Admixed population: a population that contains genes from at least two distantly related populations or species as a result of interbreeding between populations or species that have been reproductively isolated and genetically differentiated.
Allele: alternative form of a gene.
Ancestry: related to the individual, group, or species, at any distance of time, from whom an individual is descended from.
Backcrossing: when an offspring is produced by crossing a hybrid with one of its parents or an individual that is genetically similar to one of its parents (backcrossing produces an offspring that is more genetically similar to the parent).
Chromosome: a long DNA molecule found in the nucleus of most living cells, carrying genetic information in the form of genes.
Common ancestor: the evolutionary lineage from which two or more species diverged.
DNA: Deoxyribonucleic acid (DNA) is a molecule that contains the biological instructions that are passed from adult organisms to their offspring during reproduction.
Endangered species: species of plants or animals that are at risk of extinction because of natural or anthropogenic causes.
Gamete: the reproductive cells of an organism (i.e., egg cells and sperm)
Gene: a segment of DNA whose nucleotide sequence codes for protein or RNA, or regulates other genes.
Gene introgression: movement of alleles from one population or species into another through hybridization and repeated backcrossing (the reproduction of hybrids with individuals of the parental species).
Genetic drift: random changes in allele frequencies in a population between generations due to sampling individuals that become parents and binomial sampling of alleles during meiosis. Genetic drift is more pronounced in small populations.
Genetic recombination: the process that generates a haploid product of meiosis with a genotype differing from both the haploid genotypes that originally combined to form the diploid zygote.
Genome: the complete sequence of DNA in an organism.
Homologous chromosomes: a pair of chromosomes I an organism that contain the same genes (although they could have different alleles), chromosomal length, and centromere location. One chromosome of the pair comes from the father and the other from the mother.
Hybridization: mating of individuals from genetically distinct populations, or species, that results in offspring with mixed ancestry.
Hybrids: offspring produced by hybridization. Hybrids can be either first-generation (F1) hybrids or the offspring of hybrids themselves.
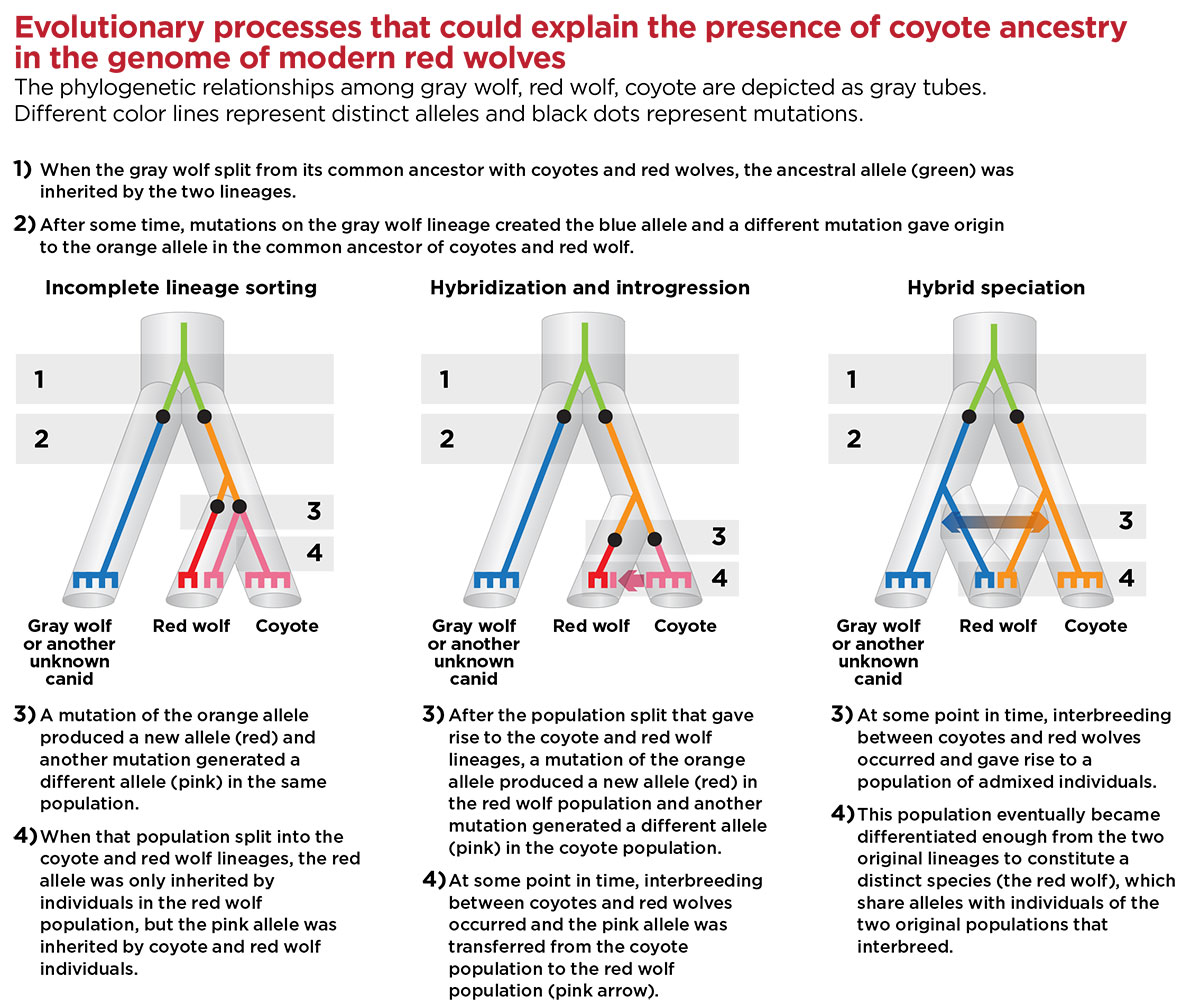
Incomplete lineage sorting: a characteristic of phylogenetic reconstruction in which the tree produced by a single gene differs from the population or species level tree because a polymorphism present in the ancestral species is retained in the descendant species.
Natural selection: differential contribution of genotypes to the next generation due to differences in survival and reproduction.
Pre-mating isolation: reproductive isolation that stops production of hybrid offspring by preventing mating.
Subspecies: a taxonomically defined subdivision within a species that is physically or genetically distinct and often geographically separated.
Species: a group of organisms with a high degree of physical and genetic similarity that naturally interbreed among themselves and can be differentiated from members of related groups of organisms.
Speciation: the evolutionary process of forming new species.
 Genomic Processes in Hybridization: Implications for
Species Taxonomy
Genomic Processes in Hybridization: Implications for
Species Taxonomy