During the second session of the workshop, panelists explored the current state of the science about human extraembryonic lineages, including how they are defined and characterized, and they discussed the impact of extraembryonic lineages on human embryo model systems. The session was moderated by Amander Clark, a professor in and the chair of the Department of Molecular, Cell, and Developmental Biology at the University of California, Los Angeles. Clark opened the session by commenting that the talks would focus on the difficult question of what gives an embryo organismal potential.
MODELING TROPHOBLAST DIFFERENTIATION USING PLURIPOTENT STEM CELLS
Mana Parast, a professor in residence of pathology at the University of California, San Diego, presented on the use of pluripotent stem cells to model trophoblast differentiation in humans. She described trophoblast stem cells (TSCs) and the process of trophoblast differentiation, highlighting two unique markers that occur in the human but not in the mouse, and she explained how her laboratory developed their stepwise model of trophoblast differentiation. She also discussed their work in deriving TSCs from human pluripotent cells and explored the role of bone morphogenetic protein (BMP) signaling.
Trophoblast Stem Cells in Human and Mouse
Parast provided an overview of trophoblast stem cells. The concept of “pluripotent stem cell–derived trophoblast” itself is somewhat contentious, she said, because the term “totipotent” instead of “pluripotent” is usually used to characterize the cells that give rise to trophoblast (see Box 3-1). Based on data primarily from mouse models, pre-implantation inner cell mass (ICM) is thought to give rise to embryonic stem cells (ESCs) that form the embryo proper and the extraembryonic endo-
derm and mesoderm. The trophectoderm (i.e., the outer cells of the pre-implantation blastocysts) are thought to contribute only to TSCs and other trophoblast lineages. In the mouse, TSCs can be derived from pre-implantation blastocyst as well as from extraembryonic ectoderm up to embryonic day 8 (Tanaka et al., 1998). Mouse TSCs respond to fibroblast growth factor 4 (FGF4) and Activin signaling; in the absence of these factors, the cells differentiate into a combination of labyrinthine trophoblast (SynT I, SynT II, and sinusoidal giant cells) and junctional zone trophoblast (spongiotrophoblast and trophoblast giant cells). Human TSCs respond to completely different growth factors, Parast said: Wnt, epidermal growth factor (EGF), and transforming growth factor beta (TGFβ) inhibitor. Human TSCs can be derived from both blastocyst-stage embryos and early first-trimester placental tissues (Okae et al., 2018). Human TSCs are thought to be derived from the cytotrophoblast (CTB) progenitor layers, which also give rise to syncytiotrophoblast (STB) and extravillous trophoblast (EVT).
Trophoblast Differentiation
Parast went on to describe the process of trophoblast differentiation. As illustrated in Figure 3-1, the trophectoderm is believed to initially proliferate and give rise to the progenitor CTB cells. In early embryonic development, the CTB cells proliferate; these cells are primarily positive for EGF receptors as well as other markers. In anchoring villi, CTB differentiate into EVT, cells that invade the uterine tissue; they are characterized by human leukocyte antigen-G (HLA-G) expression and secrete matrix metalloproteinases (MMPs). In floating villi, CTB undergo cell–cell fusion to form STB cells that secrete human chorionic gonadotropin (hCG) and are involved
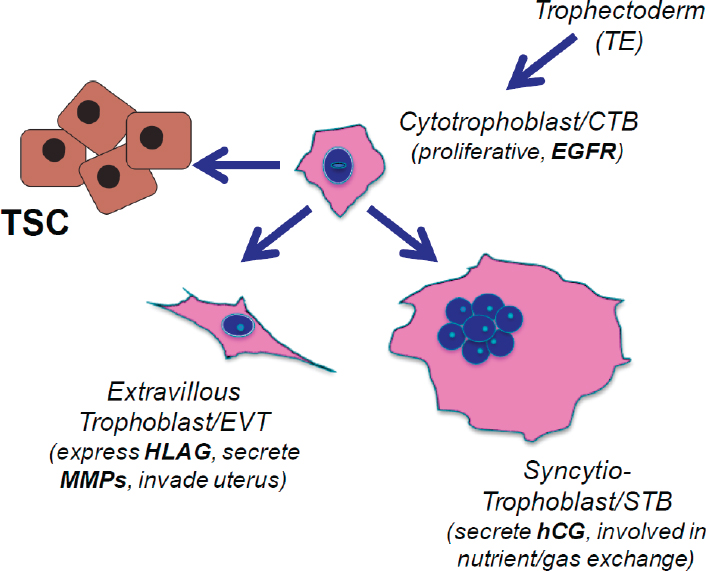
NOTE: CTB = cytotrophoblast; EGFR = epidermal growth factor receptor; EVT = extravillous trophoblast; hCG = human chorionic gonadotropin; HLAG = human leukocyte antigen-G; MMP = matrix metalloproteinase; STB = syncytiotrophoblast; TE = trophectoderm; TSC = trophoblast stem cell.
SOURCE: Mana Parast workshop presentation, January 17, 2020.
in nutrient and gas exchange. The niche for TSCs is presumed to be within the CTB layer, Parast added.
Markers of Human Trophoblast Differentiation
Two specific markers of CTB and TSCs found in the human but not in the mouse were highlighted by Parast, and she noted that there are many other differences as well. The first marker is p63, a member of the p53 family of nuclear proteins that is involved in the maintenance and differentiation of stem cells in stratified epithelia (Li et al., 2013). Unlike the mouse trophoblast, the human trophoblast is organized (particularly in the villi) as a stratified epithelium with the p63-positive CTB layer being closest to the mesenchymal layer. In humans, p63 is absent in the amnion. In the human
placenta, p63 expression is confined to trophoblasts, and it is expressed in all proliferative CTBs (Lee et al., 2007b). A more recently identified marker that is uniquely expressed in the human placenta is vestigial like family member 1 (VGLL1) (Soncin et al., 2018), which is a human homolog of the Drosophila vestigial gene that is involved in wing development. Although little is known about the function of this gene, it is known to be a binding partner for TEA domain proteins, making it a promising candidate for evaluation in the context of human trophoblast lineage specification, Parast said. Early evidence suggests that VGLL1 and TEAD4 may come together to induce p63 and trophoblast lineage in humans, much like YAP and TEAD come together to induce CDX2 and trophoblast lineage in the mouse. According to unpublished work that Parast has done with Kathy Niakan, this supposition is supported by evidence that VGLL1 and p63 are expressed in the very early trophectoderm layer of human blastocyst-stage embryos.
Modeling Human Trophoblast Differentiation
Prior to the derivation of human TSCs, models of human trophoblast differentiation were poor because the cell lines available were not fully representative of the in vivo cell type, Parast explained. Primary cells derived from the placenta are the “gold standard” in the field, but access to those cells is challenging due to issues around tissue availability. Term placentas are the most widely accessible, but term CTB can only differentiate into STB. First-trimester CTB is required to study EVT differentiation—because it can differentiate into both STB and EVT—but access to those tissues is limited and is complicated by ethical concerns. This has motivated research into the use of human pluripotent stem cells (hPSCs) to model trophoblast differentiation. One strategy used to derive trophoblast from hPSC is to culture embryoid body-derived trophoblast using 3D Matrigel (Giakoumopoulos and Golos, 2013). Another is to treat hPSC with bone morphogenetic protein 4 (BMP4) in 2D culture;1 this strategy has been used to induce cells that resemble STB (Xu et al., 2002) as well as HLA-G-positive cells (Roberts et al., 2018).
Parast’s laboratory investigates whether the treatment of human ESCs with BMP4 is a developmentally accurate model (i.e., whether the CTB cells are being obtained before the cells differentiate terminally into STB and EVT cells). Using p63 as a marker, they found that down-regulation of p63 inhibits trophoblast differentiation of human ESCs (Li et al., 2013). The cells still differentiate, but not into the terminally differentiated trophoblast that expresses HLA-G. They also observed blunted secretion of hCG and
___________________
1 BMP4 is part of the TGFβ superfamily of proteins.
hyperglycosylated hCG. Gene expression profiling showed that the cells cluster most closely with first-trimester placental tissue and CTB rather than with other fetal and placental cells and tissues.
Developing a Stepwise Trophoblast Differentiation Model
A major criticism of the BMP-induction approach to derive CTB cells is that BMP4 is known to induce mesoderm, Parast said. However, evidence suggests BMP4 induction of mesoderm is Wnt-dependent, while BMP4 induction of trophoblast appears to be Wnt-independent (Kurek et al., 2015). Based on this finding, Parast and colleagues developed a two-step differentiation model that uses an Inhibitor of Wnt Production-2 (IWP2) in combination with BMP4 (Horii et al., 2016, 2019). Initially, this facilitates a consistent and uniform differentiation of hPSCs into CTB-like cells. A subsequent re-plating of the cells on feeder-conditioned media derives a more differentiated cell type. The advantage of this approach is that they were able to separate trophoblast lineage specification from terminal trophoblast differentiation so that the cells could be compared with primary placenta-derived CTB cells, Parast said. After the first differentiation step, Parast and her colleagues had CTB-like cells that were positive for p63, CDX2, nuclear YAP, and TEAD4. After a terminal differentiation of derived CTB cells in the second step, the majority of cells were multinucleated STB-like cells and produced hCG; 5–20 percent of the cells were EVT-like and HLA-G positive. This approach may offer an improved method to evaluate trophoblast derived from hPSCs, she said. When Parast’s team compared the hPSC-derived CTB-like cells with primary CTB cells, both cell types responded to low oxygen in a similar manner (Horii et al., 2016): STB differentiation was inhibited and EVT differentiation was enhanced. Parast and her colleagues also used cells affected by trisomy 21 to demonstrate that the cells showed blunted STB differentiation that could be rescued by treatment with Activin A, just like primary CTB cells with trisomy 21.
Deriving Trophoblast Stem Cells from Human Pluripotent Cells
Another area of exploration for Parast and her colleagues involves understanding whether TSCs can be derived from human pluripotent cells. They evolved their two-step protocol by applying the media protocol used to derive human TSCs by Okae et al. (2018) to the CTB-like cells from the first step of their stepwise trophoblast differentiation model. This resulted in cells that look morphologically similar to TSCs after a few passages in the media. The cells produced by this process were positive for EGFR and found to express p63, VGLL1, GATA3, and other CTB markers. The team is currently performing ESC/trophoblast stem cell (TSC) aggregation
assays to determine if the TSCs have the potential to aggregate and form embryonic-like structures. According to unpublished work by Tony Bui and Mariko Horii, an advantage of using the Okae protocol appears to be that it increases the amount of EVT-like cells by more than 80 percent, Parast said. This addresses a limitation of the second step of the previous stepwise approach, in which just 5–20 percent of the cells were EVT-like. Parast’s colleagues used the updated protocol with several lines of induced pluripotent stem cells (iPSCs) derived from patients with preeclampsia. The phenotype observed using the previous two-step protocol was somewhat variable, but Bui and Horii’s approach reveals a distinct phenotype between the preeclamptic and normal cell types.
BMP Signaling: Beyond Mesoderm Induction
Parast considered the role of BMP signaling beyond mesoderm induction. BMP machinery has been shown to be present in human trophectoderm (Blakeley et al., 2015). BMP signaling is also known to be required for the development of extraembryonic lineages, including trophectoderm, in the mouse pre-implantation period (Graham et al., 2014). According to unpublished work by Jennie Au and Francesca Soncin, when BMP4 is overexpressed in differentiating cultures of mouse TSCs, it blunts and inhibits differentiation. This suggests that BMP4 plays a role in the maintenance of the undifferentiated state in mouse TSCs, Parast said.
Features of Trophoblast Derived from Human Pluripotent Cells
In conclusion, Parast reflected on the question of whether hPSC-derived trophoblast cells are “real” or not. Given that the human embryo and mouse embryo develop at different rates, there may be a human-specific process by which the hESCs may not have yet undergone lineage commitment. However, BMP4 has been shown to induce some trophoblast markers in mouse ESC and mouse epiblast-derived stem cells (EpiSCs). This suggests that the process may not necessarily be human-specific, she said. As to whether it is strictly an in vitro phenomenon of “transdifferentiation,” Parast countered that there may be in vivo stress conditions whereby cells derived from ESCs or EpiSCs can contribute to trophoblast. Alternatively, hPSCs or a subpopulation thereof may not even be pluripotent stem cells—they might be totipotent, because certain types of mouse ESCs have been shown to cycle between the totipotent and pluripotent state (Macfarlan et al., 2012).
NATURE OF TROPHOBLAST GENERATED FROM EMBRYONIC AND INDUCED PLURIPOTENT STEM CELLS
R. Michael Roberts, the Chancellor’s Professor of animal sciences and biochemistry at the University of Missouri, examined the nature of trophoblast generated from embryonic and induced pluripotent stem cells by describing another model system that has been used to study the formation of early trophoblast from ESCs. The model uses BMP4 and inhibitors of FGF and TGFβ signaling to drive human-epiblast-type (often called “primed”) ESCs and iPSCs to trophoblast. He addressed criticism that the hESCs do not fully differentiate to trophoblast in the model.
The first reported use of BMP4 to drive trophoblast formation was conducted by Xu et al. (2002), but there was also evidence in the microarray data for the presence of derivatives of both the endoderm and mesoderm and hence a mixed-lineage cell population. Further work by several groups has refined this model by excluding the factors that maintain pluripotency and including various factors that inhibit FGF2 and TGFβ/ACTIVIN signaling, which together cause the BMP4-primed cells to differentiate uni-directionally to trophoblast populations composed of both mononucleated small cells and clumps of multinucleated STB. A host of subsequent studies have used this approach to follow trophoblast emergence and the differentiation of the other sub-lineages, Roberts said.
Use of Stem Cell–Derived Trophoblasts as a Model for Placentation
Stem cell–derived trophoblasts can be used as a model for early placentation, Roberts said. The process is initiated by a brief treatment of the ESC/iPSC colonies with proteinases before cutting the colonies up into individual pieces of about 200 cells each, which are seeded into new medium, where each begins to form a new colony. The ESC/iPSC colonies are exposed to BMP4 and the FGF2 and TGFβ signaling inhibitors (the so-called BAP treatment) to drive the differentiation to trophoblast. By around day 4, uneven surfacing is evident in the colonies, a feature that becomes more extensive as culture proceeds and that reflects the emergence of STB. The cells produce increasingly larger amounts of hCG between days 6 and 8, probably reflecting the requirement for hCG production by the embryo as it implants, the need to ensure successful maternal recognition of pregnancy, and the continued production of progesterone by the maternal ovary. Earlier (days 2–6 of BAP treatment) one typically sees the emergence of cells that are HLA-G positive. These cells can invade a Matrigel matrix and move through pores in a membrane below, thereby
allowing their invasiveness to be measured. Roberts emphasized that in addition to invasive features, BAP-treated hESCs clearly have a phenotype that is similar to placental trophoblast and dissimilar to any other organs and tissues. Studies of the transcriptome of BAP-treated hESC demonstrate a lack of any signature corresponding to mesoderm or other lineages except trophoblast (Jain et al., 2017). Roberts went on to outline the knowledge accumulated thus far on this BAP-treated hESC model (see Box 3-2).
Nature of BAP-Generated Trophoblast
Having established that it is not mesoderm, Roberts then spoke about the nature of this BAP-generated trophoblast, which is a focus of ongoing research. He presented a principal component analysis of transcriptomic landscapes (both single-cell and whole transcriptome) of human trophoblasts obtained from placental and tissue culture origins in order to highlight the similarities and differences between the ESC-derived trophoblast and trophoblast from other sources. He concluded that the ESC-derived cells most strongly resembled trophoblast differentiated from the trophoblast stem cells described by Okae et al. (2018).
Dynamics of Early Trophoblast Development in Cultured Human Embryos
To frame his discussion of the dynamics of early trophoblast development in cultured human embryos, Roberts sketched the early stages of human placental development. The early stages of implantation are associated with the formation of an invasive STB that “burrows” into the endometrium. He suggested that BAP-differentiated cells correspond most closely to this syncytial mass associated with embryo implantation rather than to the STB associated with either mature placental villi or even the little understood, multi-nuclear trophoblast giant cells embedded deep in the endometrium (Turco and Moffett, 2019). However, some unique markers are shared between ESC-generated trophoblast and the STB associated with early placental villi during the early weeks of the first trimester of a human pregnancy.
Roberts then described a single-cell RNAseq analysis performed on cultured human blastocysts, which, between days 8 and 12 of culture, appear to develop much as they appear to do in vivo, including forming an outer shell of STB and producing large amounts of hCG (West et al., 2019). By showing a live cell microscopy movie during the workshop, Roberts demonstrated the materialization of a hitherto unknown, migratory trophoblast cell around embryonic day 10 that moves away from the embryo. These cells are positive for HLA-G and share some markers with the extravillous trophoblast cells that later in placental development invade the endometrium from the tips of anchoring placental villi. The single-cell RNAseq analysis also revealed how proliferating cytotrophoblast closer to the embryo proper give rise to two more differentiated populations of cytotrophoblast, one with some of the markers defining the motile cells, and the other carrying some STB markers shared by the large STB cells (West et al., 2019). Roberts concluded that the BAP-differentiated trophoblast generated from ESCs and iPSCs might provide ideal models for studying these early stages of human pregnancy without using human embryos.
“Failed” Criteria for ESC-Derived Trophoblast
The BAP model has been criticized on various grounds, Roberts said (Lee et al., 2016). One line of criticism is that the ELF5 promoter in the differentiating cells remains hypermethylated; however, this is perhaps not surprising because early human trophoblast appears not to express the ELF5 gene extensively, if at all. Another criticism is the lack of expression of C19-encoded miRNA; Roberts conceded that the ESCs down-regulate this locus as they differentiate, but it is unclear what this means. However, he rebutted the criticism that ESC-derived trophoblast do not express HLA-G
and have an uncharacteristic HLA profile. All papers where an adequate analysis was performed have revealed robust production of HLA-G and the full-length HLA-G transcript, he said. The perceived lack of positive trophoblast markers is also unquestionably wrong. In conclusion, Roberts maintained that BAP-treated human ESCs fully differentiate to trophoblast, although that trophoblast is different from that associated with villous trophoblast later in pregnancy.
Roberts added that there are several potential uses of trophoblast generated from ESCs and iPSCs, in addition to studying early trophoblast differentiation. These potential uses include (1) assessing toxicology and responses to foreign agents, (2) generating TSCs, (3) following trophoblast lineage divergence, (4) studying origins of placental disease (e.g., preeclampsia), and (5) evaluating the susceptibility of early trophoblast to pathogens (e.g., Zika virus) (Sheridan et al., 2018, 2019; Yang et al., 2015).
MOLECULAR INNOVATION IN THE HUMAN TROPHOBLAST LINEAGE
Paul Robson, an associate professor and the director of single-cell biology at The Jackson Laboratory in Farmington, Connecticut, discussed molecular evolutionary innovation in the human trophoblast lineage. He noted that Parast and Roberts provided evidence that it is possible to get trophoblast from pluripotent cells and that these models provide an opportunity to look at the earliest states of human trophoblast. In his presentation he emphasized the molecular differences between species by focusing on work that uses those models to study the cells’ molecular features and uses comparative genomics to look at the sequences that are functional in these cells.
Variation in Early Development Across Species
Robson opened by explaining the concept of conservation of evolution and development. As reflected in the “hourglass” model, it is thought that species-to-species differences in the developing organisms are most pronounced early in development and late in development, with the conserved part of development in the phylotypic stage (Kalinka and Tomancak, 2012) (see Figure 3-2). The phylotypic stage occurs much later in development than the blastocyst: in the human, it occurs 4 weeks post-fertilization; in the mouse, it occurs about 10 days post-fertilization. Based on the hourglass model, significant differences would be expected in comparative embryology.
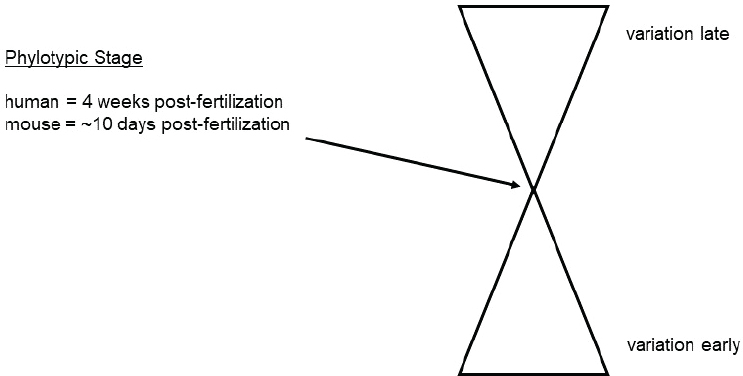
SOURCE: Paul Robson workshop presentation, January 17, 2020.
Differences Between Primates and Other Mammals in Early Development
Robson explored some of the morphological, physiological, and molecular differences between primates and other mammals in early development, noting that different features of human versus mouse embryology are well established and had been discussed by other speakers. Morphological differences between primates have been demonstrated by a cladistic analysis of the ontogeny of primate fetal membranes, which positions species accurately in the phylogenetic tree based on differential features in early embryonic development (Luckett, 1976). This type of analysis elucidates differences not only between the mouse and the human, but also between the human and old-world monkeys, for example.
Physiological and Molecular Differences Between Simians and Other Mammals
Substantial physiological differences are also evident between simians and other mammals during this early period of development, Robson said. A simian-specific feature of this developmental time period is that the endometrium goes through spontaneous decidualization2 and menstruation.
___________________
2 Decidualization refers to the process of rapid proliferation and differentiation of endometrial stromal cells into decidual cells, which is required for implantation (Haller et al., 2019).
Therefore, the implanting blastocyst is intimately interacting with this simian-specific feature of endometrial tissue. This difference motivated the working hypothesis that the primitive syncytium co-evolved with spontaneous decidualization and menstruation because it attaches, invades, and embeds in the endometrium.
Evidence also shows significant molecular differences hardwired in the genome that underpin these physiological and morphological differences, Robson said. To illustrate, he offered the example of a newly evolved system for hCG, an essential hormone for pregnancy that is simian-specific; hCG must be expressed in the implanting embryo, or the embryo will be immediately aborted. Chorionic gonadotropin (CG) is expressed by the late-stage human blastocyst and the syncytium. The CG signal is required to block menstruation; thus, it is essential to maintain pregnancy. CG is derived from anterior pituitary hormones (specifically luteinizing hormone) that went through a gene duplication 40 million years ago to form the beta peptide of chorionic gonadotropin (CGB). Both the alpha peptide (CGA) and CGB of hCG had to evolve the regulatory architecture to be expressed in the trophoblast lineage. These molecules are not expressed in the mouse trophoblast, although paralogs are expressed in the anterior pituitary.
Transgenic mice have been used to study the regulation of human CGA, Robson said. This work found that anterior pituitary expression was faithfully maintained in the mice, but there was no expression in the mouse trophoblast, suggesting that there are transcription factors in the human trophoblast that are not present in the mouse trophoblast to drive expression of this gene. This highly specific expression of CGA in the mouse system is only in the anterior pituitary. He noted that in one of the early publications on mouse epiblast stem cells treated with BMP4, one of the four genes used to try to establish similarities between mouse and human ESCs was CGA (Brons et al., 2007).
Molecular Signature of Primitive Syncytium Evident in Late-Stage Human Blastocyst
To characterize the molecular signature of the primitive syncytium, Robson and colleagues generated single-embryo RNA-sequencing data through pre-implantation development. In the 7-day blastocyst they found a signature that is clearly indicative of a syncytial cell signature. It includes the glial cells missing transcription factor 1 (GCM1)—the “master regulator” of syncytialization—as well as syncytin itself and other genes in the estrogen biosynthetic pathway (e.g., HSD3B1). This signature equates with the CCR7/GCM1+ polar trophectoderm population defined in Petropoulos et al. (2016). Robson explained that because these genes are not found in the mouse blastocyst, this work clearly indicates the emergence of the syncytial
transcriptome in the late-stage human blastocyst. This fits with the corpus of existing morphological studies, because the syncytial cell forms as soon as the human embryo implants. This model, or variations thereof, has been used extensively to show that hESC treated with BMP4 and FGF inhibitor result in blastocycstic trophoblast and primitive syncytium, Robson said (Xu et al., 2002; Yabe et al., 2016). Studies measuring the transcriptional response to BMP4 using ChIP-seq experiments clearly demonstrate that trophectoderm transcription factors are direct targets of BMP4. Classic trophoblast transcription factors such as GATA2, GATA3, and AP2A all have SMAD1/5/8 binding sites and immediately upregulate upon BMP4 induction, which explains how trophoblast is being driven from ESCs.
Markers of Primitive Syncytium Derived from Human Embryonic Stem Cells
Robson highlighted several markers of hESC-derived primitive syncytium established by RNA-sequencing data comparing the human villous placenta (i.e., later-stage trophoblast) to hESC-derived trophoblast and to the late-stage blastocyst. Although there are many similarities between the primitive syncytium and the villous STB, the specific markers that distinguish primitive syncytium from the more mature villous trophoblast include CCKBR, MRGPRX1, and syncytin-3/envP(b). He noted that unlike syncytin-1 or syncytin-2, which are also expressed, syncytin-3 is abundant and fairly specific to this primitive syncytium in the late-stage blastocysts as well as in the human ESC-derived trophoblast; it is much lower in the villous trophoblast. Thus, syncytin-3 is another example of a simian-specific trophoblast gene that has been co-opted from an endogenous retrovirus insertion 40 million years ago. Another example is the co-option of a long-terminal repeat to drive CYP19A1 expression in the human trophoblast, contributing to the activity of the estrogen biosynthetic pathway from dehydroepiandrosterone sulfate (DHEA-S) to estrogen production; this pathway is known to be active in the human placenta, but not the mouse placenta. It is also active in the primitive syncytium derived from pluripotent cells. Robson suggested that DHEA-S-derived estrogen production is a useful functional assay for the primitive syncytium or any human syncytiotrophoblast. Adding DHEA or DHEA-S to the trophoblast medium results in large amounts of estrogen being produced, which indicates that the biochemical pathway is active.
Molecular Heterochrony in a Developmental Regulator: GCM1
To look more deeply at molecular control of the primitive syncytium, Robson returned to GCM1, the classic master regulator of syncytial
formation. GCM1 has been knocked out in the mouse and is lethal to the embryo until embryonic days 8.5–9, which is when the syncytium forms in the mouse placenta, at a much later developmental stage than in humans. It has also been characterized in human placental explants, and it is functionally involved in syncytialization of the villous. Because it is expressed in the human at the blastocyst stage, which is much earlier than in the mouse, there is interest in characterizing it further. CRISPR technologies have been used to knock it out in iPSCs and to conduct gene expression using pluripotent cell differentiation protocols; when this is done, it drastically reduces syncytialization, as anticipated. However, global gene expression techniques reveal large numbers of altered genes and the apparent upregulation of CTB markers. Robson said that some of the top GCM1-bound and functional cis regulatory elements within PSC-derived trophoblast have emerged over recent evolutionary time.
Robson provided two examples of GCM1 target genes in the primitive syncytium: LMO2 and ALX4. In humans the LMO2 proximal promoter contains five GCM1 binding sites. However, the binding sites have been lost in other species, with no evidence for GCM1 cis regulatory elements in the mouse, for example. ALX4 is a similar example of a GCM1 target gene in the primitive syncytium. This gene is down-regulated in the GCM1 knockout, so it is a functional binding site. As with LMO2, there are species-specific differences that are indicative of active evolutionary change around the regulatory regions that bind GCM1 and drive expression of these transcriptional regulators in primitive syncytium.
Early Development as a Site for Rapid Mutation and Selection
Robson concluded by emphasizing the extent of the molecular and functional differences between species in the primitive syncytium and predicting that there are significant differences in other cell types at these early stages of development. He surmised that this period of early development is a window of rapid selection and mutation in the human population. Therefore, he suggested altering the hourglass model to increase the size at the bottom of the hourglass, because there is probably much more variation early in development than in later development. He added that early development probably creates selection pressure for many novelties as humans and other species evolve. In fact, there may be variation of the human population being selected for today. If selection is occurring early in development (e.g., aborting an embryo that is dysfunctional at 4 weeks, rather than leading to likely death at 25 years in an adult) it allows for an increased rate of evolution. For instance, the single-nucleotide polymorphism (SNP) in the human population that appears to be most strongly selected for is a SNP in EPAS1, the gene encoding HIF2-alpha. It appears to have been rapidly
selected for among Tibetans, presumably through the low-oxygen environment at high altitude, as this is an oxygen-sensing transcription factor. Some have suggested that it regulates oxygen concentration in adults, allowing them to live more comfortably at high altitudes. It is also well known that oxygen sensing is an important feature of trophoblast development. Of note, the lineage decision between a CTB and an STB is oxygen-regulated, and EPAS1 is the most highly abundant transcription factor expressed in these hESC-derived primitive syncytial cells.
PANEL DISCUSSION
Variability in Trophoblast-Related Terminology
Panelists were asked by Amander Clark, the session moderator, to explain why they refer to the trophoblast system as a syncytium and how that relates to the syncytiotrophoblast, CTB, mural trophoblast, or polar trophoblast. She also asked how the primitive syncytium relates to the cell types differentiated in their pluripotent stem cell models.
The terminology can be confusing, Roberts said. It was initially presumed that the syncytium that was forming was villous STB, but it later became clear that it was not. Others have claimed that it is mesoderm, but the question of what it actually is remains open. Roberts uses the term “primitive placenta,” because it represents the placenta’s function very early in development when it is developing at the expense of locally produced histrophe and attempting to produce sufficient hormones to maintain the pregnancy. The trophoblast is a complex lineage of cells with multiple subtypes, Robson replied, which gives rise to definitional challenges. Many studies have compared the pluripotent cell–derived trophoblast to villous trophoblast because it is accessible, but it comes from the placenta much later in development (7–8 weeks) and its composition is completely different. Robson characterized this as a progression: the trophoblast in the peri-implantation stage that he described in his presentation—the blastocystic trophoblast—is the first trophoblast which seals the blastocyst. The next apparent trophoblast is the primitive syncytium, a term first established in the 1960s (Boyd and Hamilton, 1966). The use of terminology in this work is variable across the disciplines of cell and developmental biology, placental researchers, and pathologists, Parast said. Efforts have been made to standardize definitions, but in the case of primitive syncytium this is challenging because the nature of the cell is still unknown. Presumably, the cell has a combination of characteristics from the STB (e.g., it is multinucleated and expresses some of the factors that induce cell–cell fusion) while also having invasive potential. There is still much to learn about the primary
cell types using the embryo and the placenta to determine the nature of the cells in culture, Parast said.
Differentiation of Trophoblast Versus Amnion Cell Lineages
A workshop participant commented that recent self-organizing models have shown the development of amnion is dependent on BMP. Given that there are many markers shared by amnion and trophoblast and that the emergence of amnion from the epiblast is more aligned with what is thought to occur during early development in vivo, the participant asked the panelists to comment on the relationship between their models and amnion differentiation. He also asked about the molecular pathways that may turn cells to trophoblast versus amnion lineages. p63 is not expressed in the amnion and is not required for amniogenesis, Parast said, because p63-null mice survive through embryogenesis. Furthermore, the initial burst of transcription factor, Brachyury, occurs when amnion is generated and does not occur during trophoblast differentiation, particularly in the presence of IWP2. A challenge is that there is currently no measurement of real amnion formation in humans, Robson said. He suggested that the absence of Brachyury expression suggests that BMP4 and FGF inhibition are directly up-regulating classic trophoblast markers. BMP is involved in multiple differentiation pathways, so it could potentially play a role in both amnion and trophoblast systems. Roberts maintained that there is no evidence that amnion produces any of these characteristic trophoblast products.
Cells Used to Direct Trophoblast Stem Cell Derivation
A workshop participant asked Parast whether the protocol she follows to direct TSC derivation used undirected pluripotent cells or BMP-derived cells. She replied the cells were already BMP-derived cells at the end of the first step. After a 4-day differentiation in the presence of BMP4 and IWP2, they switch to the Okae media for several passages. In the original Okae study, the TSCs were derived from trophectoderm of blastocyst-stage embryos as well as from early placenta villous CTB. The participant inquired about detailed molecular analysis of what the PSC-derived trophoblasts are being reverted to; Parast replied that the RNA-sequencing data will be available soon.
Syncytin-3 Expression
A workshop participant requested clarification on the identity of syncytin-3 and how it relates to the other syncytins. Syncytin-3 is in the
intron of the gene Rin3, Robson said, and it is a 40-million-year-old simian-specific insertion, so it is the same age as syncytin-2. It is immunogenic and fusogenic, according to functional studies in cell lines conducted when it was first identified by virologists (Blaise et al., 2005). Robson and colleagues found that it is significantly expressed both in the late blastocyst and in primitive syncytium derived from ESCs.
Heterogeneity in Trophectoderm
The topic of heterogeneity in the trophectoderm during the transition from pre-implantation to peri-implantation was raised by a workshop participant. The publication by Petropoulos et al. (2016) clearly shows heterogeneity and indicates a transition in trophectoderm closest to embryo (i.e., polar cells), Roberts said. This developmental progression coincides both with implantation and likely with the emergence of invasive syncytium from those cells. In the blastocyst it is not syncytial, but it becomes syncytial as it implants. Robson added that primitive syncytium probably forms at the polar end of the human blastocyst upon attachment. Parast noted that this heterogeneity also extends to post-implantation CTB. Single-cell sequencing of placental tissues at 5–7 weeks reveals multiple cell populations in the CTB that seem to differ according to their proximity to the fetal versus maternal surfaces.
Wnt Signaling in Trophectoderm
The function of Wnt signaling in the trophectoderm was raised by a workshop participant. Parast and colleagues are using Wnt inhibition to get from the pluripotent state to the CTB state; it appears that this may limit BMP4-induced mesoderm induction. In the trophectoderm itself, Wnt signaling seems to be important in maintaining the undifferentiated state, Parast said, but its role is complicated. Wnt has to be withdrawn to initiate differentiation into extravillous trophoblast, then Wnt has to be re-added to get full differentiation into mature extravillous trophoblast (Haider et al., 2018).
Trophoblast in the Peri-Implantation Window
The panelists were asked to reflect on the trophoblast in the peri-implantation window and the various differentiation approaches that are or could be used to make trophoblast from stem cells. Robson’s laboratory is deriving trophoblast directly from human ESCs. Because ESCs are also used to model other definitive lineages, he suggested that a novel strategy could be devised to derive both trophoblast and the other lineages simultaneously
in the same structure. Parast suggested further exploring the mouse system to generate chimeras and examine contributions to trophectoderm, given the ethical challenges of working with human and nonhuman primate models. Roberts argued that the current model is totally unsatisfactory and needs to be refined. For example, markers observed in trophoblast are not observed even in the early embryo; the function of BMP4 is also unknown. Further mouse research will be needed to understand and improve the system, he said. The embryo does not develop in isolation, Robson noted. Cross-communication between the embryo and endometrium—in addition to bidirectional signaling from the syncytium—is likely informing developmental decisions in ways that are not yet understood. Organoid systems are available to study the human endometrium, and co-culturing PSC-derived cells with these will be important, he said.
Role of Trophoblast Signaling in Inducing Gastrulation
The panelists were asked whether early human trophoblast must signal to the epiblast to induce the processes of gastrulation, given that (1) in humans the first trophoblast develops as primary syncytium and (2) in the mouse, signals from the primitive endoderm and signals between epiblast and trophoblast are critical for early patterning. Parast replied that they are investigating this crosstalk between trophoblast and other cells by co-culturing human ESCs and TSCs, and it looks as though ESCs remain relatively undifferentiated, at least morphologically.
This page intentionally left blank.




















