coyotes consume a similar carnivorous diet composed of white-tailed deer, rabbits, small mammals (mice, rats, shrews, and voles), furbearers (muskrats, nutrias, and raccoons), and pigs (feral and domestic). However, their diets differed in the proportions of different types of prey and in whether prey use was seasonal. While red wolves predominantly consumed white-tailed deer followed by rabbits and, to a lesser extent, small mammals, the diet of coyotes was dominated by rabbits, followed equally by deer and small mammals. Red wolf consumption of deer did not vary by season, whereas coyote consumption was greater in winter. Red wolves consumed more rabbits during the autumn (harvest season), whereas coyotes consumed more rabbits in the spring and summer (growing season). Diet was correlated with body mass, with larger breeding pairs consuming more white-tailed deer and fewer rabbits and other small mammals. This was the case even when analyzing red wolves and coyotes separately.
McVey et al. (2013) examined 228 scats collected over 14 months, overlapping with the first 14 months of the Hinton et al. (2017) study. They were able to identify 179 of the scats as coming from wolves and 64 as coming from coyotes. They reported similar overall dietary habits to those reported by Hinton et al. (2017) but found no significant differences between wolves and coyotes and no significant seasonal variations in diet. They were unable to collect identifiable coyote scat in 4 of the 14 months of the study.
In general, although there are differences in diet composition reported between the original natural and extant populations, food habits seemed to be highly dependent on prey availability and individual body size.
Finding: The diet of the red wolves in the restored population includes a greater consumption of deer than the natural population. However, this may be a function of prey availability and body size. Both red wolves and coyotes in North Carolina consume a similar diet in terms of the types of prey, but they differ in the proportions of deer, rabbits, and other small mammals in their diets and in their seasonal consumption of these prey types.
SYNTHESIS OF FINDINGS
The archaeological record of canids in eastern United States suggests the presence of a canid during the past 10,000 years that was slightly smaller than gray wolves and substantially larger than coyotes. The population showed morphological cohesiveness in this period and was compatible with the existence of a species in the eastern United States distinct from gray wolves and coyotes. The mtDNA type of this species falls within the coyote clade in historical specimens dating back to 1,900 years ago. There are three possible explanations for these observations. First, all seven ancient DNA specimens were coyotes unrelated to red wolves in the area. This does not seem a likely explanation as these specimens had wolf-like morphology. Furthermore, as argued by Brzeski et al. (2016), this would vastly expand the historical coyote distribution. This leaves two possible explanations: either the historical red wolf was a sister species to coyotes, or it was, for at least the past 1,900 years, an admixed species with a preponderance of matrilineal coyote ancestry. In either of these two scenarios, the committee, for the reasons described above, considers the red wolf to be a species with a distinct morphology, possibly as a result of specialization to the eastern forest habitat. This conclusion is also in agreement with the observation from model-based approaches that the extant population carries genetic ancestry divergent from coyotes dating back 55,000–117,000 years, although there has been substantial gene flow from both gray wolves and coyotes into the extant managed red wolf population since then.
Some might argue that the red wolf could be designated as a subspecies of coyotes. However, because of its consistently differentiated morphology, its proposed ecological specialization, and the initial divergence between red wolves and coyotes is estimated to have occurred before the divergence between the subspecies of C. lupus that are analyzed in the G-PhoCS analysis by vonHoldt et al. (2016), a species designation is more appropriate. The estimate of an initial divergence between red wolves and coyotes between 55,000 and 117,000 years ago also provides evidence for a unique genetic ancestry of the extant population. If the extant red wolves were purely a product of recent gray wolf–coyote admixture, model-based approaches should not find evidence for old ancestry in red wolves distinct from coyotes. A second line of evidence for a distinct ancestry of extant red wolves comes from the recent observation of canids on Galveston Island, which have the appearance of red wolves and which share private alleles with red wolves. These lines of evidence, together with estimates of the introgression tract length distribution, also suggest that the captive/managed red wolf population cannot be purely a product of hybridization between gray wolves and coyotes that occurred during the recent anthropogenic eastward expansion of the coyote range.
The unique allele sharing between the managed/captive red wolf population and the Galveston Island canids can be explained either by introgression of gray wolf ancestry distinct from that observed in any extant gray wolf population used as reference or else by a shared genetic ancestry with historical red wolf populations. The sharing does not in itself demonstrate continuity between the historical and the extant red wolf populations. The most substantial support for continuity comes from the estimates that the deepest divergence between extant red wolves and coyotes occurred between 55,000 and 117,000 years ago, which would be compatible with the continuity hypothesis. However, the committee notes that to fully establish continuity, genomes from historical red wolf specimen would be needed. Such genomes could also help distinguish between the hypotheses of
the historical red wolf being an ancestrally admixed species or being a mostly non-admixed sister taxon to the coyote.
The possible options for the taxonomic status of the red wolf include designation as a unique species, as a subspecies of gray wolves, as a subspecies of coyotes, or as a group of recently admixed individuals belonging to neither species. The last option is not sustainable because of (1) the estimates of deep divergent DNA in red wolves using G-PhoCS in vonHoldt (2016), (2) the estimates of an admixture time mostly predating the coyote expansion even if it is considered a hybrid species between gray wolf and coyote using ancestry tracts in vonHoldt (2011), and (3) the presence of unique alleles shared with the Galveston Island individuals and not shared with other reference population (Heppenheimer et al., 2018). A subspecies designation within the gray wolves is also inappropriate since red wolves, historically and presently, show genetic evidence of being more closely related to coyotes than to gray wolves. A subspecies designation within coyotes is also not a tenable option. There are substantial morphological and behavioral differences between coyotes and red wolves. Furthermore, pre-zygotic isolation mechanisms are at least partially maintained between red wolves and coyotes, possibly driven by size differences. Combining this observation with the genetic evidence of traces of a relatively deep divergence (G-PhoCS analysis in vonHoldt et al., 2016) and maintenance of some unique genetic ancestry, a species status, possibly as a historically admixed species, is the most appropriate designation for the red wolf. However, genomic DNA from historical red wolf specimens could help clarify the issue regarding the continuity between historical and extant red wolves.
The time scales of divergence and the amount of introgression since divergence might affect taxonomic considerations. Before approximately 11,000 years ago there were more canid lineages in North America than in existence today, but it is not clear which group or groups of canids were ancestral to the red wolf. However, even a recently emerged species can be recognized as such if it is ecologically, functionally, and reproductively separated from other species. Additionally, the extant red wolf might trace a large proportion of its genome to relatively recent admixture with coyotes as indicated by the G-PhoCS analysis in vonHoldt et al. (2016), which suggests that 48 to 88 percent of the genome could have been replaced by gene flow from coyotes since the first divergence between coyotes and red wolves. Nonetheless, the genomes of extant red wolves might still represent much of the original red wolf genome spread into fragments in different individuals. It is possible that future, more precise, genetic analyses might help determine the exact proportion of the red wolf genome that has been replaced by recent admixture and that such analyses might provide additional insight into the species status of the red wolf.
Conclusions
- Available evidence suggests that the historical red wolves constituted a taxonomically valid species.
- Extant red wolves are distinct from the extant gray wolves and coyotes.
- Available evidence is compatible with the hypothesis that extant red wolves trace some of their ancestry to the historical red wolves.
- Although additional genomic evidence from historical specimens could change this assessment, evidence available at present supports species status (Canis rufus) for the extant red wolf.
REFERENCES
Atkins, D. L., and L. S. Dillon. 1971. Evolution of the cerebellum in the genus Canis. Journal of Mammalogy 52:96-107.
Audubon, J. J., and J. Bachman. 1851. The Quadrupeds of North America, Vol 2. New York: V. G. Audubon.
Bohling, J. H., J. R. Adams, and L. P. Waits. 2013. Evaluating the ability of Bayesian clustering methods to detect hybridization and introgression using an empirical red wolf data set. Molecular Ecology 22:74-86.
Bohling, J. H., J. Dellinger, J. M. McVey, D. T. Cobb, C. E. Moorman, and L. P. Waits. 2016. Describing a developing hybrid zone between red wolves and coyotes in eastern North Carolina, USA. Evolutionary Applications 9:791-804.
Brzeski, K. E., M. B. DeBiasse, D. R. Rabon, Jr., M. J. Chamberlain, and S. S. Taylor. 2016. Mitochondrial DNA variation in southeastern pre-Columbian canids. Journal of Heredity 107:287-293.
Carlson, D. A., and E. M. Gese. 2008. Reproductive biology of the coyote (Canis latrans): Integration of mating behavior, reproductive hormones, and vaginal cytology. Journal of Mammalogy 89(3):654-664.
Gipson, P., J. A. Sealander, and J. E. Dunn. 1974. The taxonomic status of wild Canis in Arkansas. Systematic Zoology 23:1-11.
Goldman, E. A. 1937. The wolves of North America. Journal of Mammalogy 18:37-45.
Gopalakrishnan, S., M. S. Sinding, J. Ramos-Madrigal, J. Niemann, J. A. Samaniego Castruita, F. G. Vieira, C. Carøe, M. M. Montero, L. Kuderna, A. Serres, V. M. González-Basallote, Y. H. Liu, G. D. Wang, T. Marques-Bonet, S. Mirarab, C. Fernandes, P. Gaubert, K. P. Koepfli, J. Budd, E. K. Rueness, M. P. Heide-Jørgensen, B. Petersen, T. Sicheritz-Ponten, L. Bachmann, Ø. Wiig, A. J. Hansen, and M. T. P. Gilbert. 2018. Interspecific gene flow shaped the evolution of the genus Canis. Current Biology 28:3441-3449.
Graham, R. W., and E. L. Lundelius, Jr. 2010. FAUNMAP II: New data for North America with a temporal extension for the Blancan, Irvingtonian, and early Rancholabrean. FAUNMAP II Database, version 1.0. http://www.ucmp.berkeley.edu/faunmap/. Accessed January 14, 2019.
Green, R. E., J. Krause, A. W. Briggs, T. Maricic, U. Stenzel, M. Kircher, N. Patterson, H. Li, W. Zhai, M. H.-Y. Fritz, N. F. Hansen, E. Y. Durand, A.-S. Malaspinas, J. D. Jensen, T. Marques-Bonet, C. Alkan, K. Prüfer, M. Meyer, Hernán A. Burbano, J. M. Good, R. Schultz, A. Aximu-Petri, A. Butthof, B. Höber, B. Höffner, M. Siegemund, A. Weihmann, C. Nusbaum, E. S. Lander, C. Russ, N. Novod, J. Affourtit, M. Egholm, C. Verna, P. Rudan, D. Brajkovic, Ž. Kucan, I. Gušic, V. B. Doronichev, L. V. Golovanova, C. Lalueza-Fox, M. de la Rasilla, J. Fortea, A. Rosas, R. W. Schmitz, P. L. F. Johnson, E. E. Eichler, D. Falush, E. Birney, J. C. Mullikin, M. Slatkin, R. Nielsen, J. Kelso, M. Lachmann, D. Reich, and S. Pääbo. 2010. A draft sequence of the Neandertal genome. Science 328:710-722.
Gronau, I., M. J. Hubisz, B. Gulko, C. G. Danko and A. Siepel. 2011. Bayesian inference of ancient human demography from individual genome sequences. Nature Genetics 43:1031-1034.
Hailer, F., and J. A. Leonard. 2008. Hybridization among three native North American Canis species in a region of natural sympatry. PLOS ONE 3:e3333.
Hall, E. R. 1981. The Mammals of North America, Vol. 2. New York: John Wiley and Sons.
Hall, E. R., and K. R. Kelson. 1959. The Mammals of North America. New York: Ronald Press.
Heppenheimer, E., K. E. Brzeski, R. Wooten, W. Waddell, L. Y. Rutledge, M. J. Chamberlain, D. R. Stahler, J. W. Hinton, and B. M. vonHoldt. 2018. Rediscovery of red wolf ghost alleles in a canid population along the American Gulf Coast. Genes (Basel) 9(12):E618.
Hey, J., and C. Pinho. 2012. Population genetics and objectivity in species diagnosis. Evolution 66:1413-1429.
Hickman, J. E., W. D. Gulsby, C. H. Killmaster, J. W. Bowers, M. E. Byrne, M. J. Chamberlain, and K. V. Miller. 2015. Home range, habitat use, and movement patterns of female coyotes in Georgia: Implications for fawn predation. Journal of the Southeastern Association of Fish and Wildlife Agencies 2:144-150.
Hinton, J. W., and M. J. Chamberlain. 2014. Morphometrics of Canis taxa in eastern North Carolina. Journal of Mammalogy 95:855-861.
Hinton, J. W., A. K. Ashley, J. A. Dellinger, J. L. Gittleman, F. vanManen, and M. J. Chamberlain. 2017. Using diets of Canis breeding pairs to assess resource partitioning between sympatric red wolves and coyotes. Journal of Mammalogy 98:475-488.
Hody, J. W., and R. Kays. 2018. Mapping the expansion of coyotes (Canis latrans) across North and Central America. ZooKeys 7859:81-97.
Hohenlohe, P. A., L. Y. Rutledge, L. P. Waits, K. R. Andrews, J. R. Adams, J. W. Hinton, R. M. Nowak, B. R. Patterson, A. P. Wydeven, P. A. Wilson, and B. N. White. 2017. Comment on “Whole genome sequence analysis shows two endemic species of North American wolf are admixtures of the coyote and gray wolf.” Science Advances 3:e1602250.
Jiggins, C. D., and J. Mallet, J. 2000. Bimodal hybrid zones and speciation. Trends in Ecology and Evolution 15:250-255.
Kilgo, J. C., H. S. Ray, C. Ruth, and K. V. Miller. 2010. Can coyotes affect deer populations in southeastern North America? Journal of Wildlife Management 74:929-933.
Lawrence, B., and W. H. Bossert. 1967. Multiple character analysis of Canis lupus, latrans and familiaris, with a discussion of the relationships of Canis niger. American Zoologist 7:223-232.
Lawson, D. J., L. van Dorp, and D. Falush. 2018. A tutorial on how not to over-interpret STRUCTURE and ADMIXTURE bar plots. Nature Communications 9(1):3258.
Levy, S. 2012. Rise of the coyote: The new top dog. Nature 485:296-297.
Liang, M., and R. Nielsen. 2014. The lengths of admixture tracts. Genetics 197:953-967.
Martin, L. D. 1989. Fossil history of the terrestrial carnivora. Pp. 536-568 in Carnivore Behavior, Ecology, and Evolution, J. L. Gittleman, ed. Boston, MA: Springer.
McVey, J. M., D. T. Cobb, R. A. Powell, M. K. Stoskopf, J. H. Bohling, L. P. Waits, and C. E. Moorman. 2013. Diets of sympatric red wolves and coyotes in northeastern North Carolina. Journal of Mammalogy 94:1141-1148.
Mech, L. D., S. M. Barber-Meyer, and J. Erb. 2016. Wolf (Canis lupus) generation time and proportion of current breeding females by age. PLOS ONE 11(6):e0156682.
Moore, G. C., and G. R. Parker. 1992. Colonization by the eastern coyote (Canis latrans). Pp. 23-37 in Ecology and Management of the Eastern Coyote, A. J. Boer, ed. University of New Brunswick, Fredericton: Wildlife Research Unit.
Murphy, S. M., J. R. Adams, J. J. Cox, and L. P. Waits. 2018. Substantial red wolf genetic ancestry persists in wild canids of southwestern Louisiana. Conservation Letters 2018:e12621.
Nowak, R. M. 1979. North American quaternary Canis. Monograph of the Museum of Natural History, University of Kansas 6:1-154.
Nowak, R. M. 1983. A perspective on the taxonomy of wolves in North America. Pp. 10-19 in Wolves in Canada and Alaska: Their Status, Biology, and Management, L. N. Carbyn, ed. Edmonton, Alberta: Canadian Wildlife Service.
Nowak, R. M. 1995. Another look at wolf taxonomy. Pp. 375-397 in Ecology and Conservation of Wolves in a Changing World, L. N. Carbyn, S. H. Fritts, and D. R. Seip, eds. Edmonton, Alberta: Canadian Circumpolar Institute.
Nowak, R. M. 2002. The original status of wolves in eastern North America. Southeastern Naturalist 1:95-130.
Parker, G. 1995. Eastern coyote: The story of its success. Halifax: Nimbus Publishing.
Phillips, M. K. and V. G. Henry. 1992. Comments on red wolf taxonomy. Conservation Biology 6:596-599.
Phillips, M. K., V. G. Henry, and B. T. Kelly. 2003. Restoration of the red wolf. Pp. 272-288 in Wolves: Behavior, Ecology, and Conservation, L. D. Mech and L. Boitani, eds. Chicago: University of Chicago Press.
Pomidor B. J., J. Makedonska, and D. E. Slice. 2016. A landmark-free method for three-dimensional shape analysis. PLOS ONE 11(3): e0150368.
Pritchard, J., M. Stephens, and P. Donnelly. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945-959.
Riley, G. A., and R. T. McBride. 1972. A Survey of the Red Wolf (Canis rufus). Scientific Wildlife Report No. 162. Washington, DC: U.S. Fish and Wildlife Service.
Roy, M. S., E. Geffen, D. Smith and R. K. Wayne. 1996. Molecular genetics of pre-1940 red wolves. Conservation Biology 10:1413-1424.
Rutledge, L. Y., K. I. Bos, R. J. Pearce and B. N. White. 2010a. Genetic and morphometric analysis of 16th century Canis skull fragments: Implications for historic eastern and gray wolf distribution in North America. Conservation Genetics 11:1273-1281.
Rutledge, L. Y., B. R. Patterson, and B. N. White. 2010b. Analysis of Canis mitochondrial DNA demonstrates high concordance between the control region and ATPase genes. BMC Evolutionary Biology 10:215.
Schrecengost, J. D., J. C. Kilgo, H. S. Ray, and K. V. Miller. 2009. Home range, habitat use, and survival of coyotes in western South Carolina. American Midland Naturalist 162:346-355.
Sinding, M. S., S. Gopalakrishan, F. G. Vieira, J. A. Samaniego Castruita, K. Raundrup, M. P. Heide Jørgensen, M. Meldgaard, B. Petersen, T. Sicheritz-Ponten, J. B. Mikkelsen, U. Marquard-Petersen, R. Dietz, C. Sonne, L. Dalén, L. Bachmann, Ø. Wiig, A. J. Hansen, and M. T. P. Gilbert. 2018. Population genomics of grey wolves and wolf-like canids in North America. PLOS Genetics 14:e1007745.
Slice, D.E. 2007. Geometric morphometrics. Annual Review of Anthropology 36:261-281.
Smith, D. W., R. O. Peterson, and D. B. Houston. 2003. Yellowstone after wolves. Bioscience 53:330-340.
Tang, H., M. Coram, P. Wang, X. Zhu, and N. Risch. 2006. Reconstructing genetic ancestry blocks in admixed individuals. American Journal of Human Genetics 79:1-12.
Tappen, M. 1994. Bone weathering in the tropical rain forest. Journal of Archaeological Science 21:667-673.
Tedford, R. H., X. Wang, and B. E. Taylor. 2009. Phylogenetic systematics of the North American fossil Caninae (Carnivora: Canidae). Bulletin of the American Museum of Natural History 325:1-218.
Vilà, C., I. R. Amorim, J. A. Leonard, D. Posada, J. Castroviejo, F. Petrucci-Fonseca, K. A. Crandall, H. Ellegren, and R. K. Wayne. 1999. Mitochondrial DNA phylogeography and population history of the grey wolf Canis lupus. Molecular Ecology 8:2089-2103.
vonHoldt, B. M., D. R. Stahlder, D. W. Smith, D. A. Earl, J. P. Pollinger, and R. K. Wayne. 2008. The genealogy and genetic viability of reintroduced Yellowstone grey wolves. Molecular Ecology 17(1):252-274.
vonHoldt, B. M., J. P. Pollinger, D. A. Earl, J. C. Knowles, A. R. Boyko, H. Parker, E. Geffen, M. Pilot, W. Jedrzejewski, B. Jedrzejewska, V. Sidorovich, C. Greco, E. Randi, M. Musiani, R. Kays, C. D. Bustamante, E. A. Ostrander, J. Novembre, and R. K. Wayne. 2011. A genome-wide perspective on the evolutionary history of enigmatic wolf-like canids. Genome Research 21:1294-1305.
vonHoldt, B. M., J. A. Cahill, Z. Fan, I. Gronau, J. Robinson, J. P. Pollinger, B. Shapiro, J. Wall, and R. K. Wayne. 2016. Whole-genome sequence analysis shows that two endemic species of North American wolf are admixtures of the coyote and gray wolf. Science Advances 2(7):e1501714.
vonHoldt, B. M., J. A. Cahill, Z. Fan, I. Gronau, J. Robinson, J. P. Pollinger, B. Shapiro, J. Wall, and R. K. Wayne. 2017. Response to Hohenlohe et al. Science Advances 3(6):e1701233.
Waples, R. S., R. Kays, R. J. Fredrickson, K. Pacifici, and L. S. Mills. 2018. Is the red wolf a listable unit under the U.S. Endangered Species Act? Journal of Heredity 109:585-597.
Wayne, R. K., and S. M. Jenks. 1991. Mitochondrial DNA analysis implying extensive hybridization of the endangered red wolf Canis rufus. Nature 351:565-568.
Wilson, P. J., S. Grewal, T. McFadden, R. C. Chambers, and B. N. White. 2003. Mitochondrial DNA extracted from eastern North American wolves killed in the 1800s is not of gray wolf origin. Canadian Journal of Zoology 81:936-940.






















