2
Data Sharing and Data Preservation
THE BURDENS AND BENEFITS OF “LONG-TAIL” DATA SHARING
Adam Ferguson, University of California, San Francisco
Adam Ferguson, University of California, San Francisco, explained that injuries to the central nervous system (CNS) are incredibly complex, in part because the human brain has 100 trillion synapses and the spinal cord has hundreds of billions of synapses. This complexity creates a data science problem with implications for public health. Traumatic brain injuries (TBIs) cost $400 billion annually worldwide, and spinal cord injuries (SCIs) cost $40 billion annually in the United States alone. Magnifying the problem is the absence of any U.S. Food and Drug Administration-approved therapies for TBIs or SCIs alongside the abundance of tiny measures of biofunction related to TBIs and SCIs. Ferguson asserted that sharing data and making them interoperable is the best strategy to better understand these complex disorders.
Ferguson described the bottleneck that is created when researchers perform data entry and curation on enormous amounts of heterogeneous raw data. He asserted that databases are not typically equipped to handle volume, velocity, and variety of data, the last of which is particularly relevant in the study of CNS injuries. He explained that relatively little organized big data exist throughout biomedicine. Most data fall in the long tail of the distribution, where there are modestly sized data sets and many heterogeneous data sets.
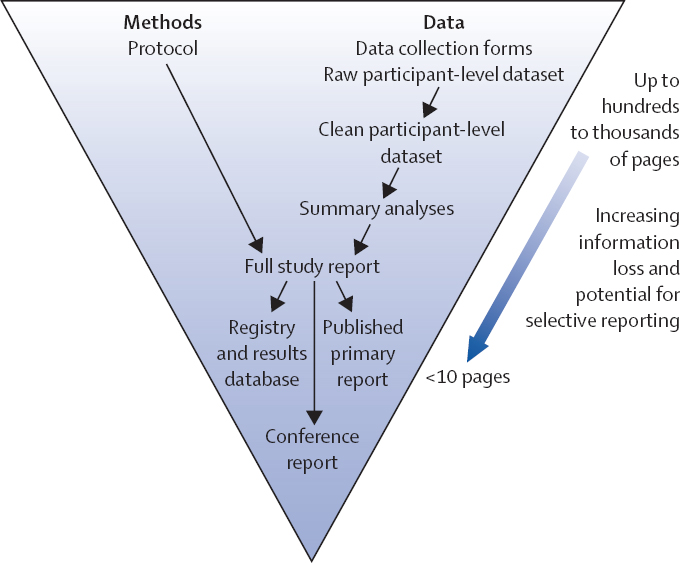
Ferguson said that published literature contains approximately 15 percent of data, which means that approximately 85 percent of data collected worldwide for biomedical research is unpublished, “dark” data. He estimated that more than $200 billion of the $240 billion worldwide annual biomedical research budget is wasted on data that are inaccessible (see Macleod et al., 2014). Furthermore, because all of the published biomedical literature contains only 15 percent of the total data collected, published research represents a biased sample of the full range of biomedical data available (see Ioannidis, 2015). Ferguson described this as a systemic problem within scientific publications, which summarize data instead of provide data. Researchers spend most of their time creating protocols, and information can be lost during the process of developing a brief high-impact paper (see Figure 2.1). This “ancient data-sharing technology” for biomedical research should be replaced with modern data repositories, he asserted.
Ferguson described an initiative to create a multispecies data repository called VISION-SCI, which contains approximately 60 million data points from 4,000 rats and mice with SCIs. These data are comingled with deidentified human medical records. This effort began with a $1 million grant for data curation but enabled access to nearly $70 million of prior research investment and data collection from the National Institutes of Health (NIH). He added that post-data collection cleaning and curation (the need for which is realized at the point of data sharing) typically requires 15–20 percent of a researcher’s total budget.
The SCI Open Data Commons initiative1 is another path forward in the field—it hosts a web portal that allows people to access VISION-SCI and to contribute and manage their own data. This initiative has expanded with the development of the TBI Open Data Commons2 and the Veterans Affairs Gordon Mansfield SCI Consortium,3 the latter of which is focused on translational SCI stem cell therapies. Ferguson also described Transforming Research and Clinical Knowledge (TRACK)-TBI4 and TRACK-SCI, which are large-scale clinical observation studies designed to generate high-quality clinical data. In July 2019, TRACK-TBI had 3,500 patients enrolled from 18 U.S. Level-1 trauma centers. He pointed out that all of these initiatives are guided by the FAIR (findable, accessible, interoperable, reusable) principles for data stewardship—biomedical research data
___________________
1 For more information about the SCI Open Data Commons, see https://scicrunch.org/odc-sci, accessed August 2, 2019.
2 For more information about the TBI Open Data Commons, see http://odc-tbi.org, accessed August 2, 2019.
3 For more information about the Veterans Affairs Gordon Mansfield SCI Consortium, see http://grantome.com/grant/NIH/I50-RX001706-01, accessed October 8, 2019.
4 For more information about TRACK-TBI, see https://tracktbi.ucsf.edu/transforming-research-and-clinical-knowledge-tbi, accessed October 8, 2019.
should be findable (i.e., data and metadata are “uniquely and persistently identifiable” as well as human and machine readable), accessible (i.e., data are “reachable” by humans and machines “using standard formats and protocols”), interoperable (i.e., “data are machine readable and annotated with resolvable vocabularies and ontologies,” which is particularly challenging), and reusable (i.e., data are harmonized with data from other contributors) (Wilkinson et al., 2016). Following these guidelines elevates biomedical data from raw material to primary work products, he explained.
The SCI community was an early adopter of the FAIR principles, hosting a workshop in 2016 on FAIR data sharing,5 a workshop in 2017 on the policy needed to execute FAIR data sharing via an open data commons,6
___________________
5 For more information, see https://www.ninds.nih.gov/News-Events/EventsProceedings/Events/Spinal-Cord-Injury-Preclinical-Data-Workshop-Developing-FAIR, accessed September 12, 2019.
6 For more information, see https://www.sfn.org/Meetings/Neuroscience-2017, accessed September 12, 2019.
and a community hackathon in 2018,7 during which participants uploaded their data to the SCI Open Data Commons. He emphasized that digital development is not possible without support from the research community. For example, the SCI community constructed a private space to make data accessible. A researcher can upload data to this private space and deposit them in the open data commons, which comingles data across laboratories. Data citation standards are then applied via SciCrunch,8 Digital Object Identifiers are issued, and the data are released under a creative commons attribution license. This framework provides a single place in which SCI researchers can organize and publish their data as well as receive primary credit for the data as a work product.
Many opportunities arise once data are organized within this framework. For example, Ferguson’s team uses Syndromic Data Integration, which suggests that any individual outcome measure is but one potential window into the underlying syndrome of a CNS injury. His team’s primary objective is to understand how an individual compares to a group of individuals on particular variables. With the help of machine learning, individuals can be clustered based on their multidimensional location within the syndromic space. This level of understanding places a renewed emphasis on precision and reproducibility, he continued.
Ferguson concluded his presentation by sharing an anecdote about researcher Jessica Nielson, University of Minnesota, who made use of the 1994–1996 Multicenter Animal SCI Study (MASCIS), which was a blinded randomized multidrug multicenter clinical trial in rats. The data from this study were dispersed across shelves at seven U.S. laboratories after the hypothesis was disproven. Nielson collected these data from binders, scanned them into PDFs, and used machine learning tools to understand why the trial failed. She learned that despite having the same biomechanical injuries, two groups in the trial had dramatically different locomotor outcomes. Animals with high blood pressure on the operating table had poor long-term prognoses. She realized that blood pressure during SCI surgery was the predictor of long-term locomotor recovery, not medication (Nielson et al., 2015). Although researchers have long focused on how to avoid low blood pressure in humans in the operating room, this evidence could now prompt clinical research studies on the effects of high blood pressure in the operating room. Maryann Martone, University of California, San Diego, wondered what prompted Nielson to collect the records from MASCIS since animal records are generally not considered
___________________
7 For more information, see https://scicrunch.org/odc-sci/about/blog/1481, accessed September 12, 2019.
8 For more information about SciCrunch, see https://scicrunch.org, accessed August 2, 2019.
to be “data.” Ferguson responded that MASCIS was organized similar to a randomized control trial, with standardized forms and binders at all seven sites. As an SCI researcher, Nielson thought that there was value in animal research when treated like human clinical data. Ferguson asserted that generating new knowledge from old data reduces the number of animals used in future clinical trials (see Neff, 2018; Chakradhar, 2017) and advances data-driven scientific discovery.
David Chu, Institute for Defense Analyses, noted the value of sample design when the cost to collect all data points is too high. He pointed out that nonstatistically significant results are often crucial for scientific understanding, and he wondered whether researchers should sample dark data instead of trying to preserve all data. Ferguson noted that, with enough data and the application of appropriate machine learning methods, it might be possible to estimate effect sizes. Further, the amount of dark data being collected in biomedicine could be reduced if the community better supported data sharing.
Robert Williams, University of Tennessee Health Science Center, said that the creation of a journal series via Jupyter Lab Notebooks could help address long-tail data issues, although this may increase publication costs for researchers. Ferguson speculated that publications in the future might resemble the Internet—one could click to source data from links within a web-based version of an article. Patricia Flatley Brennan, National Library of Medicine (NLM), noted that the need for investigator interpretation of data will always exist; it is an investigator’s responsibility to share his or her perspective on data, especially when interim artifacts (e.g., preprints, models, and protocols) could be reconstituted by another investigator. Sharing data via publications is not sustainable because there may be multiple data resources linked to one publication or multiple publications linked to a single data resource. She explained that it is critical to understand future relationships among publication, accountability, knowledge building, and the roles that archival data can play.
William Stead, Vanderbilt University Medical Center, asked how Ferguson’s approaches to data sharing could reduce costs over time and increase sustainability. Ferguson said that data are more likely to be reused if they are made digital earlier in the process and if stakeholders have direct input into the study design. Jessie Tenenbaum, Duke University and the North Carolina Department of Health and Human Services, described her desire to “make data a first class citizen” (by prioritizing curation and annotation early in the process) and wondered if doing so is practical. Ferguson agreed that data should be a “first class citizen” and said that the adoption of standards (similar to peer review) is the key. Ilkay Altintas, University of California, San Diego, wondered about the potential for machine learning and artificial intelligence to be “disruptors”
for data curation, and Ferguson proposed inserting machine learning earlier in the curation process. Altintas predicted that there would be issues due to conflicting curation procedures between private and public archives and wondered how to capture the cost of curation if it is done automatically.
In response to a question about curation costs and associated standards from Alexa McCray, Harvard Medical School, Ferguson said that data curation costs increase when many thousands of variables are present in siloed data systems. He explained that although common data elements may exist in two data sets, interoperability drifts over time as various people begin to recode variables in published papers. Sarah Nusser, Iowa State University, estimated that data curation costs are 20 percent in the fields of astronomy and physics, and any large-scale research operation requires significant preplanning around the collection of information. She wondered how researchers should think, from the front end, about sharing data. Ferguson said that prospective clinical studies have a direct analogy to astronomy and physics in that they include a substantial amount of preplanning and still have high costs. He hopes to use findings from a study of ultra-dark data in the preclinical space to build living common data elements that can be updated more rapidly. An online participant wondered if costs for curation would decrease with the use of new methodologies to collect data. Ferguson expressed his hope that as data become increasingly digital, people will start to drive costs down by doing more curation at the front end of the process.
Ferguson said that the 10–15 percent of an overall project budget being used for curation is an over-time cost as opposed to an upfront cost (i.e., it is not included in the initial funding). Ferguson added that once data are collected and stored in an informatics system, registration requires significant personnel time. Warren Kibbe, Duke University, noted that a transformational cost still exists even with digital data; thus, cleaning data for a second time (and a different purpose) has to be accounted for in the total cost.
Brennan emphasized the need to consider the costs of curation at the point of use. Brad Malin, Vanderbilt University Medical Center, said that these costs are extremely high. For example, NIH’s All of Us Research Program9 is tasked with capturing genomic data, medical records data, and survey data on 1 million Americans; harmonizing these data; and making them accessible to the public. This post-hoc harmonization of electronic medical records data is a multiyear effort, Malin explained. Defining the
___________________
9 For more information about the All of Us Research Program, see Allofus.nih.gov, accessed September 25, 2019.
Observational Medical Outcomes Partnership (OMOP)10 language was relatively simple, but, at times, it has been difficult to translate the data onto a model because many records have incomplete or incorrect data, he continued. Problems also arise when there are differences in measurement techniques. He estimated that millions of dollars of an approximately $100 million budget are being spent to get the data into a usable form. Lars Vilhuber, Cornell University, said that what Malin described could be considered secondary data acquisition (as opposed to curation at the point of use), which results in an additional substantial expense. Philip Bourne, University of Virginia, observed that culture plays a prominent role in any discussion about curation at the point of collection and at the point of use—people have false expectations for the future based on what has happened in the past.
Monica McCormick, University of Delaware Library, asked about the role of the individual university in curation: Where should the tools be developed, and where should the training occur? Ferguson responded that it would be ideal if more university libraries accepted these responsibilities. Vilhuber pointed out that Ferguson’s streamlined approach to organize and publish data places more value on the data and thus increases the incentives to share and reuse data. However, if data are protected only by a creative commons attribution license, researchers might not have control over or earn credit for how those data are used in the future. Ferguson noted that a researcher’s motivation for sharing data is typically to advance scientific discovery. He added that it is unclear what motives prevent researchers from sharing their data; however, these motives will become clearer if funders begin to enforce data-sharing policies.
PANEL DISCUSSION: RESEARCHERS’ PERSPECTIVES ON MANAGING RISKS AND FORECASTING COSTS FOR LONG-TERM DATA PRESERVATION
Margaret Levenstein, University of Michigan, Moderator
Nuno Bandeira, University of California, San Diego
Jessie Tenenbaum, Duke University and the North Carolina Department of Health and Human Services
Georgia (Gina) Tourassi, Oak Ridge National Laboratory
Robert Williams, University of Tennessee Health Science Center
Margaret Levenstein, University of Michigan, moderated a panel discussion among researchers who were asked to share their individual
___________________
10 For more information about the Observational Medical Outcomes Partnership, see https://fnih.org/what-we-do/major-completed-programs/omop, accessed December 5, 2019.
perspectives on (1) managing risks and forecasting costs for long-term data preservation, archiving, and accessing decisions, with consideration for different kinds of biomedical data (e.g., clinical data, survey data, imaging data, genomic data) and research endeavors (e.g., collecting new data and leveraging existing data assets); (2) methods to encourage NIH-funded researchers to consider, update, and track lifetime data costs; and (3) challenges for academic researchers and industry staff to implement these tools, methods, and practices. She emphasized that expanding data curation and data-sharing efforts requires cultural change. With the data revolution, the roles of both data users and data producers are changing.
Nuno Bandeira, University of California, San Diego, described his interest in the development of algorithms for interpreting mass spectrometry data in metabolomics, proteomics, and natural products. The NIH-funded Center for Computational Mass Spectrometry at the University of California, San Diego, develops algorithms for large-scale analyses of mass spectrometry data and for two prominent service platforms for sharing mass spectrometry data. One platform is the Mass Spectrometry Interactive Virtual Environment,11 which contains more than 10,000 data sets that are assigned identifiers and shared in conjunction with publications. The other platform is the Proteomics Scalable, Accessible, and Flexible environment12 with more than 80 data analysis workflows. Bandeira noted that one of the Center’s key tasks is to develop tools to analyze and increase the value of data,13 which leads to the creation of new knowledge. Computer infrastructure and communities are needed to understand how data can be used to connect researchers who might unknowingly be working on related problems.
Bandeira noted that before discussing cost, it is imperative to develop domain-specific standards to determine what constitutes high-quality reusable data. A data set’s storage value for future research can be determined by its uniqueness and its potential for additional discoveries. He added that it is important to develop platforms that will engage researchers in an ongoing curation process (i.e., the data that are available are automatically integrated into ongoing research projects, and reanalysis goes back into the curation of the data in the repository). This type of centralized platform is a departure from the traditional repository model
___________________
11 For more information about the Mass Spectrometry Interactive Virtual Environment, see http://massive.ucsd.edu, accessed September 25, 2019.
12 For more information about the Proteomics Scalable, Accessible, and Flexible environment, see http://proteomics.ucsd.edu/ProteoSAFe, accessed September 25, 2019.
13 For more information about the software tools developed at the Center for Computational Mass Spectrometry, see http://proteomics.ucsd.edu/software, accessed September 25, 2019.
and promotes the exchange of knowledge as well as crowdsourcing to annotate data.
Tenenbaum stressed the value of user-friendly interfaces, visualization, and discoverability. Having worked on a large-scale community-based biorepository and registry of data (the Measurement to Understand the Reclassification of Disease of Cabarrus/Kannapolis study14), she noted that even experts are challenged with understanding relevant standards and what it means to annotate data. Her research group extracts symptom-related terms from electronic health record (EHR) data, which can be clustered for an analysis to understand underlying mechanisms of disease. She explained that curating, interacting with patients, obtaining consents, recruiting, and following-up are all important but expensive research activities. Thus, it is crucial to leverage data that are collected through clinical care and “build a learning health care system.” Working with structured mental health data and text includes access to fully identified, highly sensitive clinical data. One error could inadvertently expose these data or cause problems in the data analysis and the resulting documentation.
Tenenbaum commented that tools for calculating potential costs for storage would be helpful for researchers. A clarification of rules and clear policies would also be beneficial—for example, experts currently disagree about what is considered to be a breach of the Health Insurance Portability and Accountability Act (HIPAA). She noted that grant budgets should include specific descriptions for tracking lifetime data costs. She also suggested an incentive system in which people who do not share data are either prevented from securing an indexed publication in PubMed or are added to a “wall of shame” (“sticks”), while people who exhibit best practices receive recognition (“carrots”). She cautioned that adding components to an already requirement-laden grant proposal process could create additional challenges.
Georgia (Gina) Tourassi, Oak Ridge National Laboratory, explained that the big data revolution in biomedical science began in radiology. She added that the U.S. Department of Energy (DOE) laboratories pride themselves on being stewards of data and tools across various domains. For example, in a partnership between DOE and the National Cancer Institute, Tourassi explores the use of high-performance computing and large-scale data analytics on cancer registry data—70 percent of data collected across cancer registries is text data that need to be curated. Artificial
___________________
14 For more information about the Measurement to Understand the Reclassification of Disease of Cabarrus/Kannapolis study, see https://globalhealth.duke.edu/projects/measurement-understand-reclassification-disease-cabarruskannapolis-study-murdock, accessed October 8, 2019.
intelligence technologies can be used to abstract information from the data, and computational models of patient trajectories can be developed. The ultimate goal is to deliver tools to data owners (in this case, the cancer registries) and to distribute and scale algorithms that will enable them to train their own data using open compute resources. She emphasized that advances in biomedical science arise from using various data modalities to present a holistic view of a patient. However, it is challenging to do scalable data-driven discovery with heterogeneous data sources, including nontraditional data sets that could provide additional insights into patient care.
Tourassi wondered about the cost of data storage versus the cost of data analysis. Although colocating compute and data can make work-flow and data management more feasible and more cost-effective, continued infrastructure investments are necessary. Questions also remain about the ownership of patient data and their derivatives as well as the legal responsibility for the deidentification of data. Hardware, software, and algorithmic approaches to enable collaboration are needed, as are increased incentives to share data. She reiterated Bandeira’s assertion that the value of data will determine cost. Tourassi concluded by emphasizing the value of sustained infrastructure investment, which can advance innovation, support scientific discovery, and improve clinical practice.
Robert Williams, University of Tennessee Health Science Center, began his career with a study of brain architecture, which demanded a global approach and the integration of data with primitive tools such as Excel, FileMaker, and FileVision. After collecting data for 7 years, he received funding from the Human Brain Project to scale up his efforts. His primary objective is to enhance precision medicine by building animal model resources (in addition to human cohorts) that allow the incorporation of genetic diversity into mice and rats. He asserted that if data sit in a silo and cannot be correlated to anything, they are not of high value. However, if data can be acquired in a multiscalar way, as Ferguson described, additional vectors of data become multiplicative; in order for this to work computationally, web services and other tools have to be developed.
Williams said that open source code tools are useful, but they can be unpredictable. He emphasized that low-cost, secure enclaves are needed for protected health information, EHRs, and genomes to capture long-tail data. Encryption technologies that still allow data to be computable are also essential. He noted that both “carrots and sticks” should be used to motivate researchers to manage data and costs. However, he acknowledged that it is difficult to make predictions and develop solutions in a highly changeable environment. He asserted that while data loss and cost are problematic, capturing missing information (i.e., initial data, metadata) is a more immediate concern. He mentioned the importance
of motivating researchers to use the InterPlanetary File System15 and to have reputable workflows (e.g., via Guix, Galaxy, Jupyter, Open Science Foundation). Ultimately, he said, researchers should push against static publications (and move to Jupyter or R Shiny), allowing the data to “breathe and breed.” The scientific culture needs to move away from primarily storytelling: Journal readers need to be able to read the narrative, see the data, and validate the narrative in real time.
Levenstein reiterated that the goal of the National Academies’ study is to help researchers and funders better integrate risk management. She pointed out that all of the panelists mentioned an element of risk related to privacy and confidentiality, and she wondered about the following:
- What do researchers need to be able to forecast risks and costs in grant proposals?
- How can researchers think simultaneously about the cost of computing and the cost of preserving data?
- What are the incentives for researchers to share data and manage risks? Which “carrots and sticks” resonate most with researchers?
- Can economic and cultural standards change so that researchers willingly take on the costs of making their data available?
Bandeira described data as “having a life of their own” after an initial narrative is published. Therefore, to accurately assess curation costs, he said that it is important to think about data as a work in progress instead of as an end product. He added that if an investigator can use the same platform for both data analysis and data sharing, incentives increase and costs decrease. The Center for Computational Mass Spectrometry’s systems offer continuous reanalysis of data; as new knowledge becomes available, it is automatically transferred to the data sets over the course of a project, thus reducing the data analysis burden for researchers and connecting them to other researchers with overlapping data sets. Levenstein championed the role of data sharing in community building, especially for interdisciplinary research.
Tourassi said that the cost of data storage is expected to continue to decrease but computing costs can vary by domain. Moving data can also require substantial investments in both time and money; thus, costs need to be evaluated early in the process, especially for projects that require continuous data movement. Capital, operational, and maintenance expenditures need to be considered when building and sustaining an infrastructure that can keep pace with evolving software and operating systems. To
___________________
15 For more information about the InterPlanetary File System, see ipfs.io, accessed December 5, 2019.
avoid burdening researchers with software engineering, she suggested bringing scientists and engineers together to create a sustainable ecosystem. An online participant commented that technologists who maintain the operating systems of infrastructure are often the same people who maintain the technological capabilities for the rest of an organization. This participant wondered about the costs for such a model and whether it is sustainable for data maintenance. The participant added that without the right incentives, organizational-level information-technology teams could become barriers instead of enablers, prompting researchers to take things into their own hands.
Levenstein noted that data are active resources that extend beyond FAIR principles, thus creating both challenges and opportunities for the future. Tenenbaum asserted that financial risks and privacy risks are not mutually exclusive—when a breach occurs, both types of risk are realized—and noted that data provenance is another area in which to consider risk. Altintas emphasized the need for NLM to consider the risks and costs associated with data sharing and supported the development of a neutral cross-agency strategy to address health and privacy risks for the future of science. She asked panelists about their visions for the future. Tenenbaum suggested increased public–private partnerships; however, she recognized that this would add another layer of complexity to an already complicated process. Tourassi described the challenges of working with data across registries, including registry-specific restrictions as well as multiple memoranda of understanding, data use agreements, and business associate agreements. Additional complexities arise in private partnerships, particularly in terms of the ownership of intellectual property. A neutral entity could deploy centralized models to each registry to enable data sharing across registries (i.e., knowledge would be shared without each registry seeing the others’ data). Bandeira pointed out that it is difficult to analyze heterogeneous data from different siloes, which in turn affects the data’s value. He said that it is not necessary for one repository to have both the expertise to manage a particular data type and the ability to store and compute those data. There should be one place where all of the data can be stored, but the different entities that manage each data type and repository are still research endeavors that should be awarded separately. This approach dissociates the compute and storage capabilities from the infrastructure needed to organize, process, and connect data. An interoperable platform would bring the tools, data, and computing capabilities together, better connecting tool developers with the research. Because privacy will always be a concern, Tenenbaum suggested the increased use of application programming interfaces (APIs) to share information (as opposed to data). Tourassi noted that in her partnership with the National Cancer Surveillance program, tools are being built
and deployed in the form of APIs. Such an ecosystem makes it possible to offer both open and restricted access. She added that it is important to consider the potential for adversarial use of algorithms; a benchmarking process could help determine the accuracy and vulnerability of algorithms to this misuse.
In response to a question from Martone, Bandeira said that any proteomics data that were assigned identifiers can be found in the ProteomeXchange consortium. With the emergence of tools that allow for joint analysis, there is an influx of transcriptomics data surfacing alongside the proteomics data. Martone wondered how repositories could communicate and coordinate in the absence of a centralized entity, and Bandeira noted that replication would be unavoidable.
Martone asked the panelists how career and grant cycles drive costs for and decisions about data. Tenenbaum said that the cycle itself provides the “carrot” for researchers to document metadata, which makes it possible for a project to continue even after a graduate student or other researcher departs. Williams added that his team typically prioritizes generating high-quality data over creating a narrative. Tourassi suggested employing a data manager and a software engineer for each project. Creating a culture of good practices requires support from the top down, she continued. Bandeira said that for long-term projects, data sets should be available for follow-up analysis and independent reanalysis. With the right tools to enable data sharing, he said that data will improve with age and become enduring resources for the research community.













