1
Introduction
The taxonomic status of the modern red wolf, Canis rufus, has been controversial. The controversy extends beyond the scientific realm because the red wolf has been the subject of extensive conservation and recovery efforts that are not universally popular. Once common throughout the eastern United States, red wolves were extirpated from the wild by 1980. In 1987, individuals from a captive breeding program were used to re-introduce red wolves into a protected area in North Carolina.1 The re-introduced individuals established a population, and the U.S. Fish and Wildlife Service (USFWS) now manages this population.
To found the captive breeding colony, the USFWS had selected a small number of individuals from a population of canids discovered in western Louisiana. The USFWS selected those individuals based on their resemblance to wolves in morphology and vocalizations, supported by analyses of genetic variation in enzymes, which are alternate forms of the same protein. Subsequent genetic studies of the managed population indicated that these wolves displayed evidence of significant coyote ancestry. This discovery raised the question of whether extant captive and managed red wolves (also referred to in this report as extant red wolves or extant red wolf populations) was a valid species that was eligible for conservation and recovery. Opponents of the USFWS effort to restore the red wolf have used questions about the ancestry of the extant populations to argue that the restoration program is flawed.
In 2019, the National Academies of Sciences, Engineering, and Medicine issued a report titled Evaluating the Taxonomic Status of the Mexican Gray Wolf and the Red Wolf (NASEM, 2019), which addressed the taxonomic status and evolutionary history of the red wolf. The report, which had been prepared upon the request of the USFWS, addressed three questions about red wolves. First, is there evidence that the historical population of red wolves was a lineage distinct from other
___________________
1 Captive refers to the 245 red wolves maintained in 43 breeding facilities all throughout the United States. Managed refers to the approximately 20 red wolves comprising a nonessential experimental population in the Alligator River National Wildlife Refuge in North Carolina.
canids? Second, is there evidence that the extant populations of red wolves in the United States represent a lineage of canids distinct from modern gray wolves (C. lupus) and coyotes (C. latrans) (Figure 1-1)? Third, is there evidence for genetic continuity between the historical red wolf populations in the eastern United States and the extant populations of red wolves?
The authors of the report answered “yes” to the first two questions. They concluded that, with respect to the third question, there was evidence that the extant populations traced some of their ancestry to the historical red wolf. They concluded that, taken as a whole, the available evidence supported species status (C. rufus) for the extant red wolf. This finding retained the existing taxonomic designation of the red wolf and reinforced the validity of conserving and restoring red wolves. However, the authors acknowledged that additional genomic evidence from historical specimens could change this assessment by changing the answers to one or more of the three questions.
While the report was being prepared, Murphy et al. (2019) described gene markers indicating red wolf ancestry for a wild, unidentified canid population in southwestern Louisiana. In addition, Heppenheimer et al. (2018) discovered alleles in a canid population in Galveston Island, Texas, that had not been observed previously in coyotes or extant red wolves. These alleles, called “ghost alleles,” could represent genes characteristic of historical red wolves that were lost in the selection of the founders of the breeding colony. If, as these discoveries suggest, these two populations are remnant red wolf populations and harbor genetic information unique to red wolves, then the evolutionary history of the red wolf needs to be re-examined and the taxonomic and conservation status of these populations re-assessed.
PURPOSE OF THE STUDY
This study begins where the 2019 National Academies report concluded. The USFWS requested this study, prompted by the discoveries of the Gulf Coast canid (GCC) populations mentioned above. The USFWS specifically requested the National Academies to appoint an ad hoc committee that would: (1) solicit and review applications to carry out research to determine the taxonomy of the canid population in southern Louisiana,2 and (2) develop a research strategy to further assess the taxonomy of the red wolf (see Box 1-1). The first request was completed in January 2020.3 This report addresses the second request from the USFWS.4

SOURCE: B. Bartel, U.S. Fish and Wildlife Service (left) and Melba Coleman (middle and right).
___________________
2 The request for applications (RFA) was issued on November 4, 2019. The deadline for submission of applications was December 3, 2019. The RFA is available at http://nas-sites.org/dels/red-wolf-study (accessed on April 14, 2020).
3 National Academies of Sciences, Engineering, and Medicine. 2020. Evaluation of Applications to Carry Out Research to Determine the Taxonomy of Wild Canids in the Southeastern United States. Washington, DC: The National Academies Press. https://doi.org/10.17226/25661.
4 The full statement of task (Part 1 and Part 2) is located in Appendix G of this report.
WHY DO MORE RESEARCH ON THE RED WOLF?
Limitations in Existing Data
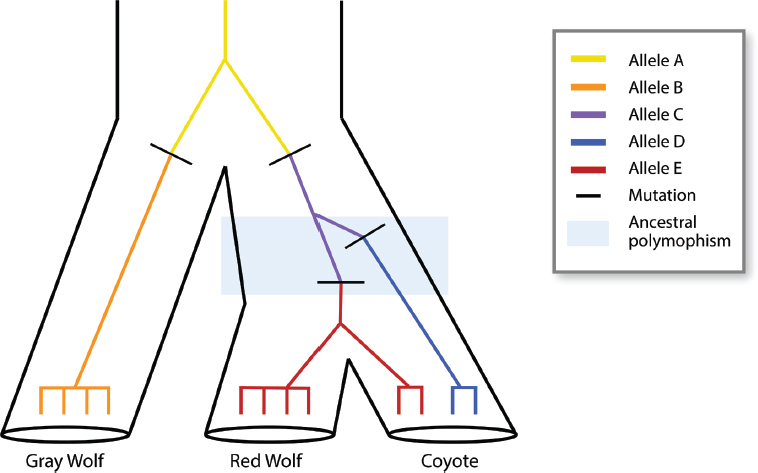
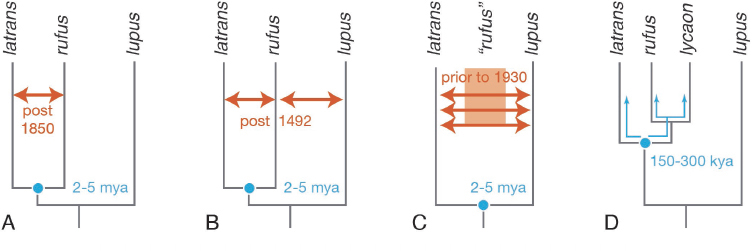
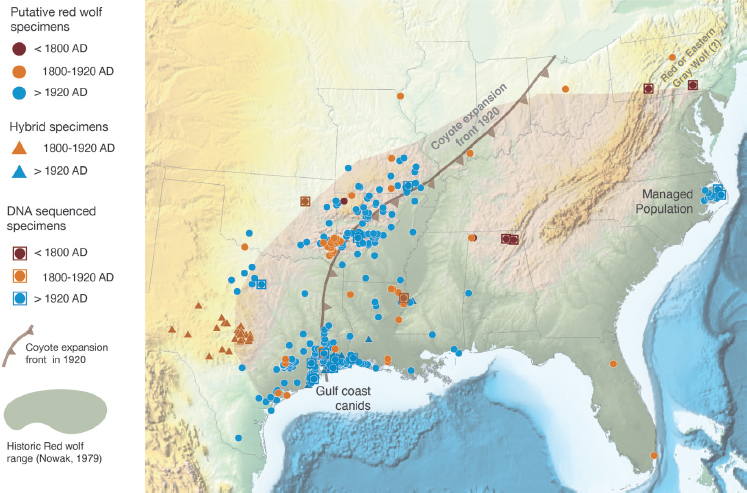
The status of the red wolf has been a contentious issue for many years. For one reason, the USFWS selected the progenitors of the extant captive and managed red wolves from a population with significant levels of coyote ancestry. There are three possible explanations for this sharing of alleles between red wolves and coyotes, which are discussed in more detail in NASEM, 2019. Briefly, the first possible explanation is that the extant red wolves may be hybrids between coyotes and either gray wolves or red wolves. Hybridization may have occurred long ago or in more recent years. Evidence suggests that some hybridization certainly occurred in the 20th century as red wolf populations dwindled in size while coyotes expanded their range eastward (Box 1-2; Figure 1-2). Second, the extant red wolves may have diverged from coyotes recently enough that they continue to share alleles, even though each species has its own forward evolutionary trajectory—a situation known as incomplete lineage sorting (Figure 1-3). Incomplete lineage sorting is a situation in which a polymorphism (two more alleles of a gene) that was present in an ancestral species persists in descendent species. It is more likely to occur when two species have diverged quite recently from a common ancestor. Incomplete lineage sorting is a challenge for reconstructing phylogenetic relationships, because a phylogenetic tree based on a shared polymorphism in red wolves and coyotes will differ from a tree based on other genes for which each species has a unique allele. Third, hybridization and introgression of coyote genes into red wolves could reflect historical phenomena associated with the ancestral origin of the red wolf. In this case, the presences of coyote alleles would be the result of hybridization between coyotes and gray wolves (or another ancient canid) that gave rise to the red wolf, in which some level of reproductive isolation exists, but is incomplete. These three possible explanations for shared alleles between coyotes and red wolves are not mutually exclusive. Resolving them requires tracing the ancestry of extant red wolves and ascertaining when coyote genes entered the lineage of red wolves (Box 1-3; Figure 1-4).
A second reason for the controversy over the status of red wolves is the paucity of data from ancient specimens that could serve as a standard for the red wolf before hybridization with coyotes in the 20th century. There are no morphological data from ancient specimens, defined here as specimens dated prior to 1800 (Appendix B). The year 1800 represents the approximate time when American colonists moved in large numbers across the Appalachian Mountains into what is now Kentucky, Tennessee, Alabama, and other areas in the southern United States and began to alter the forest habitat in those areas on a large scale. There also are no nuclear DNA evidence from ancient specimens. There are data for mitochondrial DNA (mtDNA) sequences from only four ancient
specimens, all of which are archaeological specimens that are undated or more than 1,000 years old (Appendix C). The mtDNA data indicate a close relationship between coyotes and red wolves, but not the nature of that relationship.
The data from historical specimens, defined here as those specimens collected between 1800 and the start of the eastward expansion of coyotes in 1920, do not resolve questions about the ancestry of red wolves. Analyses of morphological data from historical specimens indicate that specimens identified as red wolves are distinguishable from specimens of gray wolves and coyotes. However, those specimens identified as red wolves have many intermediate characteristics of gray wolf and coyote specimens, which is consistent with either a recent divergence of red wolves from coyotes and gray wolves or a hybrid origin of red wolves. The analyses of mtDNA from historical specimens of putative red wolves place them in a lineage with coyotes, but these data are from specimens collected at the western edge of the historical range of red wolves, where the range abutted that of coyotes and where hybridization could have occurred (Figure 1-2; Appendix D). As a result, whether the mtDNA from historical specimens originates from coyotes or red wolves cannot be resolved without an analysis of nuclear DNA from the same specimens.
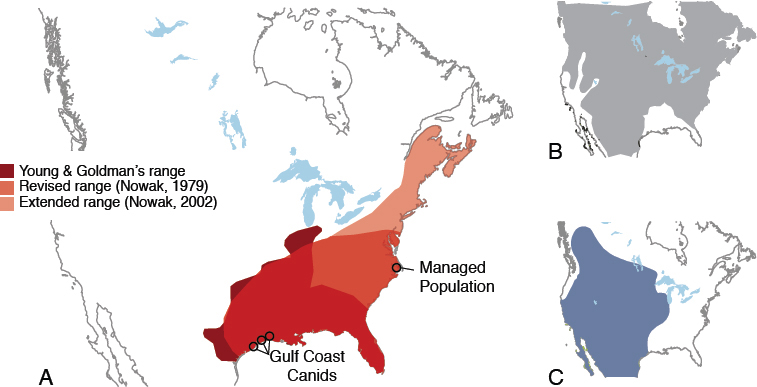
Genetic and genomic data from extant putative red wolves come from populations at the southwestern edge of the historical range (western Louisiana and eastern Texas) and the managed population in North Carolina (Figures 1-2 and 1-5; Appendix D). The populations at the southwestern edge of the historical ranges are the ones from which the progenitors of the captive and managed populations were selected. It is unclear how well those populations represent what was once a numerically large, geographically widespread population of canids.
The evidence for past hybridization events in the history of the red wolf has led some geneticists to question the validity of the red wolf’s species designation. However, evidence of historical
hybridization in a species does not automatically invalidate its taxonomic status as a valid species. While evidence of historical hybridization can indicate a lack of reproductive isolation between the species represented in hybrid individuals, there are other possible outcomes of hybridization events. One outcome is that one species can be mostly absorbed into another, as seen in the brown bears of Alaska (Cahill et al., 2013). A very different outcome is the evolution of a new species via hybridization between two other species. Although such an occurrence was once considered unlikely, biologists have described repeated cases of species formation via hybridization in a broad range of groups—plants, insects, fish, and birds (Schumer et al., 2016; Marques et al., 2019).
The Opportunities for New Research
Advances in technology and statistical methods have created new opportunities to address the taxonomy of the red wolf. These advances include techniques for extracting DNA from ancient specimens (referred to hereinafter as “ancient DNA”), techniques for powerful statistical analyses of genomic DNA, and techniques for capturing and analyzing morphological data from skeletal specimens that go far beyond the methods that have been used thus far in studies of red wolves. This report describes these methods and how they can be used to collect new data from existing specimens and from the canids in southwestern Louisiana, Galveston Island, and any other canid populations discovered in these areas.
The guiding principle for the research strategy described here is that the research must address the relationships among ancient red wolves, historical red wolves, the extant red wolves, and the unidentified canids along the Gulf Coast. These relationships will indicate whether red wolves are, or once were, a distinct lineage (a species sharing a common ancestor with either coyotes or gray wolves or a species that has an ancient, hybrid origin) or whether “red wolves” are recent hybrids between gray wolves and an expanding coyote population. They will also clarify how the extant red wolves are related to the GCC populations. Clarifying all of these relationships is necessary to align red wolf taxonomy with modern evolutionary biology. To clarify those relationships, new research should employ three types of data: ancient DNA, genomic data from the unidentified canids and the extant red wolf populations, and morphological data from ancient specimens and extant animals. Prior research has used genetic data from extant canids and morphological data from ancient specimens. An approach that integrates these three types of data is now possible, and that integration is critical for resolving the status of the red wolf.
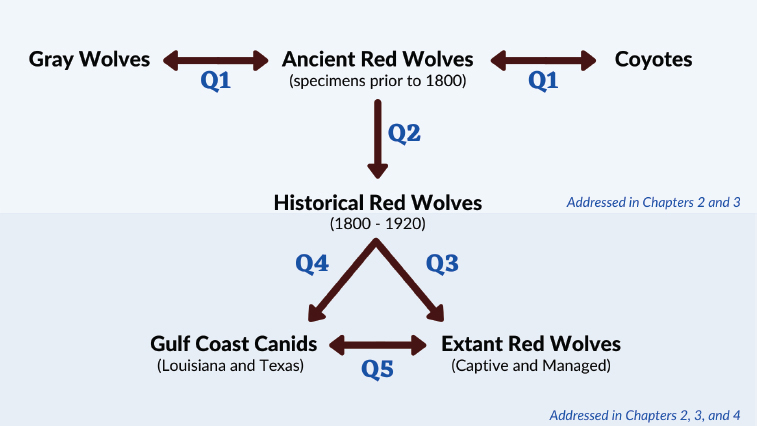
Five Research Questions and the Data They Require
The three conceptual questions outlined in the previous report lead to five operational research questions:
- Are the ancient red wolves (i.e., specimens dated prior to 1800) a distinct lineage from the lineages of gray wolves and coyotes?
- Are the historical red wolves (i.e., those in the 19th century, before the eastward expansion of coyotes) a continuous lineage with ancient specimens in the eastern United States?
- Are the red wolves from the extant captive and managed populations a continuous lineage with those in the eastern United States before the coyote expansion in the 20th century?
- Are the Gulf Coast canids part of the distinct lineage of canids that lived in the eastern United States before the coyote expansion?
- Is the genetic diversity of the extant red wolf populations a subset of the diversity of the Gulf Coast canids?
These questions address the relationships among the various canid groups associated with red wolves. Figure 1-6 outlines the five questions, the canids to which each question pertains, and where this report describes the necessary data.
1. Are the ancient red wolves (i.e., specimens dated prior to 1800) a distinct lineage from the lineages of gray wolves and coyotes?
An affirmative answer would give the USFWS the confidence to call the red wolf a distinct species that is not the product of a recent hybridization between gray wolves and coyotes. If this distinction were to be established, then the red wolf, at least in theory, would be a species eligible for conservation and restoration efforts if endangered. Chapter 2 describes the morphological evidence, and Chapter 3 describes the genomic evidence necessary to answer this question.
2. Are the historical red wolves (i.e., those in the 19th century, before the eastward expansion of coyotes) a continuous lineage with ancient specimens in the eastern United States?
An affirmative answer would imply that the red wolves still existed as a distinct lineage before the coyote expansion to the eastern United States. If this is so, then the red wolves found in the eastern United States in the 20th century will have had genetic continuity with ancient red wolves
and retain their eligibility, at least in theory, for conservation and restoration efforts. Chapter 2 describes the morphological evidence important for this question, and Chapter 3 describes the genomic evidence that would address this question
3. Are the red wolves from the extant captive and managed populations a continuous lineage with those in the eastern United States before the coyote expansion in the 20th century?
An affirmative answer would imply that, despite evidence of hybridization with coyotes, the captive and managed populations have retained some of the genetic ancestry of the red wolves. If this is the case, then the captive and managed populations represent at least a subset of the original red wolf lineage and retains its eligibility for conservation and restoration. Chapter 2 describes the morphological data necessary to answer this question, and Chapters 3 and 4 describe the genomic data necessary to answer this question.
4. Are the Gulf Coast canids part of the distinct lineage of canids that lived in the eastern United States before the coyote expansion?
An affirmative answer will imply that, despite hybridization with coyotes, the GCC populations have retained some of the genetic diversity of the red wolves. If this is the case, it implies that the canids found along the Gulf Coast, from which the founders of the captive and managed populations were selected, represent the remainder of the red wolf lineage after a long period of
population decline. This would strengthen the argument that the extant populations can be considered as legitimate red wolves. Chapter 2 describes the morphological, behavioral, and ecological evidence relevant to this question, and Chapters 3 and 4 describe the genomic data necessary to answer this question.
5. Is the genetic diversity of the extant red wolf populations a subset of the diversity of the Gulf Coast canids?
An affirmative answer to this question would imply that the extant population of red wolves is part of a larger lineage of red wolves and not a separate lineage. The small number of founders used to establish the captive and managed populations most likely contained only a fraction of the genetic diversity found in the historical population of red wolves. A further reduction in genetic variation from the historical population is likely because those founders were themselves a subset of a carefully selected group of individuals. If the GCC populations are indeed a remnant of the original red wolves that lived in the area before the coyote expansion in the 20th century, then its members should share genetic variation with the captive and managed population. This would reinforce the conclusions from an affirmative answer to question 4. The GCC populations may also harbor genetic variation shared with historical red wolves that is found in the managed population. The GCC populations and the managed population likely share variation with coyotes due to hybridization. Chapter 2 describes the morphological, behavioral, and ecological data necessary to answer this question, and Chapter 4 describes the genomic data necessary.
COMMITTEE’S APPROACH TO ITS CHARGE
Information Gathering Activities
To address its charge, the committee held a total of five meetings (three were virtual, and two were face-to-face meetings) and two webinars between October 2019 and June 2020. The first meeting (virtual; held on October 18, 2019) was spent discussing the two tasks requested by the USFWS and obtaining clarification from the USFWS representative. The second meeting (held on December 8-9, 2019, in Washington, D.C.) was closed in its entirety and was focused on the assessment of applications submitted in response to the request for applications, which was issued on November 4, 2019. The committee’s third meeting (February 20-21, 2020, in Irvine, California) included an open session that featured presentations from experts in hybridization, statistical analyses of genetic data, paleogenomics, red wolf ancestry in the southeastern United States, and research strategies and methodologies to study mammalian evolution. Two committee webinars were held on March 2 and March 12, 2020, and featured experts in introgression and the behavioral ecology of coyotes, respectively. The committee meetings held in May and June 2020 were virtual; both were closed in their entirety and were focused on report preparation. The agendas for the open session of the committee meeting and the webinars are provided in Appendix A of this report. All materials presented at the open session and in the webinars are available at the study website.5
Throughout the study the committee also solicited input from the public via the study website. All submitted comments and documents were added to the study’s public access file, which is available on request from the National Academies’ Public Access Records Office.6
___________________
5 See https://www.nationalacademies.org/red-wolf-research.
6 Requests can be directed to PARO@nas.edu.
Interpretation and Syntheses
The committee assessed the information in the meetings described above and in additional closed teleconferences. The committee performed these as a whole and by dividing into three groups that focused on individual topics. The groups each drafted a chapter for the report; all members of the committee then read these chapters and suggested revisions. The committee did not re-analyze existing data but did examine existing data to determine which new data would provide the most useful information and complement the existing data.
ORGANIZATION OF THE REPORT
This report comprises five chapters. Chapter 1, this chapter, introduces the study, provides the general background for the study and statement of task for the committee, and outlines the research strategy devised by the committee. Each of the next three chapters describes one type of data necessary to resolve the status of the red wolf (Figure 1-1). Chapter 2 describes the ways in which new morphological, behavioral, and ecological data can inform the diagnosis of the taxonomic status of red wolves. In particular, the chapter distinguishes informative data from uninformative data in order to guide future research into the most profitable paths. Chapter 3 describes the value and uses of genomic data from historical and ancient specimens. It describes the methodological challenges in obtaining genomic data from those types of samples and the processes needed to ensure that those data are indeed the genomic data of the specimen under scrutiny, without contamination from other sources of DNA. Chapter 4 describes the types of analyses that are possible when the ancient DNA is combined with whole-genome data on extant canids. The chapter describes why whole-genome data are necessary and which technical methods are best for obtaining those data. It also describes the best statistical approaches to those data for answering questions about the status of red wolves. Chapter 5 presents overarching principles for conducting these studies and summarizes the committee’s recommendations.
REFERENCES
Cahill, J. A., R. E. Green, T. L. Fulton, M. Stiller, F. Jay, N. Ovsyanikov, R. Salamzade, J. St. John, I. Stirling, M. Slatkin, and B. Shapiro. 2013. Genomic evidence for island population conversion resolves conflicting theories of polar bear evolution. PLOS Genetics 9 (3):e1003345.
Chambers, S. M., S. R. Fain, B. Fazio, and M. Amaral. 2012. An account of the taxonomy of North American wolves from morphological and genetic analyses. North American Fauna 77:1–67.
Heppenheimer, E., K. E. Brzeski, R. Wooten, W. Waddell, L. Y. Rutledge, M. J. Chamberlain, D. R. Stahler, J. W. Hinton, and B. M. vonHoldt. 2018. Rediscovery of red wolf ghost alleles in a canid population along the American Gulf Coast. Genes 9(12):618.
Hody, J. W., and R. Kays. 2018. Mapping the expansion of coyotes (Canis latrans) across North and Central America. Zookeys 759:81–97.
Marques, D. A., J. I. Meier, and O. Seehausen. 2019. A combinatorial view of speciation and adaptive radiation. Trends in Ecology & Evolution 34(6):531–544.
Murphy, S. M., J. R. Adams, J. J. Cox, and L. P. Waits. 2019. Substantial red wolf genetic ancestry persists in wild canids of southwestern Louisiana. Conservation Letters 12(2):e12621.
NASEM (National Academies of Sciences, Engineering, and Medicine). 2019. Evaluating the taxonomic status of the Mexican gray wolf and the red wolf. Washington, DC: The National Academies Press. https://doi.org/10.17226/25351.
Nowak, R. M. 1979. North American quarternary canis. Monograph of the Museum of Natural History, University of Kansas 6:1–154.
Nowak, R. M. 2002. The original status of wolves in eastern North America. Southwestern Naturalist 1:95–130.
Nowak, R. M. 2009. Taxonomy, morphology, and genetics of wolves in the Great Lakes region. In A. P. Wydeven, T. R. VanDeelen, and E. J. Heske (eds.), Recovery of gray wolves in the Great Lakes region of the United States: An endangered species success story. New York: Springer. Pp. 233–250.
Schumer, M., R. F. Cui, D. L. Powell, G. G. Rosenthal, and P. Andolfatto. 2016. Ancient hybridization and genomic stabilization in a swordtail fish. Molecular Ecology 25(11):2661–2679.
USGS (U.S. Geological Survey), National Geospatial Technical Operations Center. 2017. USGS National Boundary Dataset (NBD) Downloadable Data Collection. https://nationalmap.gov/boundaries.html.
Wayne, R. K., and S. M. Jenks. 1991. Mitochondrial DNA analysis implying extensive hybridization of the endangered red wolf Canis-rufus. Nature 351(6327):565–568.
Wilson, P. J., S. Grewal, I. D. Lawford, J. N. M. Heal, A. G. Granacki, D. Pennock, J. B. Theberge, M. T. Theberge, D. R. Voigt, W. Waddell, R. E. Chambers, P. C. Paquet, G. Goulet, D. Cluff, and B. N. White. 2000. DNA profiles of the eastern Canadian wolf and the red wolf provide evidence for a common evolutionary history independent of the gray wolf. Canadian Journal of Zoology 78(12):2156–2166.
Wolfram Research, Inc. 2020. Mathematica, Version 12.1. Champaign, IL: Wolfram Research Inc.
Young, S. P., and E. A. Goldman. 1944. The wolves of North America. Washington, DC: American Wildlife Institute and Dover Press.












