3
Microbial and Genetic Movements Across the One Health Domains
During session I of the workshop, speakers and discussants explored microbial and genetic movements across health, agriculture, and environmental compartments. The session’s first half, moderated by Rima Khabbaz, deputy director of infectious diseases at the U.S. Centers for Disease Control and Prevention (CDC), covered knowledge gaps and opportunities to strengthen the evidence base, with a focus on surveillance of antimicrobial resistance. It opened with an overview of the National Antimicrobial Resistance Monitoring System (NARMS) by Patrick McDermott, director of NARMS at the U.S. Food and Drug Administration (FDA). Paula Cray, professor and head of the Department of Population Health and Pathobiology at North Carolina State University-Raleigh, followed with a review of the quality of antimicrobial surveillance across countries at differing levels of development. James Tiedje, university distinguished professor of microbiology and molecular genetics and of plant, soil, and microbial sciences at Michigan State University, discussed environmental surveillance for antimicrobial resistance and identifying horizontal gene exchange as the point of control. The effect of antimicrobials in the environment was the focus of the session’s second half, moderated by Jeffrey Silverstein, deputy administrator of animal production and protection with the U.S. Department of Agriculture (USDA) Agricultural Research Service. Lance Price, professor at The George Washington University Milken Institute School of Public Health, explained how resistance determinants on microbes are transmitted between human and animal hosts. Ed Topp, principal research scientist at Agriculture and Agri-Food Canada, described how human activities can potentiate antimicrobial resistance within the
environmental reservoir of microorganisms. Lisa Durso, a microbiologist with the USDA Agricultural Research Service, evaluated management and mitigation strategies for reducing the transfer of antimicrobials to the environment. The session’s final speaker, Steve Brooks, vice president of environment, health, and safety at Pfizer Inc., provided a manufacturing perspective and outlined the biopharmaceutical industry’s road map to reduce the environmental impact of antibiotic production.
STRENGTHENING THE KNOWLEDGE AND EVIDENCE BASE
National Antimicrobial Resistance Monitoring System
McDermott explained that NARMS was established in 1996 as a collaboration among CDC, FDA, and USDA. At its outset, NARMS was tasked with integrated surveillance of antimicrobial resistance in foodborne bacteria, which he defined as
the coordinated sampling and testing of bacteria from food animals, foods, and clinically ill humans and the subsequent evaluation of antimicrobial resistance trends throughout the food production and supply chain using harmonized methods.
In the years since, he said, the concept of integrated surveillance has shifted to One Health surveillance, which includes an environmental component. He described the key function of each agency in NARMS today. USDA carries out randomized, nationally representative sampling of food animals at slaughter, which is providing new insights into the ecology of resistance. FDA works with state partners to test samples of the retail meat supply annually. CDC performs susceptibility sampling on 5 percent of the nontyphoidal Salmonella samples collected by participating state public health departments, with plans to begin annual whole genome sequencing on every isolate collected.
Value of One Health Surveillance
One Health surveillance is foundational to all national action plans and to the World Health Organization (WHO) recommendations for combating antimicrobial resistance, said McDermott. He emphasized that the approach has value across multiple dimensions. It is used to establish baseline levels of pathogens and resistance in different reservoirs and to describe the spread of resistant bacterial strains and genes across ecosystems. When temporal and spatial resistance trends are identified, they underpin hypotheses about the sources and reservoirs of resistant bacteria, he said. Link-
ing those sources and reservoirs to specific antibiotic-use practices helps to shape more informed and targeted interventions through risk analysis. One Health surveillance also provides information about the burden of illness, he noted, including the risk factors and clinical outcomes related to resistant infections versus antibiotic-susceptible ones. Furthermore, he added, it generates critical data to inform decisions about actions taken to mitigate resistance when emerging trends are identified. McDermott said that NARMS data are central to FDA’s regulatory processes (see Box 3-1 for information on a new FDA policy) and other evidence-based policies regarding judicious antibiotic use. Pre-harvest surveillance data support risk analysis of foodborne antimicrobial resistance hazards as part of the qualitative risk assessment process for preapproving new animal antibiotics. Post-harvest data contribute to identifying interventions to contain resistance and to evaluating the effectiveness of antibiotics after approval.1 Finally, he said that One Health surveillance data are used to evaluate
___________________
1 As examples, he cited FDA’s withdrawal of fluoroquinolones for use in poultry and its prohibition of extra-label use of third-generation cephalosporins; the latter has had a measurable and fairly immediate effect in both human and animal isolates, he said.
whether an evidence-based intervention has achieved its intended effect—that is, establishing a new baseline—at which point this cycle begins anew.
Potential Challenges of One Health Surveillance
Based on his experience with NARMS, McDermott outlined a set of challenges faced in One Health surveillance. He noted gathering accurate information and bacterial isolates is expensive, laborious, and requires sustainable commitment from government and public health sectors. Maintaining a sound sampling scheme along the food chain and environment is challenging, albeit critical, for valid trend analysis, he added. McDermott said that silos hinder collaboration and data sharing among the agriculture, industry, and public health sectors, as well as among microbiologists, epidemiologists, and other specialists within and across sectors. In that vein, he reflected, fostering international harmonization and cooperation is becoming increasingly important. Publishing complex findings in a manner that is timely and appropriate for different audiences is a continuous challenge, he said. A related challenge is using the data to formulate sound public health policy, because there are conflicting opinions about how much evidence is sufficient to act upon. He noted that calls for more data represent a major cause of delay.
Lessons Learned from NARMS
McDermott reflected on lessons learned from the 20-year history of NARMS. He explained that some nontyphoidal Salmonella are more adept than others at acquiring multidrug-resistant plasmids; therefore, resistance to critically important antibiotics differs by serotype.2 He reported that since NARMS began, the overall susceptibility picture for all serotypes on its current 15-drug panel has steadily improved. Resistance to three or more classes of drugs (multidrug resistance) among human isolates declined between 1996 and 2014 in the United States (see Figure 3-1). Among the critically important antibiotics—ceftriaxone, azithromycin, and ciprofloxacin—only ceftriaxone resistance is material in human isolates. According to McDermott, the rates of resistance to quinolones and third-generation cephalosporins in human nontyphoidal Salmonella isolates in the United States are comparable to the best susceptibility situations in the European Union. Among nontyphoidal Salmonella isolates in broiler meat
___________________
2 Antimicrobials are deemed critically important if they are (1) sole therapies or one of few alternatives to treat serious human disease, and (2) used to treat diseases caused by either organisms that may be transmitted to humans from nonhuman sources, or human diseases causes by organisms that may acquire resistance genes from nonhuman sources (WHO, 2012).
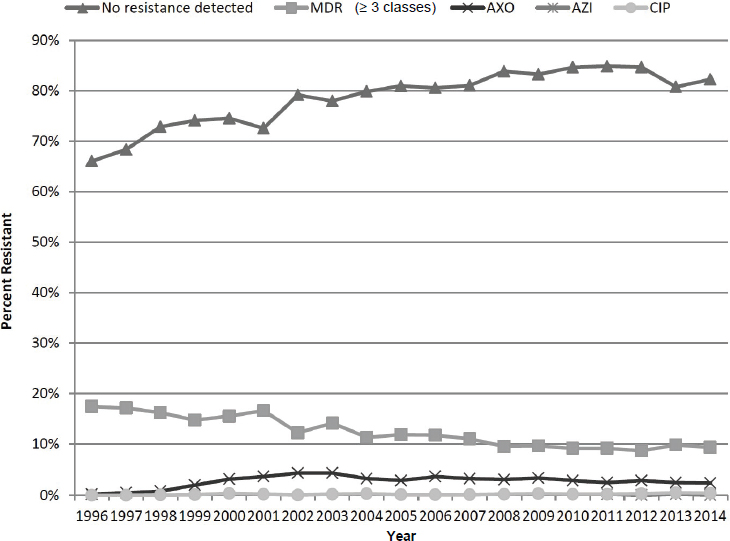
NOTE: AXO = ceftriaxone; AZI = azithromycin; CIP = ciprofloxacin; MDR = multidrug resistance.
SOURCE: McDermott presentation, June 20, 2017.
supply in the United States, antibiotic resistance is roughly on par with the European Union.
Whole genome sequencing is affecting a sea change in surveillance, observed McDermott. It provides comprehensive genomic information on resistance-related genes and is thus proving to be a good surrogate for traditional in vitro susceptibility testing, per several recent studies on resistance genotype–phenotype correlations for target food-borne pathogens. For example, a clinically resistant strain of Salmonella enterica will have a known resistance determinant (genotype–phenotype correlation) in around 99 percent of cases (Zankari et al., 2013; McDermott et al., 2016). A One Health approach is essential because resistance is a problem that transcends national borders, cautioned McDermott. Plasmids are transmitted readily around the world, and those that accumulate resistance genes do so rapidly and spread to other pathogens within the same family. To illustrate, he noted that the history of accumulated resistance is recapitulated in some
modern strains. One particular plasmid backbone (IncA/C2 plasmid backbone 113,320 bp), now fairly widespread in U.S. agriculture, is 99 percent identical to a plasmid from a child in Madagascar who had plague in 1997. McDermott said that NARMS provides comprehensive genomic information on its website,3 and it is preparing to launch a data dashboard tool for real-time reporting that will enable resistance tracking.
McDermott concluded with reference to some existing gaps in One Health surveillance. NARMS needs to incorporate programs for food animal and companion animal pathogen surveillance, he said, as well as for on-farm testing to assess husbandry practices on resistance. He said that an environmental surveillance piece is needed to complete the One Health platform and to better understand the movement of pathogens and resistance genes, both in the United States and worldwide. He predicted that the tide will shift toward microbiome-type surveillance, but he cautioned against allowing the sheer volume of data generated by new genomic approaches to impede or delay the most critical step—taking appropriate action to mitigate resistance.
Quality of One Health Surveillance in Developed and Developing Countries
Cray reviewed the quality of antimicrobial surveillance across the One Health domains, with a focus on the respective challenges faced by countries at differing levels of development. She is involved in developing guidance documents to frame the minimal requirements for establishing a surveillance system for antimicrobial resistance. Efforts in developing countries may face different challenges than efforts in developed countries, she said. To illustrate the practical reality of data collection fieldwork in parts of some developing countries,4 she described work carried out by her graduate students in Uganda. Cray noted they tend to face lengthy delays caused by impassable roads, frequent power outages, and a poor laboratory infrastructure that renders bacterial culture curation and procurement of supplies challenging. Very few translators are available to enable communication across Uganda’s 14 different dialects and to help surmount cultural differences for her students, she added.
___________________
3 For more information, see www.cdc.gov/narms/index.html (accessed July 31, 2017).
4 Marcos Espinal, director of communicable diseases and health analysis at the Pan American Health Organization, commented to Cray that her generalization about developing countries may be sending the wrong message—not all countries in Latin America, Asia, and other areas lack infrastructure and standards. Rather, the main problem in many of those countries is the lack of political will to build capacity. She responded that if time permitted, she would have included a third category for transitioning countries and regions.
Potential Opportunities to Improve Surveillance Data Integration
Most current surveillance, especially in developing countries, primarily involves country-based and human-focused systems, said Cray. However, there is an ongoing shift from site-specific monitoring toward continuous, long-term, multisectoral monitoring systems. She noted that this shift has underscored the need for improved data collection and integration across the human, animal, retail, and environmental spheres. This research gap is evident in the “infinitesimally small” number of total publications on antimicrobial surveillance systems—relative to its critical importance—in the past 45 years, she noted. Sampling methodologies for antimicrobial susceptibility testing are not consistent within and between different countries, said Cray, so improving quality control and harmonizing culture-sampling methodologies will be crucial for better data integration. Current activities aimed at this goal include global- and national-level action plans led by WHO and its Advisory Group on Integrated Surveillance of Antimicrobial Resistance,5 by the Food and Agriculture Organization of the United Nations (FAO), and by the World Organisation for Animal Health (OIE). WHO’s Global Antimicrobial Resistance Surveillance System is working to coordinate how data are captured,6 she said, but only for human surveillance and not for animal, veterinary, retail, or environmental surveillance at this time. Other immediate needs, she added, include harmonizing the ways that data are analyzed and reported as well as supporting and enhancing the WHO, OIE, and FAO programs.
Cray called for creating a real-time global databank of existing data sets that could include, for example, antimicrobial resistance, infectious disease, sequencing, climate, wildlife migration, and migratory birds. The databank could be compiled, assimilated, and analyzed to identify gaps, promote innovation, and take collaborative action against antimicrobial resistance. She recommended framing this work within a new paradigm—the Collective Antimicrobial Resistance Ecosystem (CARE) (see Figure 3-2). The paradigm is predicated on continual exposures to multiple types of resistance determinants at the interface of humans, animals, and the environment. To conclude, she suggested that hurricanes, floods, droughts, fires, and volcanic eruptions represent unique opportunities to study antimicrobial resistance after the environment has been “reset,” observing: “Once we have homeostasis again in the environment, if we begin sampling, can we then watch at a true evolutionary development of resistance and bacterial gene movement over time?”
___________________
5 For more information, see www.agisar.org (accessed July 31, 2017).
6 For more information, see www.who.int/antimicrobial-resistance/publications/surveillancesystem-manual/en (accessed July 31, 2017).
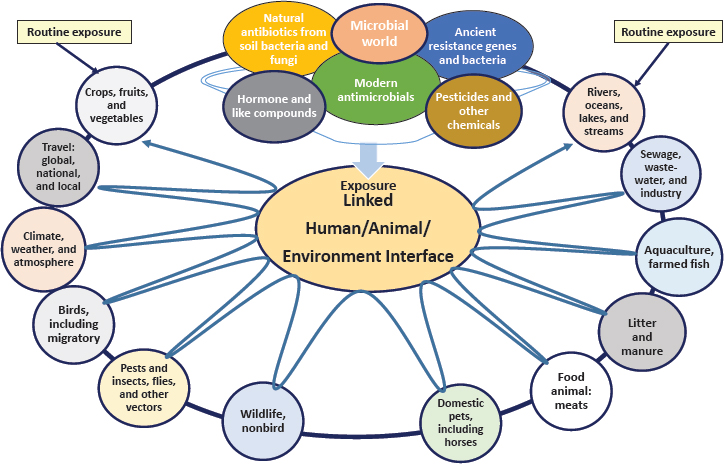
SOURCES: Cray presentation, June 20, 2017; adapted from USDA, 2014.
Antimicrobial Surveillance in the Environment
According to Tiedje, better understanding of antibiotic resistance genes and antibiotic-resistant bacteria in the environment is a high-priority knowledge gap. This includes their survival rates, survival conditions, and treatment methods, as well as their transport, growth substrates, growth conditions, and the microbial ecology of manure and commensal hosts. In a commensal relationship between bacteria and host microorganisms, the bacteria benefit from the relationship while the hosts remain unaffected. Further research on horizontal gene exchange is a specific priority for Tiedje, because it is the point of control of antimicrobial resistance.
Horizontal Gene Exchange
To explain the process of horizontal gene exchange, Tiedje described the continuum through which multidrug-resistant organisms emerge. Every gram of soil in the native resistome, which is a collection of all the antimicrobial resistance genes in a microbial environment, contains antibiotic resistance in its microbes, which can be steered in a problematic direction
by anthropogenic factors such as general pollution, antibiotic production, wastewater treatment plants, animal agriculture, and aquaculture. Horizontal gene exchange occurs in the “organismal soup” that arises out of this environmental selection, he said. The soup contains commensals that can carry antibiotic resistance genes, mobile genetic elements, and pathogens. However, the elements in the soup that are critical in driving the creation of multidrug-resistant organisms are selection, growth conditions, cell density, and cell contact, Tiedje added. Managing those elements, he said, is the key to minimizing horizontal gene exchange and the creation of resistant pathogens. Ample evidence demonstrates this process of horizontal gene exchange, Tiedje said. He cited a study on the abundance of antibiotic resistance genes in the metagenomes of different environments (Li et al., 2015). The abundances in natural environments, such as sediments, soils, and river water, were up to three orders of magnitude smaller than total abundances found in environments that were seriously affected by anthropogenic environmental selection, such as feces and wastewater from animal agriculture.
Resistance Clusters and Coselection
Resistance clusters and coselection for antibiotics, heavy metals, and disinfectants are key parts of the resistance problem, said Tiedje. Evidence suggests strong correlations between certain antibiotic resistance genes and mobile genetic elements, he said. Clusters of identical sequences found in three different pig farms in different regions of China indicate that genes are transferring globally and in particular clusters (Johnson et al., 2016). He noted that the same study analyzed the “growth” of one cluster in compost from a single Chinese farm. In manure, it appears that coselection occurs for the growth of organisms with certain types of resistance genes, he said, which seem to be genetically linked in a resistance cluster.
Strategies for Environmental Surveillance
Tracking the volume of antibiotic use, production, and emission around the world will be critical for enacting a targeted strategy for action, Tiedje argued. He quoted Rai Kookana, an expert from Commonwealth Scientific and Industrial Research Organisation in antibiotic quantification, as attributing the majority of the world’s production to China and India. Kookana suggested that strategies target Asia, because the necessary components for horizontal gene exchange and selection are highly prevalent in that region (Kookana et al., 2014) (see Figure 3-3).
Tiedje cautioned that each sector, such as dairy, pig, chicken, fish, wastewater treatment plants, and antibiotic production facilities, as well
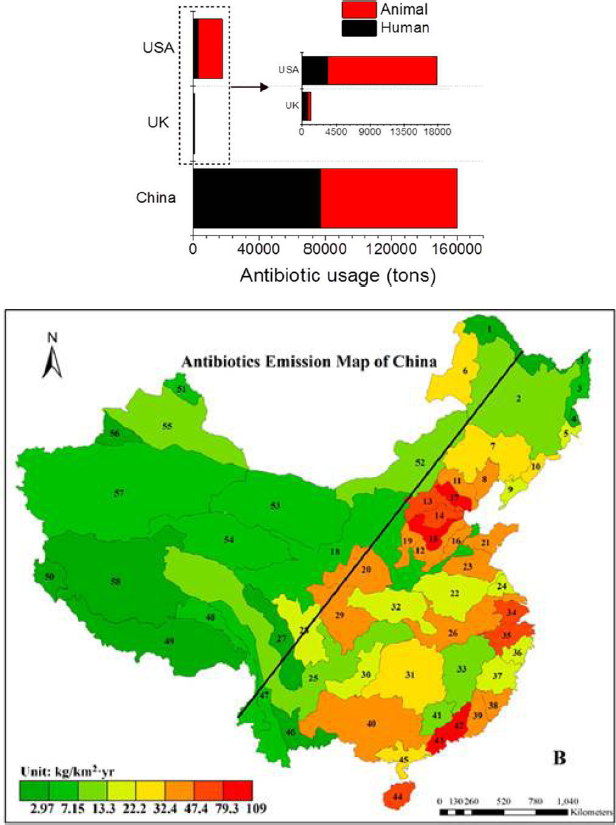
Top: Comparison of antibiotic usage among China, the United Kingdom, and the United States.
Bottom: Density of antibiotics emissions in river basins of China.
SOURCES: Tiedje presentation, June 20, 2017; Ying et al., 2017; Zhang et al., 2015. Reprinted (adapted) with permission from Ying, G. G., L. Y. He, A. J. Ying, Q. Q. Zhang, Y. S. Liu, and J. L. Zhao. 2017. China must reduce its antibiotic use. Environmental Science and Technology 51(3):1072–1073. Copyright (2017) American Chemical Society; Reprinted (adapted) with permission from Zhang, Q. Q., G. G. Ying, C. G. Pan, Y. S. Liu, and J. L. Zhao. 2015. Comprehensive evaluation of antibiotics emission and fate in the river basins of China: Source analysis, multimedia modeling, and linkage to bacterial resistance. Environmental Science and Technology 49(11):6772–6782. Copyright (2015) American Chemical Society.
as each region needs a tailored strategy for surveillance, because each faces different issues with respect to development, income level, rural or urban settings, and level of antibiotic production. Operations are rapidly changing owing to technological advancements, for example, that allow larger-scale farmers to use management strategies including infrared infection detection, saliva monitoring, and behavioral monitoring to quickly isolate and treat animals. He explained that surveillance data will help to validate the relationships among quantitative polymerase chain reaction data, metagenomic data, isolate data, and residue data. Another of Tiedje’s actionable priorities was sector-specific education and training about resistance, such as the “Pork Checkoff” campaign in the United States, which provides resources for young people on antimicrobial use and regulations among other educational tools.7
Given the volume of molecular data becoming available, Tiedje said, improvements in data integration, data mining, and epidemiology are needed for developing quantitative risk-assessment models to guide action against resistance. Furthermore, he argued that there is a strong economic incentive for a NARMS-style system that provides real-time data monitoring (cow-side, for example) that can be linked to risk and allow users to make better decisions, to save money, and to promote less resistance. Tiedje also suggested that integrating environmental surveillance data with clinical data from hospitals would help to identify links and perform real-time risk analyses. As an example, he noted that Stedtfeld et al. (2016) have developed an antimicrobial resistance dashboard application designed to geospatially map antibiotic resistance genes and antibiotic-resistant bacteria from environments and clinics.
DISCUSSION
David Rizzo, chair of the Department of Plant Pathology, University of California, Davis, asked about the practice of spraying antibiotics on plants and its impact on soil. Cray said that her model plans to capture the use of antimicrobials and pesticides on plant crops, because there is particularly selective pressure at the bacterial level. Tiedje commented that such use on plants is very low in the United States, at 0.1 percent of total use.
Jeffrey Duchin, health officer and chief of Communicable Disease Epidemiology and Immunization Section for Public Health for Seattle and King County, Washington, asked if there are drivers of resistance in humans, besides the administration of antibiotics, that can be identified through the food production chain, such as pre- or postharvest animal husbandry prac-
___________________
7 For more information, see www.pork.org/production-topics/antibiotics-resource-center (accessed September 18, 2017).
tices, and if there are key points at which the resistance problem is exacerbated that are analogous to horizontal gene transfer hot spots. McDermott said that genomic data can reveal links with antimicrobial resistance traits that the surveillance system does not test for, such as quaternary ammonium compounds used in carcass washing during animal processing, so those types of processing steps warrant closer consideration. Tiedje noted that copper and zinc resistance are important co-selectors being used more widely as the use of antibiotics decreases. Cray added that practices for moving animals could even be drivers: it can increase animals’ stress and thus the recrudescence of shedding, which increases the distribution of bacteria in the population.
Peter Daszak, president of EcoHealth Alliance, asked if the livestock industry still uses growth promoters that are clinically significant antimicrobial drugs in human health. McDermott clarified that in the United States, the use of any antibiotics that are classified as medically important can no longer be used for growth promotion as of January 2017; however, that does not include ionophores, which are a class of antibiotics used to slow the growth and reproduction of parasites in animals. Tiedje suggested that tetracycline, used very heavily in animals, could be a critical driver of coselecting the important human resistances. Cray noted that tetracycline use is common in human dermatology; in Denmark, an increase in one bacterial population’s resistance to tetracycline was attributed primarily to increased human use (Lomholt and Kilian, 2014).
David Relman, professor of medicine at Stanford University, suggested designing specific studies to better understand the microenvironments in which gene flow is most significant and asked about the best possible study design for capturing and interpreting genetic information. McDermott recommended using longitudinal studies that capture national use data as well as biological (and ultimately metagenomic) samples. When coupled with data on variable practices nationwide, he predicted, those studies will help clarify which of those practices are driving resistance. Cray reiterated the value of conducting a study after the environment has been reset—for example, after a flood affecting agricultural land, sewage treatment plants, and hospital wastewater treatment plants.
ANTIMICROBIALS IN THE ENVIRONMENT
Interface and Pathways of Gene Transfer
Many current antibiotic-resistant infections are the result of very rare genetic events, said Price, so the overarching goal should be reducing the opportunity for these rare genetic events to occur, for example, in billions of food animals worldwide. He added that intervening effectively requires
microbial-level understanding of how resistance determinants can be transmitted between hosts.
Transmission of Resistance Determinants
Some resistance determinants can be “hardwired” into a microbe’s genome, said Price, and some resistance determinants are located on mobile elements that can move around within the genome of that organism. Furthermore, some types of mobile elements can jump around within an organism’s genome, while others sit on different kinds of mobile elements that can be passed between bacteria. Both hardwired and mobile elements are implicated in amplifying resistance in food animals and ultimately threatening human health, he said. Bacteria with hardwired resistance determinants can emerge in food animals and jump to humans, causing antibiotic-resistant infections. The same transmission scenario holds for bacteria with mobile resistant determinants. In another transmission scenario, bacteria with mobile resistance determinants can emerge in food animals, jump to humans, and transfer their resistance determinants to another bacterium that causes antibiotic-resistant infections. Finally, he said, bacteria with hardwired resistant determinants can emerge in food animals, jump to humans, and transfer the resistance determinant via transformation or transduction.
Resistance Transmission in Salmonella and Campylobacter Bacteria
New multidrug resistance determinants in Salmonella have been associated with the introduction of antibiotics to animals, said Price. The reverse scenario, rapid decrease in resistance, has also been observed when antibiotics have been removed from food animal production. For example, when the Canadian government asked Quebecois broiler producers to stop injecting broiler chicken eggs with cephalosporins, Price said, there was an immediate and precipitous decrease in cephalosporin-resistant Salmonella in poultry products.8 In parallel, he said, there was a direct positive effect on human health: a precipitous decrease in cephalosporin-resistant Salmonella infections in people and a decrease in cephalosporin-resistant Escherichia coli. In another example, ciprofloxacin-resistant Campylobacter infections increased rapidly in humans after the introduction of enrofloxacin (the animal version of ciprofloxacin) in broiler chicken production, according to Price. This evidence spurred FDA to block the use of enrofloxacin
___________________
8 For more information, see www.canada.ca/content/dam/canada/public-health/migration/publications/drugs-products-medicaments-produits/antibiotic-chicken-industry-surveillance-resistance-antibiotique-industrie-poulet/alt/pub-eng.pdf (accessed July 31, 2017).
in broiler chicken production,9 Price said, but removing the drug did not cause a rapid decrease in resistance (Nelson et al., 2007). The rates merely flattened out, he explained, because resistance was coded by a single-point mutation in a housekeeping gene hardwired in the genome. Because there was no measurable metabolic cost to the organism to carry the mutation, there was no counterselection to eliminate it.10
Resistance Transmission in Colonizing Opportunistic Pathogens
As of 2013, CDC estimated that there were about 410,000 drug-resistant infections by Salmonella and Campylobacter each year in the United States (CDC, 2013b); however, this figure has not been sufficient to induce aggressive policy actions. He surmised that “body counts” are more powerful than infection statistics for catalyzing action among policy makers, and the number of deaths for those two bacterial infections was estimated at less than 1,000. Furthermore, he said, infectious disease doctors in general do not focus on Salmonella and Campylobacter in the context of superbugs, but rather tend to think about colonizing opportunistic pathogens (COPs) instead. According to Price, these COPs include
- Klebsiella pneumoniae, including carbapenem-resistant Enterobacteriaceae (CRE);
- Extraintestinal pathogenic Escherichia coli, including CRE;
- Enterococcus, including vancomycin-resistant Enterococcus; and
- Staphylococcus aureus, including methicillin-resistant Staphylococcus aureus (MRSA).
COPs are more difficult to study than foodborne pathogens, said Price, because they cause “insidious epidemics” spanning human, animal, and environmental hosts. They also have indefinite and asymptomatic colonization periods and are transmitted silently (asymptomatically) from person to person.
Next-generation DNA sequencing and other new tools are revealing more about coresistance and horizontal gene transfer in COPs, said Price. An example from the Netherlands illustrates how coresistance can fuel the spread of resistance in the absence of profligate antibiotic use. In 2004, a new strand of livestock-associated MRSA (ST398) was found in an infant and traced to the family’s pig farm, and a survey of livestock throughout
___________________
9 For more information, see www.fda.gov/AnimalVeterinary/SafetyHealth/RecallsWithdrawals/ucm042004.htm (accessed July 31, 2017).
10 He cautioned that in Spain, they continued to use cipro-enrofloxacin until virtually all of their Campylobacter was resistant to ciprofloxacin.
Europe found the strain was already spreading rapidly (Voss et al., 2005). Whole-genome sequencing was applied to understand the evolutionary history of its epidemiology, which originated as methicillin-susceptible Staphylococcus aureus (MSSA) that had made a host jump from humans to animals. Heavy antibiotic use mediated the transition from human MSSA to pig MRSA, which began spreading back to people working in the agricultural industry, mainly through direct exposure. Price warned that farmers, farm workers, and veterinarians—and their families—are on the frontline of livestock-associated MRSA, which can be fatal to a susceptible host. An important cautionary example of horizontal gene transfer, said Price, is mobilized colistin resistance (see Box 3-2).
Future Directions for Research in Resistance Transmission
Going forward, said Price, whole-genome phylogenetic analyses will help to trace host jumps and the natural history of mobile element acquisition. Bayesian molecular clock analyses can be used to estimate the point of time in prehistory when two or more life forms diverged; he suggested that it will also help to estimate the timing of when mobile elements jump
between animals and people. However, mobile genetic elements are often noisy, with much recombination and little reliable phylogenetic signal. He said that longer-read sequencing methods may help address this challenge, but it will require significant investment to define the clouds of genetic diversity that can exist even on a single farm. Furthermore, it is not yet possible to quantify the proportion of antibiotic-resistant human infections that are caused by antibiotic use in livestock production, be it historic or contemporary. Addressing antibiotic resistance will require ensuring transparency, obtaining source samples, closing the routine disease prevention loophole, and raising animals and people in ways that promote health and obviate the need for antibiotics, he concluded.
Environmental Reservoirs of Antimicrobial Resistance and Effects of Antibiotic Residues
The One Health framework has three key elements, said Topp: humans who receive antibiotics; animals and fish that receive antibiotics; and the terrestrial and aquatic environments. Microorganisms in the environment represent a reservoir of genes that confer resistance to antibiotics, he explained, and those genes can be recruited into pathogens of significance to human or animal health. The environment also represents a way for these antibiotic-resistant organisms to transmit to humans, to animals, and between humans and animals. Pathogenic organisms can carry antibiotic resistance genes, he said, but commensal organisms can also carry antibiotic resistance genes that ultimately can be transmitted to pathogens. He presented Figure 3-4, which articulates the sources and transmission pathways of antimicrobial resistance within the One Health framework.
Impact of Anthropogenic Activities on the Environmental Reservoir
From a One Health perspective, Topp said, a major concern is that anthropogenic activities are potentiating an environment conducive to resistance transmission. Researchers are working on multiple fronts to mitigate this effect. Most work is focused on the increasing abundance of antibiotic resistance genes in the environment, which entails an undesirable increase in exposure to these genes through the environment. Another concern is the promotion of resistance to new antibiotics as they are brought to market, he said. Evidence suggests that bacteria in environments exposed to certain chemicals have an accelerated evolution rate. Another focus is whether the association between antibiotic resistance genes and elements that confer mobility is increasing, which would accelerate the speed at which human pathogens recruit those genes. Yet another focus is whether resistance genes get “stacked,” thus increasing the probability of resistance to multiple
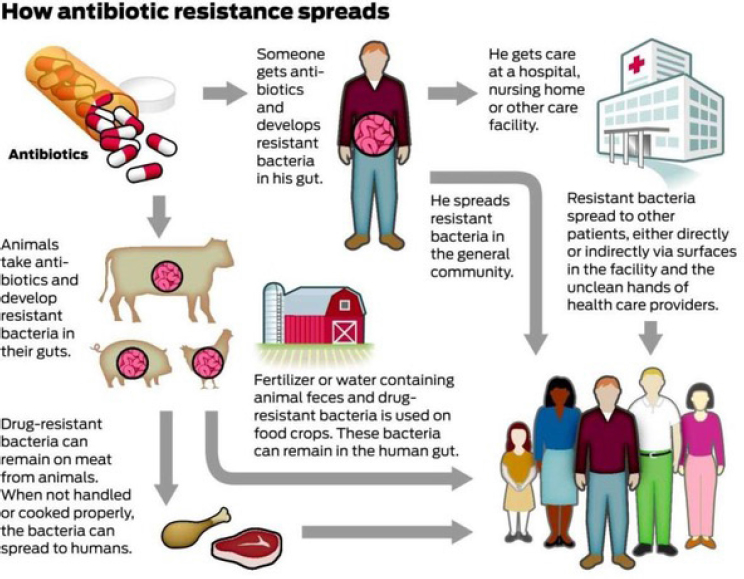
SOURCES: Topp presentation, June 20, 2017; Todd Trumbull/San Francisco Chronicle/Polaris; adapted from CDC, 2013a.
classes of antibiotics. Genes conferring multiple types of resistance that are dispersed in different bacteria are of less concern than those on a single bacterium that can be exchanged to other bacteria, he said.
Being able to identify the source of waterborne antimicrobial resistance is particularly critical in low-income countries without adequate infrastructure for potable water or wastewater treatment, Topp said. Some bacteria, such as Escherichia coli O157:H7 bovine, have reservoirs that are specific enough to trace if detected in water. Water can contain fecal source markers as well. Caffeine or human/veterinary pharmaceuticals are human-specific chemical markers, he said, and biological markers include host-specific DNA from Bacteroidales bacteria or mitochondrial DNA from fecal material.
Biological Contaminants in the Resistome
Many anthropogenic activities can potentiate the environmental resistome, said Topp. People who are medicated excrete antibiotic residues
and bacteria that have been selected in their digestive tracts, so human waste streams are a primary threat. Human waste streams also include effluents from wastewater treatment plants that enter directly into aquatic environments, such as through irrigation with reclaimed water or land application of recovered biosolids. Animal waste streams from livestock production systems also contain excreted antibiotic residues and resistant bacteria selected within the animals, he said. This can potentiate the environmental resistome when manure is recycled onto crop production ground. In aquaculture, direct application of drugs to water is a major concern. Manufacturing plants that fabricate antibiotics in many parts of the world also produce effluents that contaminate the environment with high concentrations of drugs, he said. Establishing minimal selective concentrations of antibiotics, said Topp, will require better understanding the relationship between environmentally relevant concentrations of antibiotics and whether those concentrations are high enough to select for resistance.
Chemical Contaminants in the Resistome
In addition to antibiotic-resistant bacteria from biological contaminants, Topp explained, waste streams also contain organic or inorganic chemicals such as copper and zinc that can coselect for resistance (Song et al., 2017). Mounting evidence shows that livestock animal fecal matter is enriched for antibiotic-resistant bacteria and that crop ground fertilized with these materials become enriched with antibiotic resistance genes (Marti et al., 2013). Researchers are investigating whether those genes will appear in crops grown on those soils, he said, and whether consumption of those crops by humans or animals represents a route of transmission from the environment.
Environmental Persistence of Antibiotic Resistance Genes
Antibiotic resistance genes can persist for at least months in crop ground through commercial-scale application of animal manures, said Topp. Whether all antibiotic resistance genes in soil behave consistently with respect to their persistence in dynamics remains unknown, but evidence suggests that certain proxies or sentinel DNA markers may represent many antibiotic resistance genes, he said. For example, class 1 integrons seem to be highly correlated with the abundance of many antibiotic resistance genes (Gillings et al., 2015). Topp explained that when antibiotic residues are applied to the ground, their persistence varies widely. For example, beta-lactams are quickly destroyed in soil or water, but fluoroquinolones are quite persistent because they are “sticky” to soil and their
bioavailability is reduced.11 Environmental factors such as aeration and tilling can affect persistence, he said, as can different exposure scenarios (e.g., punctual manure application versus constant effluent from wastewater).
Topp concluded by recommending a shift in focus from exposure to risk:
In the realm of environmental science related to antimicrobial resistance, the state of the science right now is exposure assessments. We can measure, we can quantify antibiotic resistance genes or bacteria, but we really have very little understanding of the significance of that to human health in a risk assessment context. So we really need to make the leap from exposure to hazard to risk.
Some Management Strategies for Reducing the Transfer of Antimicrobials to the Environment
Durso opened her presentation by drawing a conceptual distinction between the term resistance as used in environmental versus clinical settings. In clinical settings, she described, antibiotic resistance is a function of a pathogenic bacterial isolate and is often linked directly to treatment failure. In environmental settings, she said, the term is not consistently defined and it can be applied either to an isolate or to an entire community of bacteria. Relationships are generally indirect; most bacteria are not pathogens and pathogenicity of zoonotic and opportunistic bacteria is assumed, but not known. Durso’s first priority is the need for more precise vocabulary for discussing environmental antibiotic resistance, both within the field and across the One Health triad. This would strengthen problem-solving efforts and allow for resources to be allocated where they will have the most impact, she suggested.
Evaluating Strategies for Mitigating Antimicrobial Resistance
To evaluate the efficacy of mitigation strategies to reduce the transfer of antibiotic resistance, Durso said, three categories of targets can be used: drugs and bioactive drug breakdown products, bacteria that can grow in a predetermined drug concentration, or any part of any target gene or resistance determinant. Different targets tell different stories, she warned. Different conclusions about resistance can be drawn from the same sample, because each of those categories has multiple subtargets that can
___________________
11 Among pharmaceutical chemical contaminants in a Canadian sample, Topp reported, the biocide triclocarban had the highest concentration of dry weight in biosolids. Ciprofloxacin’s concentration was also high, as were concentrations of other fluoroquinolones, tetracyclines, and macrolide antibiotics.
be measured and there are various ways to present and analyze the data. Establishing a reasonable goal for reduction, she said, requires being able to assess native or naturally occurring (background) resistance and reference (baseline) levels of resistance prior to a mitigation treatment. Data from manure, soil, water, and air reveal that the relationship between drugs and antibiotic resistance is complex, she said. Sometimes there are clear links between drugs and the resistance measure, in line with the current assumption that “more drugs equal more resistance” (Zwonitzer et al., 2016). However, because there are other drivers of resistance in the environment, other studies have found that increases in antibiotic drug concentrations are not necessarily correlated with increasing measures of the resistance target being measured (Dalkmann et al., 2012).
Drivers of Resistance Beyond Drug Use
Durso explained that antibiotic resistance transfer from agricultural to human settings is an issue deeply entwined with—and confounded by—the idea that agricultural antibiotic drug use is the primary or sole driver of resistance. However, studies carried out in organic and drug-free systems may help to begin disentangling these two separate questions. She cited a study reporting that resistance persisted in organically raised swine even in the absence of farm use of antibiotics (Stanton et al., 2011). A growing body of evidence from this and other studies, she said, suggests that reducing resistant bacterial populations will require strategies in addition to prudent use of antibiotics, such as the manure management strategies described in the next section. Realistic mitigation targets for drug residues, bacteria, and genes should reflect the fact that resistance occurs both naturally and as a result of historical and current anthropogenic activities, she argued. She further argued that from the perspective of long-term risk reduction, the source of the target is of little importance to achieving the goal of reducing transfer within and outside of agricultural systems. From the regulatory perspective, however, she conceded that identifying the source of the target does matter. The working assumption that antibiotic use is the primary driver of resistance on farms and in the surrounding water, soil, and air is untested, she noted. While drug use is a strong driver of antibiotic resistance in the animal gut, she said, increasing evidence suggests the need to revisit that assumption and identify the factors—other than drug use—that drive the transfer of antimicrobial-resistant drugs, bacteria, and genes out of the agricultural system.
Manure Management Strategies
Durso surveyed current management options for reducing resistance transfer in manure. The main types of manure are ground-deposited feces
from grazed beef and dairy animals, solid manure collected from the surface of feedlot pens or from inside of poultry houses, and physically contained liquid manure slurries. Application methods include liquid irrigation, slurry injected beneath the soil surface, and dry product applied to the soil surface. She reported that several manure management strategies have shown promise for mitigating some measure of antibiotic resistance, including wood chip bioreactors, composting, land application, and anaerobic digestion (for more detail, see Box 3-3).
A Manufacturing Perspective on Reducing the Environmental Impact of Antimicrobials
Brooks provided a manufacturing perspective on reducing the environmental impact of antimicrobial production. He began his presentation by highlighting that maintaining a supply of affordable, accessible antibiotics is essential to global public health and yields huge societal benefits. Manufacturing and the (proper or improper) use and disposal of medicine,
he continued, are potential sources of antimicrobials in the environment, but there are many other sources as well (see Figure 3-5). While antimicrobial resistance in the environment is ancient and predates the industrial-scale use of antibiotics (D’Costa et al., 2011), Brooks acknowledged that the phenomenon at hand may indeed be accelerated, and many stakeholders in the industry recognize the elevated levels of antimicrobials in environmental samples.
The industry’s supply chain for established antibiotics is complex and global, he said, stating that it has a significant footprint; it has also raised concerns of environmental pollution from some drug manufacturing companies in emerging markets such as China and India. A recent influential study on antimicrobial resistance asserted a link between manufacturing pollution and antimicrobial resistance and called for better control of manufacturing effluent (Review on Antimicrobial Resistance, 2016). Brooks explained that active pharmaceutical ingredients can be found in
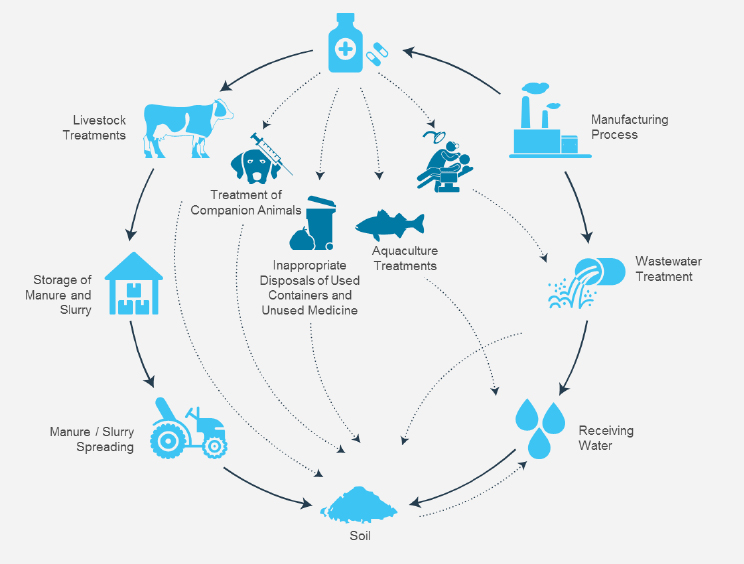
SOURCES: Brooks presentation, June 20, 2017; adapted from Boxall, 2004.
two main types of manufacturing waste streams.12 Solid waste generated by the manufacturing process, such as sewage sludge, must be managed to prevent soil and ground water contamination. Wastewater from manufacturing plants requires effective controls to minimize the concentration of active pharmaceutical ingredients in the receiving water. To ensure the effectiveness of those controls, he indicated, research is needed to measure wastewater concentrations, to establish “safe” discharge concentrations, and to better understand the role of other coselective agents such as metals, biocides, and cleaning agents.
Road Map to Reduce Environmental Impact of Antibiotic Production
Leading companies and industry organizations have publicly committed to address the risk of antimicrobial resistance by releasing the Declaration on Antimicrobial Resistance at the World Economic Forum in Davos, Switzerland, in January 2016,13 with signatories including more than 80 biopharmaceutical companies. It calls on key stakeholders to take collective action to address antimicrobial resistance, including governments to commit to allocating the funds needed to create a sustainable and predictable market for new antibiotics and diagnostics while also implementing the measures needed to safeguard the effectiveness of antibiotics. The signatory companies commit to reducing the development of antimicrobial resistance, including “measures to reduce environmental pollution from antibiotics.”
Because that is the only specific language related to environmental pollution in the declaration, he said, a group of 13 leading companies signed an industry road map in September 2016. The document further details their practical commitments to implement measures to reduce the environmental impact of antibiotic production. Brooks explained the road map includes other commitments to promote research and development through new collaborations and incentives; to improve access to antibiotics, diagnostics, and vaccines; and to ensure antibiotics are only used by people who need them. For the environmental piece, the road map signatories commit to the following:
- Reviewing (individually) their own manufacturing and supply chains to assess good practices in controlling releases of antibiotics into the environment;
___________________
12 He noted that there are also less direct sources; for example, if a manufacturing plant has contamination around it, then storm water runoff could contain antibiotic material.
13 Full text of the Declaration on Antimicrobial Resistance is available at www.ifpma.org/wp-content/uploads/2016/01/Industry_Declaration_on_Combating_Antimicrobial_Resistance_Jan2017.pdf (accessed July 30, 2017).
- Establishing a common framework for managing antibiotic discharge by 2018;
- Working with stakeholders to develop a practical mechanism to transparently demonstrate that their supply chains meet the standards in the framework;
- Working with independent technical experts to establish science-driven, risk-based targets for discharge concentrations for antibiotics and good practice methods to reduce the environmental impact of manufacturing discharges by 2020; and
- Supporting calls for the establishment of a high-level coordinating mechanism to provide global leadership, mobilize resources, set goals, and measure progress toward them.
Brooks reported that the Antimicrobial Resistance Industry Alliance Environmental Working Group is currently in the process of developing the environmental framework, establishing science-driven standards and risk-based targets for discharge concentrations, influencing other companies through outreach efforts to take appropriate action, and sharing best practices and environmental assessment programs with one another. The process is supported by collaboration with relevant experts and stakeholders, he said, and progress will be transparently reported. However, he emphasized that widespread promulgation of these standards and practices in the industry—among both innovators and generic manufacturers—beyond the 13 signatories will be critical to reduce the overall manufacturing contribution to antibiotics in the environment.
DISCUSSION
Duchin asked if traditional toxicologists have been engaged to help address challenges in risk assessment. Durso replied that the toxicological framework is being widely adopted in the environmental realm, with antibiotic-resistant bacteria and antibiotic resistance genes classified as contaminants. There may also be room for incorporating a conceptual framework from infectious disease modeling to inform thinking about environmental resistance, she added. Duchin suggested the food production industry could follow the pharmaceutical industry’s lead in creating a road map to monitor antimicrobial drug resistance, as it already monitors for bacteria considered dangerous to the food supply. Price agreed that a regulatory infrastructure should be established, given the existence of potentially untreatable bacteria in the food supply that are not classic food-borne pathogens but that can transmit resistance and cause disease (such as mcr-1).
George Poste, chief scientist of the Complex Adaptive Systems Initiative at Arizona State University-SkySong, asked about new categories of tech-
nologies that could be deployed in compound destruction and elimination. Brooks replied that there are a variety of technologies in use, constrained by cost and practicality. Many biologic wastewater treatment plants use a “zero liquid discharge” system to evaporate liquid discharge through multistage evaporators; other technologies include dose analysis and tertiary carbon treatment. He explained that the industry traditionally looks at more typical ecotoxicological end points to assess the safety of discharge from plants, but surrogate resistance-based end points are an area of ongoing research. Brooks predicted that a wider network of publicly owned wastewater treatment plants will be necessary, especially in large cities, to handle other nonmanufacturing sources of resistance, such as excreted metabolized antibiotics.
Gerald Keusch, associate director of the National Emerging Infectious Diseases Laboratory at Boston University, added that clinicians rarely consider the excretion of a drug from a patient. He suggested initiating medical educational programs about excreted active metabolites and the disposal of unused antibiotics. Price added that a perfect system for the amplification of resistance arises in resource-limited settings, where water systems contain antibiotic manufacturing effluents, people drink untreated water, and human waste is untreated. But with the advent of globalization, he warned, this is not a localized problem. Even countries with excellent antibiotic stewardship practices have travelers bringing home drug-resistant bacteria. “We need education to amplify the message of the value of these drugs, but we also need these physical interventions to prevent this amplification and dissemination,” according to Price.
A webcast participant asked if, from a One Health perspective, there is any evidence that the accumulation of antibiotics in soils and water systems is modifying natural microbial diversity and associated ecosystem functions. Price replied that he is more concerned about decreasing diversity by the inhibition of bacteria by antibiotics. Topp noted active research in the area suggests that antibiotic exposure in aquatic and terrestrial systems is having some impact, but it is not yet clear whether those changes are within normal operating ranges. Durso added that at least one study supported by the USDA Agricultural Research Service is specifically addressing the effect of antibiotics on the functioning of nitrogen cycling in soil.
This page intentionally left blank.


























