HISTORY OF LIFE DETECTION APPROACHES
Gerald A. Soffen
Goddard Space Flight Center
National Aeronautics and Space Administration
Abstract
The history of life detection (search for extraterrestrial life) began with the invention of the telescope. Astronomer Perceval Lowell stimulated public interest in the canals of Mars from his personal observations. With the advent of the National Aeronautics and Space Administation (NASA) in 1958, the search for extraterrestrial life became a U.S. national priority. Telescopic and spectroscopic observations were made of planets and interstellar space for detection of evidence for life in the 1960s. The development of numerous in situ instruments was supported by NASA in the period 1960-1975. These involved detection of form, function, and chemistry. The Viking experiments to Mars in 1976 carried numerous instruments and performed observations and experiments on the surface and atmosphere to directly detect living organisms or their organic products or debris. Terrestrial meteorites that have been recovered have been examined for possible signs of life. The Search for Extraterrestrial Intelligence (SETI) program has been listening for coherent radiation, searching for signals from other technological civilizations.
Introduction
NASA was initiated in 1958, in response to the Soviets having launched their Sputnik. On May 21, 1961, President Kennedy announced a decision to “put a man on the Moon before the end of the decade,” despite the urgings of a number of U.S. scientists who were arguing for instrumented planetary missions. Homer Newell headed the first Space Science Office (OSS) and realized the needs of the new agency to invoke some practical as well as pure biology into the program. “NASA, (is) concerned with life sciences in a variety of ways . . . medical support for manned spaceflight, . . . life support systems, . . . aviation medicine . . . [and] exobiology (the search for and the study of extraterrestrial life) . . . Only space biology and exobiology could be regarded as pure science . . . ” is quoted from a chapter entitled “Life Sciences: No Place in the Sun” in Newell's book on the history of space science. The decade of the 1960s was used by the exobiologists mostly to develop techniques of life detection.
Life Detection
The exobiologists agreed that Mars is the most likely planet in the solar system, other than the Earth, to support living organisms, and that if there was any martian life it would most likely be microbial in nature. At that time, there was serious attention to what was called “the wave of darkening” on Mars, which some planetologists interpreted as possible biological microorganisms that were quiescent during the dry part of the season and became active when water became available. Another spectroscopic observation was the “Sinton Bands,” absorption at 3.58 µm and 3.69 µm, which suggested possible organic material; the bands were subsequently discovered to be caused by deuterium in the Earth's atmosphere.
Wolf Vishniac is credited to be the first to invent a device for monitoring microbial growth on another planet. Under a NASA grant in 1961, Vishniac built a laboratory model of a machine that would self-inoculate a small soil sample into liquid growth media. This was illuminated, and the subsequent increase in the number of organisms was measured by the changing opacity of the growing inoculation. This technique of measuring cloudiness by forward light scattering (nephelometry) to measure growth rate had been previously shown. Changes in pH as a function of time were also measured. The device called the “Wolf Trap” was developed and field-tested on Earth, but was not included onboard the Viking missions that landed the first life detection experiments on Mars in 1976.
Another life detection instrument funded by NASA utilized a radioactive tracer, 14C, to measure the respiratory products of a growing culture of microorganisms. Gilbert Levin, having used this technique for the rapid
detection of coliform organisms in sewage, modified the growth media and the inoculation technique and built the first 14C detector instrument for measuring respiration and growth of microbes on a foreign planet. He named this instrument “Gulliver,” and the concept was used as one of the three life detection experiments onboard the 1976 Viking missions. This was called the “labeled release” (LR) part of the experiment. The results are still subject to interpretation and strongly debated.
A number of other concepts were developed during the 1960s. These included the Abbreviated Vidicon Microscope, gas chromatography and mass spectroscopy (GC-MS), J-band (detection of a reaction between an organic dye and organic macromolecules such as proteins or nucleic acids), ultraviolet spectroscopy (absorption at 1800 angstroms due to peptide linkages), optical activity, Multivator (photomultiplier to detect biochemistry of biological activity using substrates), adenosine 5'-triphosphate (ATP) detection using luciferase to measure light, detection of redox potential, 18O and 15N as tracers in metabolic reactions, gas exchange (to measure metabolism), and 14CO2 uptake (to measure organic formation or photosynthesis). There was no shortage of ideas, but the actual development of these into practical experiments to be carried out on Mars required a more directed project than is normally done under research and development fiscal constraints.
In 1968, NASA decided that Mars was the most likely planet to have developed indigenous life. The agency commissioned the Viking missions to perform instrumented exploratory missions on Mars to take pictures of the surface, determine the nature of its indigenous organic compounds, perform life-detection experiments, analyze the atmosphere, and perform measurements of the meteorology and seismometry of Mars.
Venus was considered too hot (temperatures of over 600°C) for the existence of living organisms. The Moon, with its intense ionizing irradiation, was not considered a likely site. Also, there were plans for lunar surface samples to be returned to Earth by the astronauts in 1969.
In 1969 the Viking missions to Mars (two identical spacecraft) were planned. Each of the dual spacecraft consisted of an orbiter, to serve as both the bus and the communication relay, and a soft lander. The lander contained cameras, life detection experiments, chemical analysis of the atmosphere and soil, and other instruments to measure the physical nature of the surface and atmosphere. In anticipation of finding organic material on the martian surface, great emphasis was placed on the organic analysis of collected surface samples as it might bear on the question of martian biogenesis.
Besides the camera, which might have detected macroscopic life, there were three experiments performed on the Mars surface and subsurface samples to detect viable microbial activity. Since Mars is nearer the asteroid belt (where organic-bearing carbonaceous chrondrites come from), it was logical that the Mars surface must contain organic material coming from the asteroid belt. The question then was to distinguish the indigenous organic materials on Mars and place them in two categories:
-
Biological or chemical through de novo synthesis; or
-
Deposition by meteoric infall.
The three life detection experiments on the Viking missions were to detect metabolism, growth, or organic synthesis. They used 14C tracers and gas exchange techniques. LR inoculated soil samples with a dilute solution of nutrients (formic, glycolic, and lactic acids, glycine and alanine in both of their optical isomers). A second, called PR (for pyrolytic release), inoculated Mars soil samples with gaseous 14CO and 14CO2, while exposing this to simulated martian sunlight. If these gases were incorporated into the putative life, then by measuring the pyrolisate following exposure and subsequent combustion of the soil sample for 14C, this would be a test for metabolic processes (perhaps photosynthesis!). The third test, GEX (for gas exchange), was to examine the gases evolved from a rich mixture of nutrients, which was inoculated with Mars soil samples.
Each of these experiments was tested repeatedly on both of the Mars landing sites in the northern hemisphere, 7,000 km apart. The organic analysis (related to the biological search) of the Mars soil sample was performed with sensitive pyrolytic GC-MS. The device had a mass range from 12 to 250 atomic mass units and sensitivity in the range of parts per billion. It sampled the martian atmosphere and soils similar to those used for the biological tests. Despite the success of the operation of this instrument, no organic material was found on Mars at either of the landing sites.
In the case of the three biology experiments, all gave results, but the interpretation of the results was inconclusive. The PR experiment gave one anomalous unrepeatable result and several negative ones. The LR experiment yielded some evolution of CO2 but not enough to be explained by reproducing organisms. The GEX evolved O2 from the soil sample when exposed to water vapor, but no metabolic gases when the nutrient was added. This result led to an interpretation that the Mars surface includes a superoxide material over most of its surface deposited by the global dust storms. This oxidizing material (highly reactive) presumably is formed by the action of unfiltered solar ultraviolet radiation splitting the H2O and freeing the oxygen to form metal peroxide. This would account for the absence of organic material (from meteorite deposition) due to its destruction by the martian peroxide.
The conclusion reach by the Viking Biology Team was that “there is not conclusive evidence for life on Mars.” They left open the door to other experiments in other places and the possibility of fossils, should there be no contemporary life on Mars. In the succeeding decade the interest in life on Mars waned, and those interested in exobiology turned to focusing on a better understanding of how life got started on Earth.
One high-risk, high-payoff effort in exobiology is the search for extraterrestrial intelligence. This employs the use of large antennas to listen for coherent signals of extraterrestrial origin. The argument for this is that other living systems may have developed a technological capability of communicating, using the electromagnetic spectrum, that we should be able to detect by listening to the wavelengths being used. Given the dimensions of space, a search of the whole cosmos would be extremely ambitious. An unquestioned directed coherent signal detected would be considered by many as one of the greatest discoveries of mankind! Through SETI, there have been a number of efforts over the past several decades to detect coherent signals that are extraterrestrial. To date, none have been detected. Since they require unusual types of technology, most of those efforts have been supported through nongovernment funds.
Acknowledgment
NASA has initiated and developed the program of extraterrestrial life detection over the past 40 years. The work has been done by scientists and engineers in the NASA laboratories, university laboratories, and scientists working in the private sector. Some efforts were performed by scientists in the Soviet Union and other countries, but that work is poorly documented.
Bibliography
W.A. Bonner, N.E. Blair, and F.M. Darbis, “Experiments on the Abiotic Amplification of Optical Activity,” Origins of Life 11:119-134, 1981.
J. Chela-Flores and F. Raulin (eds.), Exobiology: Matter, Energy, and Information in the Origin and Evolu tion of Life in the Universe, Proceedings of the Fifth Trieste Conference on Chemical Evolution , Kluwer Academic Publishers, Dordrecht, Boston and London, 1998.
C. Chyba, P.J. Thomas, L. Brookshaw, and C. Sagan, “Cometary Delivery of Organic Molecules to the Early Earth, Science 249:366-373, 1990.
H.S.F. Cooper, The Search for Life on Mars: Evolution of an Idea, Holt, Rinehart and Winston, New York, 1976.
D. Deamer and G.R. Fleischaker (eds.), Origins of Life: The Central Concepts, Jones and Bartlett Publishers, Boston, 1994.
S.J. Dick, Life on Other Worlds: The 20th Century Extraterrestrial Life Debate , Cambridge University Press, Cambridge, Massachusetts, 1998.
E.C. Ezell and L.N. Edzell, On Mars, Exploration of the Red Planet 1958-1978, Scientific and Technical Information Branch, NASA SP 4212, Washington, D.C., 1984.
D. Goldsmith and T. Owen, The Search for Life in the Universe, Benjamin/Cummings Publishing Co., Menlo Park, California, 1980.
J.B.S. Haldane, “The Origin of Life” ( 1929) reprinted in J.D. Bernal, The Origin of Life, Weidenfeld and Nicolson, London, 1967, pp. 242-249.
N.H. Horowitz, To Utopia and Back: The Search for Life in the Solar System, W.H. Freeman and Co., New York, 1986.
H.P. Klein, “The Viking Biological Investigation: General Aspects,” Journal of Geophysical Research 82:4677-4680, 1977.
H.E. Newell, Beyond the Atmosphere: Early Years of Space Science, Scientific and Technical Information Branch, National Aeronautics and Space Administration, NASA SP-4211, Washington, D.C., 1980.
A.I. Oparin, The Origin of Life, Dover Publications, Inc., New York, 1938.
W.J. Schopf, The Cradle of Life, Princeton University Press, Princeton, New Jersey, 1999.
G.A. Soffen et al., “Scientific Results of the Viking Mission,” Science 194:1274-1353, 1976.
G.A. Soffen et al., “Scientific Results of the Viking Project,” Journal of Geophysical Research 82:3959-4681, 1977.
Space Studies Board, National Research Council, The Search for Life's Origins, National Academy Press, Washington, D.C., 1990.
THE NATURE OF BIOCHEMISTRY IN THE UNIVERSE
Norman R. Pace
Department of Molecular, Cellular, and Developmental Biology
University of Colorado
Abstract
The search for life beyond Earth requires understanding the basic chemical requirements for life and the fundamental molecular structures upon which life is likely to be based. Life on Earth is a self-replicating, evolving system based on the element carbon, and life elsewhere is highly likely to be based on carbon as well. Life anywhere will be based on macromolecules (polymers) because of the high degree of specificity required in carrying out reactions in self-sustaining living organisms and the demand for storage of large amounts of information required for living organisms. Because of their ubiquity and simplicity, amino acids, purines, and pyrimidines are likely to be the universal monomeric foundation of polymers upon which life is based. Differences in evolutionary systems will lie at the higher-order levels, the structures of the large molecules assembled from the simple units, and the mechanisms through which they are assembled and in which they participate. Techniques for detecting life beyond Earth could take advantage of the expected universality of the foundations of biology, but must account for likely variations associated with diverse evolutionary pathways.
Introduction
Humans have long speculated about the possibility of life in settings other than Earth. Only in the past few centuries, however, have we been able to conceive of the specific nature of such settings; other planets around our own Sun, and solar systems like our own elsewhere in the physical universe. Speculation on the nature of life elsewhere generally has paid little heed to constraints imposed by the nature of biochemistry, however. A century of fanciful science fiction writings has resulted not only in social enthusiasm for the quest for extraterrestrial life, but also in fanciful notions of the chemical and physical forms that life can take, what the nature of life can be.
At the current stage of the exploration of life in the solar system we are, for the first time, confronting realistically the simple question: How to detect life regardless of its nature and origin? As we undertake detection of extraterrestrial life on nearby bodies in the solar system such as Mars and Europa, or terrestrial life on outbound or returning missions, it is instructive to try to put constraints on what the nature of life can be. The search for life elsewhere than Earth will be conducted at the chemical level, so we need to try to understand what are the basic chemical requirements for life and what are the forms that life can take.
What Is Life?
An early question that we must pose, indeed one which in the last analysis requires definition, is: What is life? Most would agree that self-replication is the fundamental goal of the life process. Most would also agree that the definition of life should include the capacity for evolution. Indeed the mechanism of evolution, natural selection, is a consequence of the competing drives for self-replication that are manifest in all organisms. The definition based on those processes, then, would be that life is any self-replicating, evolving system.
The processes of self-replication and evolution are not reliably detectable, however, so we need to incorporate into the definition of life information on the nature of the chemical reactions that provide the basis for self-replication and, consequently, evolution. Based on the expected properties of molecules likely to be needed to replicate an evolving entity, life that we encounter anywhere in the universe, and can recognize, is likely to be composed of organic chemicals that follow the same general principles as our own terrestrial kind of organic-based life. The operational definition of life then becomes the following: Life is a self-replicating, evolving system based on organic chemistry. This is what we need to search for.
Why Organic Chemistry?
The basic drive of life is to make more of itself. In order to replicate, life must capture energy and transform that energy into the chemistry of new life.1 The many processes required to faithfully propagate a free-living organism necessarily require high degrees of specificity in the many interactions of the molecules that carry out the reactions. Such specificity requires information, in the form of complex molecular structure—large molecules. The molecules that serve terrestrial organisms typically are very large macromolecules, such as DNA, the repository of genetic information, and proteins. It is predictable that life, wherever we encounter it, will be composed of macromolecules.
Only two of the natural atoms, carbon and silicon, are known to serve as the backbones of molecules sufficiently large to carry biological information. Carbon, however, because of its electronic structure is unique beyond silicon in many ways. One important reason is that carbon, unlike silicon, can engage in the formation of chemical bonds with many other kinds of atoms, to thereby gain the chemical versatility required to conduct the many types of chemical reactions required for biological metabolism and propagation. Beyond carbon as a backbone atom, life—at least as we know it—requires hydrogen, oxygen, nitrogen, phosphorus, sulfur, and a host of metals, such as iron, magnesium, zinc, and others. The various organic functional groups and metals provide the enormous diversity of chemical reactions necessarily catalyzed by a living organism. Silicon, in contrast, is limited in the different atoms with which it interacts, and the large silicon molecules are monotonous compared to the infinitely combinatorial universe of organic macromolecules.
In addition, the electronic properties of carbon, unlike silicon, allow carbon to share readily with other atoms so-called double bonds or even triple bonds, chemical bonds that allow the capturing and delocalization of electronic energy. Carbon-containing compounds, therefore, can be highly polarized and thereby capture energy, and transfer this chemical energy in order to do work or to produce new chemicals in a catalytic manner. The potential polarizability of organic compounds contributes also to the specificity of intermolecular interactions, since ionic and van der Waals complementarities can shift to mesh with or to repulse one another.
The likelihood that life throughout the universe is carbon based is further encouraged by the fact that carbon is one of the most abundant of heavier elements. Astronomical studies find complex organic compounds strewn throughout interstellar space. More specifically, the common occurrence of carbonaceous meteorites speaks to an organic-rich beginning of our own solar system. Since life depends on the properties of elements heavier than hydrogen and helium, it is expected to occur only in association with second- or later-generation stars. This is because virtually all elements other than hydrogen and helium, including carbon, are manufactured within stars and must be expelled at the end of a star's lifetime.
The Universal Nature of Biochemistry
Life builds simple organic molecules such an amino acids and sugars from simple inorganic molecules: carbon dioxide, water, ammonia, phosphate, sulfur, and metals. The simple organic molecules then are used as building blocks for large molecules. Amino acids, for instance, are used to construct the long chains of proteins; simple sugars combine with purine and pyrimidine bases and phosphate to construct the nucleic acids. It seems logical that evolution of any organic-based life form would similarly result in the construction of complex molecules as repeating structures of simple subunits. Indeed, it seems likely that the basic building blocks of life anywhere will be similar to our own, in the generality if not the detail. Thus, the 20 common amino acids are the simplest carbon structures imaginable that can deliver the functional groups used in life, with properties such as repeating structure (the peptide unit), reactivity with water, and intrinsic chirality. Moreover, amino acids are formed readily from simple organic compounds and occur in extraterrestrial bodies such as meteorites, so they are likely to form in any setting that results in the development of chemical complexity necessary for life.
Similarly, the five-carbon sugars used in nucleic acids are likely to be repeated themes, perhaps in part because those are the smallest sugars that can cyclize and thereby confer spatial orientation on other molecules, for instance, the purines and pyrimidines that comprise the genetic information of terrestrial organisms. Further, because of unique abilities of purines and pyrimidines to interact with one another with particular specificity, those subunits, too, or something very like them, are likely to be common to life. Differences in evolutionary systems
likely will lie at the higher-order levels, the structures of the large molecules assembled from the simple units, and the mechanisms through which they are assembled and in which they participate.
Themes that are necessarily common to life extend beyond the building blocks. Energy transformation is a critical issue. The processes of life require capture, from physical or chemical processes, of adequate energy to conduct the chemical transformations requisite for life. There are only two such energy-obtaining processes that can support “primary productivity,” the production of biological materials from inorganic carbon dioxide.
One general process, termed “chemoautotrophy,” involves the oxidation and concomitant reduction of geochemical compounds. For instance, methanogenic organisms gain energy for growth by use of hydrogen as a source of high-energy electrons that are transferred to carbon dioxide, forming the waste product methane. Other microbes might use hydrogen sulfide as an energy source, respiring with oxygen to produce sulfuric acid. The earliest life on Earth probably relied on chemoautotrophic metabolism.
The second general process for obtaining energy, photosynthesis, uses light energy to generate energetic electrons that can be used to accomplish biochemical tasks. Photosynthesis arose early in the history of terrestrial life and probably drives most primary productivity on Earth today. The actual extent of terrestrial primary productivity remains unknown, however, since there currently is little information on chemoautotrophic metabolism that may be distributed through Earth 's crust, wherever the physical conditions permit.2
The requirements of biological energy-gathering strategies constrain the sites where life is to be sought. For example, a setting appropriate for chemoautotrophic life might be indicated or contraindicated by the occurrence or lack of an appropriate mix of oxidized and reduced chemicals. A setting for photosynthesis-based life requires sufficient light of appropriate quality. The light must be sufficiently energetic to support biosynthesis, but not so energetic as to be chemically destructive. These considerations constrain photosynthesis-based life to the spectral zone of about 300 to 1500 nm in wavelength. Below that habitable wavelength zone the light energy is sufficient to destroy organic molecules. Above that zone the light energy is probably insufficient to drive biological reactions.
Although terrestrial life and life that might arise independently of Earth are expected to use many similar, if not identical, building blocks, they also are expected to have some biochemical qualities that are unique. This expectation is because even the different evolutionary lines of terrestrial evolution also have engendered novelties unique to those lines. Thus, Archaea invented the biochemistry of methanogenesis, and the property of chlorophyll-based photosynthesis was invented among the phylogenetic domain Bacteria. Considering the variety of Earth's life, novelty, as well as commonality, must be expected elsewhere.
The expected similarity of chemistry in life's processes, regardless of the setting and origin of the life, assists in life detection in the extraterrestrial setting because it predicts that terrestrial chemicals are useful targets for analysis in any setting. On the other hand, the expected similarities of terrestrial and alien life forms complicate to some extent the interpretation of positive results. Thus, analyses of simple terrestrial-like biochemical compounds possibly could not distinguish clearly between an alien life form and a terrestrial contaminant or some organic compounds that might arise abiotically. Discrimination between different and the same origins for organisms would require analysis of macromolecules and genes. Particularly, the nature of the genetic information and the details of the information would be telling.
A Genetic Definition of Terrestrial Life
All terrestrial life is genetically related in an evolutionary past extending to 4 billion years ago or longer. All life on Earth is derived ultimately from a common ancestry. We see this relatedness in the many common structural and mechanistic features that constitute the molecular core of all cells. This relatedness is quantitatively explicit in the now-emerging maps of the course of evolution, phylogenetic trees based on gene sequences. Even in the absence of other biochemical information, genetic sequences could provide criteria to identify the evolutionary source of DNA-containing organisms and to distinguish terrestrial organisms from one another or from extraterrestrial.
The gene sequence-based overview of terrestrial biological diversity is embodied in universal phylogenetic trees such as that shown in Figure 1.3 The construction of such a relatedness diagram is conceptually simple. Pairs
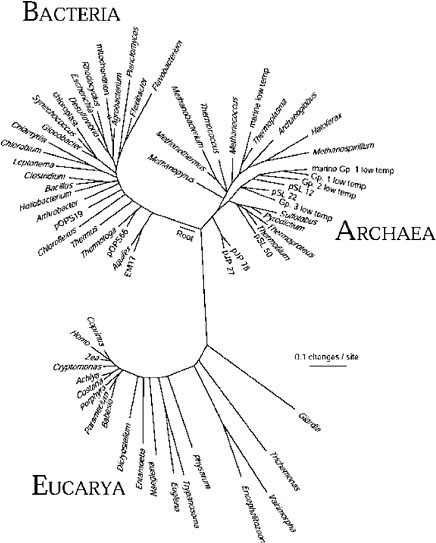
FIGURE 1. Universal phylogenetic tree based on small-subunit ribosomal RNA sequences. Sixty-four rRNA sequences representative of all known phylogenetic domains were aligned and a tree was constructed using the program FASTDNAML, correcting for multiple and back mutations. That tree was modified to the composite one shown by trimming lineages and adjusting branch points to incorporate results of other analyses. Evolutionary distance (sequence difference) between the species shown is read along line segments. The scale bar corresponds to 0.1 change per nucleotide. SOURCE: N.R. Pace, “A Molecular View of Microbial Diversity and the Biosphere,” Science 276:734-740, 1997.
of corresponding sequences from different organisms are aligned, and the number of differences between the pairs of sequences is taken to be some measure of the “evolutionary distance” that separates the organisms that contributed the sequences. Pair-wise differences between many organisms can then be used to infer relatedness trees, maps of sequence change on the evolutionary paths leading to the modern-day sequences.
The phylogenetic tree shown in the figure is based on small-subunit ribosomal RNA (rRNA) sequences, but the same topology results from comparing sequences of any other genes involved in the central, nucleic acid-based information-processing systems of cells. On the other hand, phylogenetic trees based on metabolic genes, those involved in manipulation of small molecules and in interaction with the environment, commonly are not congruent with the rRNA version. Significant incongruities in phylogenetic trees made with different molecules are generally thought to reflect lateral transfers of genes or even the intermixing of genomes in the course of evolution.4
The intrinsic commonalities in sequences of fundamental genes that occur in all organisms provide for the detection and incisive identification of organisms of terrestrial origin. This is because organisms with the same evolutionary origin are expected to contain the sequences that are universally present in the relatedness group. Conversely, organisms with independent origin are unlikely to have evolved identical genetic sequences, even if the chemical structures of the subunits that comprise the genetic information were identical. Thus, in gene sequences we can recognize the terrestrial kind of life and distinguish it from life derived from a different evolutionary origin even in the face of substantial biochemical similarity. This will become an important issue if life is discovered on another body in the solar system.
It is now clear from meteorite studies that bodies can be transported from one planet to another—for instance, from Mars to Earth—without excessive heating. In principle, life of the terrestrial kind, such as spores and cysts of microbes, could have survived such transport and seeded the solar system over the eons, wherever conditions occur that are permissible to life.
Biochemistry and Molecular Biology in Life Detection
Techniques in biochemistry and molecular biology are applicable to the detection and evaluation of life, regardless of its evolutionary origin. Thus, knowledge of the chemical requirements for metabolism can point to chemical settings where life may be found. Highly sensitive methods for the detection of simple biochemical compounds potentially are useful in detecting signatures of life and metabolism, and thereby for directing further analyses.
The interpretation of results based on simple organic compounds can be compromised by abiotic sources, however. For example, carbonaceous meteorites contain amino acids and other chemicals that might generally be considered as indicative of life. Tests for suites of simple biochemical compounds or assays for chirality could remove potential ambiguity. Tests for specific macromolecules such as proteins or nucleic acids can be unambiguous in the detection of life. Available methods for detecting macromolecules are, however, generally far less sensitive than methods for detection and identification of small molecules and, in any case, are entirely dependent upon a general knowledge of the kind of life to be detected.
Techniques of molecular biology are based primarily on the properties of nucleic acid sequences. Thus, analytical tools require a clear notion of the nature of the target. Molecular probes based on terrestrial gene sequences are unlikely to be applicable to organisms of extraterrestrial origin. On the other hand, techniques such as the polymerase chain reaction can provide exquisitely sensitive tests for terrestrial organisms, and so serve for analysis of contaminants on spacecraft bound for potentially habitable sites. Nucleic acid-based tools are potentially useful not only for detection, but also for procedures that require identification, such as to track sources of contamination.
Steven A. Benner
Departments of Chemistry and Anatomy and Cell Biology
University of Florida
Abstract
Experimental and theoretical work in organic and biological chemistry suggests the following four general principles for detection of life in nonterrestrial environments:
-
A universal genetic molecule acting in water must have a repeating charge, a structural feature of an organic molecule that is easily detected by simple probes.
-
A single biopolymer life form, presumably characteristic of all new life, will recruit cofactors with structures suited for binding to it. These can be used as confirmatory signatures of nonterrestrial life.
-
Intermediates in metabolic pathways have distinctive thermodynamic relationships, and these can be recognized in a nonterrestrial sample.
-
Organic molecules that serve as biosignatures undergo predictable diagenesis. On Mars, we may already have found some of these.
Introduction
Life is defined as a self-sustaining chemical system capable of undergoing Darwinian evolution. Despite this, organic chemists and their expertise have too often been absent from the NASA effort to define the search for life. This paper reviews four topics where an understanding of the reactivity of organic molecules can contribute to our search for life on other planets.
Universal Biosignatures Based on the Chemistry of Genetic Molecules
To sustain Darwinian evolution, a biopolymeric system must have two properties. First, it must be capable of directing its own reproduction. Second, it must be able to suffer mutation without disrupting the physical and chemical properties that are essential for reproduction.
Polymers that display the second property have come to be known as COSMIC-LOPER (“Capable of Suffering Mutation Independent of Concern over Loss of Properties Essential for Replication”).5,6 COSMIC-LOPER behavior is scarce in organic chemistry. Typical biopolymers (proteins, for example) generally change, often dramatically, their physical properties (solubility, for example) even with small numbers of changes in their sequence.7,8 The “textbook” case is sickle cell hemoglobin, where a single amino acid substitution has created a variant that undergoes partial precipitation.
Experiments with structurally modified nucleic acids have suggested that the polyelectrolyte (polyanion or polycation) structure of DNA and RNA, derived from the repeating phosphate group in the backbone, gives them their COSMIC-LOPER behavior.9
Phosphate groups force the interaction surface between strands as far distant from the backbone as possible, to the Watson-Crick “ edge” of the nucleobases. In nonionic analogues of DNA, sugar-sugar interstrand interactions, sugar-backbone interstrand interactions, interactions between the sugar and backbone groups of one strand and the Hoogsteen edge of the nucleobases on the other, Hoogsteen-Hoogsteen interstrand interactions, and Watson Crick-Hoogsteen interstrand interactions, all become important. This means that nonionic oligonucleotide analogues have rich intermolecular conformational properties (just like peptides). Conversely, the repeating polyanion in DNA constrains interstrand recognition to a small number of possible interactions, permitting the rule-based nature of strand-strand interaction represented by the rules “A pairs with T; G pairs with C.”
Phosphates keep the DNA molecule from folding on itself, allowing it to act as a template. The statistical mechanical theory of polymers suggests that the polyanionic backbone will cause natural oligonucleotides to adopt an extended structure.10 Nonionic oligonucleotide analogues have rich intramolecular conformational properties, folding like peptides.
Electronic distribution in a molecule is described as an infinite series (monopole + dipole + quadrupole + ...), with the first nonvanishing term dominating. The repeating monopole (charge) in DNA makes dipolar interactions (hydrogen bonding) secondary to its global physical properties. The physical behavior of a DNA molecule is therefore largely the same regardless of its sequence. An encoding molecule needs physical properties that are largely independent of its sequence. One does not want to mutate a gene to get a better protein, only to discover that the mutant DNA gene precipitates.
From these experiments has come a working hypothesis that may guide our search for universal chemical structures in single-biopolymer systems and in encoding biopolymers in all forms of life: As a universal chemical characteristic, living systems must contain at least one biopolymer having a repeating charge, either polyanionic or polycationic (in water), because these structures are mutable without creating dysfunctional physical properties (such as precipitation). This biopolymer will perform both catalytic and repository roles (in a one-biopolymer system) and the information repository role (in a two-biopolymer system).
This hypothesis is particularly useful because it suggests a way to detect chemical remnants of life in non-terrean samples: One searches for biopolymers or their fragments with regularly spaced positive or negative charges. This turns out to be a rather simple structural feature to search for experimentally. A chip having an absorbent with a regularly spaced negative or positive charge will bind the polycation or polyanion (respectively) more tightly than competing monocations or monoanions, and provides a convenient analytical tool for detecting such materials.
Models for the Origin of Life
Modern life on Earth is based on two biopolymers, one specialized to do genetics (DNA), and the other specialized to do catalysis (proteins). Virtually all models for how life emerged are based on the notion that a single-biopolymer performed both roles in early life forms. A decade ago, we suggested that the transition from a single biopolymer life form to a two-biopolymer system might have been slow on Earth, and discussed evidence for this hypothesis that could be found in contemporary metabolism.11 This suggests that single-biopolymer life forms would be more abundant in the universe.
The need for the polymer in a single-biopolymer Darwinian system both to be capable of searching mutation space independent of concern over the loss of properties essential for replication (COSMIC-LOPER) and to confer fitness through catalytic activity, generates competing (and occasionally contradictory) demands on the physical behavior and reactivity of the biopolymer, specifically:
-
A biopolymer specialized to be a catalyst optimally has many building blocks, so that it can display a rich versatility of chemical reactivity; a biopolymer specialized to store information should have few building blocks, as a way of ensuring faithful replication. The inverse relation between fidelity and the number of building blocks is suggested both by theory12 and experiment.13
-
A biopolymer specialized to be a catalyst must fold easily so that it can form an active site; a biopolymer specialized to store information does not fold easily, so that it can serve as a template.
-
A biopolymer specialized for catalysis must be able to change its physical properties rapidly with few changes in its sequence, enabling it to explore “function space” during divergent evolution; a biopolymer specialized to encode information must have physical properties largely unchanged even after substantial change in its sequence, so that the polymer remains acceptable to the enzymes required for replication (the COSMIC-LOPER property).
The contradiction between the structures or properties required for catalysis and those required for information storage creates problems. At the very least, any biopolymer forming a single-biopolymer Darwinian system
must generate a compromise between these goals. A two-biopolymer system allows the two biopolymers to specialize for genetic and catalytic roles, respectively. Thus, if an RNA world existed and if it had more than the four standard building blocks, one would have expected the number of building blocks to have been reduced after the breakthrough to translation to allow specialization of nucleic acids for encoding function. Further, once invented, proteins would have been expected to acquire functionality rapidly to match that not already available in RNA cofactors. The “palimpsest” of modern metabolism can be read exactly as such.14
Reconstruction of the metabolism of the putative RNA world on early Earth suggests that cofactors having structural features analogous to the structure of the catalytic biopolymer might be one way to solve these contradictions. Should non-terrean samples become available and a putative genetic biopolymer identified based on its repeating charge, a rational chemical approach to confirm the suspicion of non-terrean life would be to identify small organic molecules that are plausible building blocks of the biopolymer carrying appended reactive groups not found in the biopolymer itself. Using the RNA world model, such molecules would include reduced nicotinamide-adenine dinucleotide (NADH), flavin-adenine dinucleotide (FAD), S-adenosylmethionine, coenzyme A, and adenosine 5'-triphosphate (ATP), inter alia, presumed to be vestiges of a time when terrean life was struggling to resolve the contradicting demands between genetics and catalysis in RNA as its primordial single biopolymer life form.
Alternatives to Terrean Molecular Biochemistry
The disadvantages of a single-biopolymer life form center on the difficulty of finding a single molecular system that can satisfy competing and contradicting demands imposed by its genetic and catalytic roles, respectively. Single biopolymer life forms have advantages, however. For example, they require fewer resources; one can, for example, imagine an RNA-based organism that requires fewer sulfurs than a two-biopolymer organism. Single-biopolymer organisms can also be very small since they do not need the translation machinery, which occupies over 70 percent of the volume of a classical microorganism.
Single biopolymer life systems can also be sustained by very small metabolic repertoires. We recently proposed a 50-step metabolism that could support an autotrophic RNA organism.15 Should samples be available from non-terrean life and a putative genetic biopolymer identified based on its repeating charge, those with an understanding of organic chemical reactivity would be able to suggest pathways to generate its building blocks, in much the same way as an understanding of organic reactivity was used to hypothesize metabolic pathways in terrean organisms during the great classical period of chemistry that dominated the biochemical sciences from 1930 until about 1960, and included such figures as Robinson, Krebs, Woodward, and Bloch. Intermediates in these pathways could then be sought using the mass spectrometric analytical tools of the new millennium.
Two features are expected to distinguish biochemical pathways composed of steps catalyzed by macromolecules from nonbiological pathways. First, steps catalyzed by macromolecular catalysts surmount problems with regio- and stereochemistry with far greater ease than steps catalyzed by smaller, nonbiological catalysts. This is transparently obvious in the “design” displayed by metabolic paths. They do not “select” intermediates with much of a concern of competing reactivity.
The high specificity of macromolecular catalysts also permits the thermodynamics of metabolic pathways to be different from nonbiological chemical pathways.16 Except for those catalyzing regulated steps or the last step in a reaction sequence, metabolic steps can be approximately isoenergetic. This allows natural biochemical pathways to be more efficient than multistep synthetic sequences not involving enzymes. Because most of the steps in a natural biochemical pathway have equilibrium constants near unity, the starting materials and reagents need not be extremely reactive. Further, an enzyme-based reaction sequence can be extended indefinitely with essentially no extra energetic cost, allowing major rearrangement of the atoms in a starting material in one pot to yield a product with an entirely different structure. Because the final step in a biosynthetic pathway is exergonic, the yields are close to quantitative. The high specificity of natural enzymes allows such pathways to proceed without the chemical chaos that would result should the enzymes catalyze even small amounts of undesired side reactions.
Should non-terrean samples become available, a putative genetic biopolymer be identified, the associated cofactors found, and a metabolic pathway identified, that pathway might be identified as being “biological ” by its design without major concern for regio- and stereoselectivity issues. The thermodynamics of the pathway could then be identified to see if they fit the biological model.
Organic Molecules on the Surface of Mars
Ultimately, the search for organic vestiges of life must involve experiments on non-terrean material. In designing these experiments, we must recognize that organic matter of nonbiogenic origin is abundant and may generate a large signal beneath which a smaller signal of biogenic organic material must be sought. When trying to identify this signal, an understanding of organic chemical reactivity will be needed to predict the diagenesis of nonbiogenic organic material.
On Mars, for example, meteorites almost certainly deliver a large amount of nonbiogenic organic material to Mars. Gas chromatography-mass spectrometry (GC-MS) on the Viking 1976 Mars missions did not detect these.17 This suggested that the martian regolith might hold a potent oxidant that converts all organic molecules to carbon dioxide rapidly relative to the rate at which they arrive. A different conclusion emerged in light of what is known about the oxidation of organic compounds generally and the nature of organics likely to come to Mars via meteorite. 18 In particular, nonvolatile salts of benzenecarboxylic acids, and perhaps oxalic and acetic acid, should be metastable intermediates of meteoritic organics under oxidizing conditions. Salts of these organic acids would have been largely invisible to GC-MS.
Approximately 2 kg of meteorite-derived mellitic acid may have been generated per square meter of martian surface over 3 billion years. How much remains depends on decomposition rates under martian conditions. Since available data do not require that the surface of Mars be very strongly oxidizing, some organic molecules might be found near the surface of Mars, perhaps in amounts sufficient to be a resource. Future missions should seek these.
However, large amounts of organic molecules will almost certainly complicate the search for organics from (an entirely hypothetical) martian life. In principle, the experimental procedure will begin by creating an inventory of all organic molecules in a martian sample. Those that can be attributed to diagenesis of meteoritic organics are then subtracted from the sample. The residuals are candidates for being Mars derived. As models for the diagenesis will be sensitive to the nature of inorganic species on the planet that might catalyze various transformations, the list of possible diagenesis products will almost certainly change as more is learned about the chemical structure of the martian regolith. Additional nonbiological processes for making reduced carbon (such as the ultraviolet-derived fixation of carbon monoxide via ultraviolet-based photochemistry) must also be considered, together with the fact that biological molecules on the surface of Mars have presumably also undergone diagenesis.
Acknowledgment
Work described in this paper was supported in part by the National Aeronautics and Space Administration through its Exobiology and Astrobiology programs.
David W. Deamer
Department of Chemistry and Biochemistry
University of California, Santa Cruz
Abstract
A fundamental property of life is the capacity for polymer synthesis in a confined space, using free energy and nutrients available in the environment. In contemporary cellular life, two polymers are central to this process: Nucleic acids store and express genetic information, and proteins have structural and catalytic functions. In order to have the capacity for evolution, the two polymers are necessarily linked through a genetic code and translation process that couples mutational changes to catalytic function. The origin of cellular life presumably occurred by self-assembly of organic compounds on the prebiotic Earth into encapsulated molecular systems capable of catalyzed polymer synthesis. Although it is unlikely that nucleic acids and proteins as such were components of the first living systems, analogous polymers were produced by an as yet unknown synthetic pathway, which were capable of interacting in such a way that evolution was possible. Laboratory models of such systems offer a promising approach to test hypothetical scenarios for the origin of cellular life.
Introduction
Nucleic acid replication, transcription, and protein synthesis are characteristic of all contemporary forms of life, but such highly evolved processes could not have played a role in the steps leading to the origin of molecular systems having the properties of the living state. Instead, we must consider simpler physical processes that are collectively referred to as self-assembly. Certain organic molecules have properties that allow them to spontaneously organize into larger structures, a common example being the self-assembly of amphiphilic soap molecules into soap bubbles. All living cells are defined by membranes, and the same forces that act between soap molecules also stabilize membrane boundaries between the cytoplasm and the external environment.
Two other self-assembly processes play central roles in all life today. The first is the folding of amino acid polymers into highly ordered structures of functional proteins. This folding process occurs when amino acids are linked into proteins on ribosomes, and the folded state is stabilized by physical forces acting between the amino acids that compose the polymer. If protein-like molecules were somehow produced on the early Earth, they would also have the capacity to fold into a variety of structures, some of which could perform catalytic functions in primitive forms of life.
The second basic self-assembly process involves base pairing in nucleic acids, which is stabilized by hydrogen bonding between complementary bases. The resulting intra- and intermolecular forces produce structures such as hairpins and helices in DNA and RNA. Base pairing also plays a role in catalyzed DNA replication when nucleotides in solution bind specifically to complementary bases in templates. All life today depends on such self-assembly processes, and the earliest forms of life must have had primitive versions incorporated in their molecular systems.
Our current understanding of self-assembly processes in contemporary cellular life leads to a variety of hypothetical scenarios about how life can begin on a planetary surface.19 Given the presence of liquid water, there is little doubt that mixtures of organic compounds present on prebiotic Earth would become organized into more complex systems by self-assembly. Such microscopic molecular systems can be thought of as countless natural experiments that would occur globally for tens of millions of years prior to the origin of life. The next step toward life would take place when a few such systems happened to contain the particular set of molecules that allowed capture of energy and nutrients from the environment to be used for polymer synthesis. As noted earlier, polymer synthesis defines growth in all living systems today, and a molecular system capable of such reactions is well on its way toward the living state. Significantly, if one of the growing systems contained molecules that could be used as templates to direct further growth, a second polymeric molecule could be synthesized that was a replica of the
first molecule, thereby passing the information content of one molecule to a second molecule. This would represent the origin of replication, an essential feature of the definition of life.
The last step in the origin of life would occur when a growing, replicating system began to use the sequence of monomers in a molecule similar to a nucleic acid, to direct the sequence of monomers in a second kind of molecule such as a protein. This represents the origin of the genetic code and translation, and the beginning of life as we know it today. It also marked the beginning of biological evolution, because encapsulated molecular systems containing two different interacting molecules such as nucleic acids and proteins have the potential to undergo mutational change followed by selection.
Laboratory Simulations of Self-Assembled Protocells
How can we test the set of conjectures outlined above? One approach is to carry out laboratory simulations of molecular systems that might plausibly be present at some stage leading to the origin of life. If a simulation happened to produce a self-assembled molecular system with certain properties of the living state, the result would guide our thinking about pathways by which life could have begun on early Earth. In this brief review, one such system will be described in which amphiphilic molecules produce encapsulated microenvironments containing protein catalysts and nucleic acid templates.
Amphiphiles are defined as molecules having both hydrophilic and hydrophobic functional groups. Examples include fatty acids, detergents, and virtually all lipids such as phospholipids and cholesterol that are components of biological membranes. All amphiphiles are surface active, forming monomolecular layers at air-water interfaces, and some amphiphiles have the additional property of assembling into bilayers, which provide the permeability barrier that is essential to all biological membranes. For instance, when phospholipids are isolated from membranes and dispersed in aqueous phases, they produce spherical lipid bilayer vesicles called liposomes, which are in the size range of small bacteria.20 Standard liposome preparations are able to capture macromolecules such as enzymes and nucleic acids, but their bilayers are relatively impermeable to polar and ionic solutes. Because growth and reproduction require the efficient transport of nutrients across the cell membrane, contemporary cells employ complex protein assemblies to catalyze the transport process.
Before such proteins had evolved, what mechanism was available to transport the nutrients required for cell growth? This has been a conceptual barrier to research progress on the origin of cellular life, but new approaches have at least partially resolved the problem. Modern lipids are highly evolved products of several billion years of evolution and typically contain two hydrocarbon chains 16 to 18 carbons in length. However, much simpler amphiphilic molecules, including fatty acids, can also form reasonably stable vesicles composed of bilayer membranes.21,22 Furthermore, permeability is strongly dependent on chain length, so that shortening the chains of a given membrane lipid dramatically increases permeation rates of ionic solutes.23 Such liposomes can encapsulate enzymes, nucleic acids, and other macromolecules, yet they are sufficiently permeable to allow influx of smaller solutes that are potential substrates. This property has permitted systems of encapsulated catalysts and nucleic acids to be used as models of primitive cell-like structures.24,25
Before going on, it will be useful to define the properties of such model systems, referred to here as protocells. An ideal experimental model of a protocell should have the following properties:
-
There must be a simple mechanism for encapsulating macromolecules that could plausibly occur on the prebiotic Earth.
-
An encapsulated polymer such as a nucleic acid must be capable of replication by a template-directed polymerization process. The replication must be imperfect so that errors can occur in the genetic information, thereby allowing selection of variations that lead to evolution within the system.
-
A catalytic activity must be present that is somehow linked to the replication process, so that variations in replication affect the rate or efficiency of the catalyzed reaction. The catalyst must also be part of the growth and replication process.
-
The replicating catalytic system must be maintained within a membrane-defined volume so that selection of variations can lead to “speciation” of the encapsulated genetic material.
-
The boundary membrane itself must be able to grow. This could be accomplished either by accumulation of amphiphiles from the environment or by conversion of precursor molecules into amphiphiles. Furthermore, the growth must somehow be coupled to the internal replication process so that it neither lags behind nor gets too far ahead of polymer synthesis.
-
There must be a mechanism that allows the system to separate into two or more smaller structures at some point in the growth process, and the smaller structures in turn should incorporate the capabilities of the larger system.
Anyone with a biological background would recognize that this list of properties could also be used to define a living cell. However, the system remains a model, even though in its final version it would approach the definition of life. The reason is that virtually no metabolism occurs. Instead, all of the substrates and energy required for the growth process are provided by the experimental conditions. Even with this limitation, a laboratory version of a protocell would be a useful self-assembled molecular system that incorporates most of the processes defining life.
Progress Toward a Model Protocell
In early experiments, an RNA polymerase called polynucleotide phosphorylase (PNPase) was encapsulated either in liposomes composed of dimyristolyphosphatidylcholine (DMPC)26 or in vesicles prepared from oleic acid.27 This enzyme does not depend on a template to synthesize RNA. Instead, it can use nucleotide diphosphates such as adenosine 5'-diphosphate (ADP) as both an energy source and a monomer to be incorporated into RNA. In a typical experiment with DMPC, the enzyme was captured in liposomes by a simulated tide pool cycle in which a mixture of the enzyme and lipids was first dried, then rehydrated in the reaction medium.28 Under these conditions up to half of the original enzyme present can be encapsulated in the lipid vesicles that are produced upon rehydration. DMPC contains relatively short acyl chains, 14 carbons in length, and bilayers composed of DMPC have transient defects that allow ionized solutes such as ADP to cross the membrane barrier and supply the encapsulated polymerase with substrate. When ADP was provided to the encapsulated enzyme, vesicles containing the polymerase synthesized substantial amounts of RNA that could be detected as labeled bands in gels. The RNA could also be visualized inside the liposomes when stained with the fluorescent dye ethidium bromide and then observed by fluorescence microscopy. RNA was synthesized even if a protease was present in the medium, which demonstrated that the lipid bilayer protected the polymerase in the vesicle interior. The experiments with oleic acid vesicles show that even the simplest known membrane-forming lipid—a pure fatty acid—is able to function as a bounded environment for the enzyme. Again, the ionic substrate ADP is able to penetrate the bilayer membranes fast enough to supply the enzyme with substrate for RNA synthesis.
The encapsulated RNA polymerase provides a partial answer to the question of substrate permeation in a primitive cell. That is, a model cell composed of lipids maintain a catalyst and its RNA product in a protected environment, yet still provides access to a external nutrient and chemical energy in the form of ADP. Similar problems of substrate permeation must have been solved by the earliest forms of cellular life.
The next obvious step toward a laboratory model of a protocell is to encapsulate a system capable not only of simple polymerization, but also transcription. The viral enzyme T7 RNA polymerase can use DNA as a template to produce RNA with a transcribed sequence. Using the same conditions described above for PNPase, T7 RNA polymerase was encapsulated together with a specific DNA template in liposomes. Upon addition of four triphosphonucleotides, the encapsulated polymerase was able to synthesize RNA with a sequence transcribed from the template DNA.
In the future, it should be possible to build on this progress by capturing both a reverse transcriptase and the T7 RNA polymerase in liposomes. This represents a significant hurdle, since it requires two functional enzymes, two primers, and a template to be present in the same liposome. It also requires eight different substrates (four ribonucleotide triphosphates and four deoxyribonucleotide triphosphates) to cross the membrane barrier at a rate sufficient to supply the encapsulated enzymes with substrates. Although this is a challenging problem, the payoff of a functioning encapsulated polymerase will be significant, because ribozymes could then be included as
templates for the reaction. Ribozymes have the potential to act both as catalysts and as carriers of genetic information, and have been proposed as a primitive genetic material.29 In one laboratory model of a replicating RNA system, a reverse transcriptase first copies a DNA strand from a specific ribozyme, and a second polymerase makes multiple copies of the RNA from the DNA, thereby amplifying the original RNA strand thousands of times as the cycle is repeated. Significantly, the ribozyme itself can also evolve under these conditions when faced with a suitable selective pressure. For example, Beaudry and Joyce found that it was possible to produce a specific catalytic site on a ribozyme by continuously selecting for that site with biochemical hurdles, a kind of molecular breeding carried out in the test tube.30 Wilson and Szostak went on to show that a specific catalytic site could be selected from a mixture of trillions of random RNA sequences, similar to the kind of selection that would have occurred in a population of early molecules competing for a resource.31 In such systems, the test tube acts as a macroscopic encapsulated environment that maintains the components in contact with one another while the investigator adds monomers and an energy source (nucleoside triphosphates) through the “channel” at the top of the test tube.
Further investigations of encapsulated replicating catalytic systems will help us to better understand what happened over 3.5 billion years ago as self-assembled molecular systems first began to grow, reproduce, and evolve toward the earliest forms of microbial life. It must be noted here that we have no information yet about what kinds of polymers and catalysts in the prebiotic environment could have played the roles of the nucleic acids, enzymes, and lipids that are components of the laboratory models described above. This is where our knowledge ends, and further progress in understanding the origin of life must wait until we know more about what monomers, polymers, and polymerization reactions are plausible under prebiotic conditions.
Relevance to Life Detection
Given that terrestrial life has a system of replicating polymers at its core, it is reasonable to assume that life elsewhere might also incorporate linear polymers as a structural and functional framework. It follows that life detection, for instance, in the europan ocean, could involve a search for linear ionic polymers that have been released into the environment by extant life, just as plasmids and nucleic acid fragments are released by many terrestrial microorganisms. The signal would be unambiguous, because there are no inorganic examples of linear ionic polymers resembling nucleic acids or proteins.
Until recently, instruments capable of detecting and identifying single linear ionic polymers in solution have not been available. However, recent advances in nanopore technology offer this promise, and the paper by Meller and Branton in Session 3 provides a detailed description of the remarkable sensitivity of a nanoscopic pore (nanopore) for characterizing linear polymers in solution. Earlier work established that individual single-stranded DNA and RNA molecules can be detected by a nanopore that is linked to appropriate amplification and signal processing capabilities. 32−34 The prototype nanopore used in these studies is α-hemolysin, a 33-kD protein isolated from Staphylococcus aureus, which self-assembles in lipid bilayers to form a channel with a relatively large pore.35 The limiting aperture of the nanopore—1.5-nm diameter—is formed by a ring of alternating lysine and glutamate amino acid residues at the top of the stem, and this is followed by a 2.2-nm stem that penetrates the lipid bilayer (Figure 1 A, adapted from Song et al.36).
To produce a nanopore, a single α-hemolysin channel is embedded in a planar lipid bilayer.37 Unlike most membrane channels, α-hemolysin remains open at neutral pH ranges in 1.0 M KCl, and passes a steady ionic current in the range of 120 picoamperes (pA) with an applied voltage of 120 mV. When a linear molecule is driven through the pore by an electric field, it produces a characteristic blockade of the ionic current. The amplitude, duration, and modulation of the blockade all provide information about the molecule.
Figure 1B shows a single strand of poly(dC) (a synthetic DNA hompolymer) being translocated through the nanopore. The DNA molecule nearly fills the pore and inhibits the free flow of ions being driven by the imposed voltage.
Typical blockades of poly(dC) and poly(C) are shown in Figure 1C. It is clear that small changes in the structure of the DNA would likely have detectable effects on the amount of ionic current during a blockade and thereby produce a modulation of the blockade amplitude or duration. Furthermore, because of the tight fit,

FIGURE 1. DNA and RNA blockades of the hemolysin pore. A pore in a lipid bilayer membrane separates two compartments containing a salt solution such as 1.0 M KCl. A voltage imposed across the bilayer causes an ionic current to flow through the pore of the channel. When anionic DNA is added to the cis side (negative) it can be captured by the large electrical field at the pore and driven through to the trans side. Figure 1A (adapted from L. Song, M.R. Hobaugh, C. Shustak, S. Cheley, H. Bayley, and J.E. Gouaux, “Structure of Staphylococcal α-Hemolysin, a Heptameric Transmembrane Pore,” Science 274:1859-1866, 1996) illustrates the hemolysin pore in cross section with main dimensions given in nanometers. Figure 1B shows a single strand of poly(dC) DNA traversing the pore. Ionic current blockades produced by single molecules of poly(dC) and poly(C) 100-mers are shown in Figure 1C. The blockade reduces the open channel current by approximately 90 percent (I/Io = 0.1), and for poly(dC) the duration ranges from 100 to 200 microseconds, or ~ 1 microsecond per nucleotide to traverse the length of the pore. Note that the poly(C) blockades have significantly longer durations, suggesting that RNA experiences greater molecular “friction” as it traverses that nanopore.
variations in primary and secondary structure would also be likely to affect the duration of the blockade, as demonstrated experimentally by Akeson et al. and Meller et al.38,39 These modulations of amplitude and duration provide a signature by which approximately a variety of DNA and RNA molecules have been detected and identified with a high degree of accuracy. If similar signals were produced by a nanopore instrument analyzing melted ice from the europan ocean, it would be difficult to explain the results as anything other than a positive biomarker for extant life.
Summary
Terrestrial life is fundamentally polymeric, and life elsewhere would be likely to incorporate organic polymers for information storage, catalytic, and structural functions. It follows that detection of linear ionic polymers by a suitably sensitive instrument would provide an unambiguous signature of extant life. Nanopore technology offers a new approach to single-molecule detection. A nanopore is able to detect single molecules of linear ionic polymers such as nucleic acids, and to identify them according to unique signatures related to electrical signals that are generated when the molecule is driven through the pore. Nanopores can also be designed to detect particles ranging in size from nanoscopic to microscopic, and a nanopore instrument would therefore be useful for extant life detection missions to Europa and Mars.
James F. Kasting
Department of Geosciences
Pennsylvania State University
Abstract
NASA's proposed Terrestrial Planet Finder (TPF) mission (and the European Space Agency's [ESA's] Darwin mission) may eventually provide the capability of obtaining low-resolution thermal-infrared spectra of extrasolar planet atmospheres. One of the primary goals of these missions is to determine whether such spectra provide indirect evidence for life. A TPF-like interferometer looking at Earth from a distance would be able to detect CO2, H2O, and O3. O3 (ozone) is a potential indicator for life, since it is formed photochemically from O2, and most of Earth's O2 comes from photosynthesis. Exceptions to this rule (i.e., planets with high abiotically produced O2 levels) are possible and must be considered. CH4 (methane) is a potential bioindicator in atmospheres resembling that of early Earth, prior to the rise of O2. Methane is an ambiguous indicator of biological activity, however, because it could also have significant abiotic sources from impacts and volcanism. Hence, other indicators of life on early-Earth-type planets need to be identified.
Introduction
A possible strategy for detecting life spectroscopically is to look for examples of extreme disequilibrium in planetary atmospheres. Sagan et al. credit Lederberg with suggesting this idea originally. 40,41 Lovelock made the concept more concrete by pointing out that in Earth's atmosphere, O2 is many orders of magnitude out of thermodynamic equilibrium with reduced gases like CH4 and N2O.42 All three of these gases are produced either exclusively or primarily by the biota. Lovelock argued that biological fluxes are needed to maintain this degree of disequilibrium. One has to be careful with this argument as planetary atmospheres are always out of thermodynamic equilibrium because they are being constantly irradiated by high-energy ultraviolet photons from their parent stars. Mars, for example, has 0.1 percent O2 in its atmosphere, which is not in equilibrium with CO2 and N2, and Venus has clouds of sulfuric acid, which are not in equilibrium with SO2 and H2O. The idea is still useful, however. It just needs to be backed by detailed photochemical modeling to verify that certain atmospheric compositions are truly indicative of biological forcing.
Possible Indicators for Life
If one were to look at our own solar system from a great distance using an interferometer such as those proposed for TPF or Darwin, one would see a great difference between the atmospheres of Venus, Earth, and Mars. At low spectral resolution, Venus and Mars show only the strong 15-µm band of CO2. By contrast, Earth exhibits bands of both CO2 and H2O, along with a pronounced absorption band at 9.6 µm caused by O3. As argued by several authors,43−45 O3 is formed photochemically from O2, and most of our O2 comes from photosynthesis. Thus, under most circumstances, detection of O3 by itself is fairly strong evidence for life. Two possible exceptions to this conclusion have been identified.46 The first is a runaway greenhouse planet like Venus, where rapid loss of hydrogen from a water-rich atmosphere could result in abiotic O2 buildup. The second is a frozen planet like Mars that also lacks volcanism. The absence of liquid water at the surface would inhibit oxygen loss by weathering, whereas the lack of volcanism would eliminate oxygen loss by reaction with reduced volcanic gases. Oxygen left behind by H2O photodissociation followed by hydrogen escape could therefore accumulate indefinitely in such a planet's atmosphere. As mentioned previously, Mars itself has 0.1 percent O2 in its atmosphere as a result of this process. Mars might have even more O2 if the planet were slightly larger, so that it did not lose oxygen to space, but still not so large that it would have active volcanism. Despite assertions to the contrary,47 there is no theoretical upper limit to the amount of O2 that could build up in this manner.
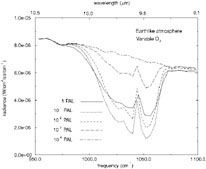
FIGURE 1. Synthetic spectra of an Earth-like planet containing various amounts of atmospheric O2. SOURCE: T.L. Schindler and J.F. Kasting, “Synthetic Spectra of Simulated Terrestrial Atmospheres Containing Possible Biomarker Gases,” Icarus 145:262-271, 2000.
One attractive point about using O3 as a bioindicator is that it is a very sensitive barometer for O 2. A substantial ozone layer can develop at relatively low O2 levels because of the nonlinear nature of ozone photo-chemistry. 48Figure 1 shows simulated spectra in the 9- to 10-µm region for an Earth-like planet with various amounts of O2. The 9.6-µm ozone band is quite strong for all O2 levels exceeding 10−2 PAL (times the present atmospheric level). These calculations are preliminary because the stratospheric temperature profile has not been calculated self-consistently. Nevertheless, it appears that ozone should be a good indicator of O2 down to very low O2 levels.
Figuring out what biogenic gases to look for in an atmosphere resembling that of present Earth is relatively simple. A more difficult task is to determine what gases to look for in an early-Earth type atmosphere. Most authors,49−52 but not all,53,54 believe that atmospheric O2 levels increased dramatically some time between 2.0 Ga and 2.4 Ga. This presumably represents the time at which net O2 production from photosynthesis followed by organic carbon burial exceeded the rate of supply of reduced volcanic gases.55 If so, the atmosphere prior to that time should have been almost completely anoxic. Some O2 should have been present at high altitudes as a consequence of CO 2 photolysis, but the ground-level concentration should have been vanishingly small.56 The column depth of O3 would have been much less than is required to produce a strong 9.6-µm band.
In the absence of O2, however, certain reduced biogenic gases could have been much more abundant. N2O is not one of these. It photolyzes rapidly at the same wavelengths at which O2 absorbs and, hence, may have been less abundant in the distant past than it is today.57 CH4 is a much better candidate. Its photolysis cutoff is at ~145 nm, so it is relatively stable against photolysis even in a low-O2 atmosphere. (Most of the solar photons capable of dissociating CH4 are in one particular line, Ly α, at 121.6 nm.) Today, CH4 is destroyed mostly by reaction with the hydroxyl radical, OH. Since the atmosphere contains abundant OH, the lifetime of CH4 is only about 12 years, so the abundance of CH4 is relatively small, ~1.6 ppmv (parts per million by volume). In an anoxic atmosphere, OH would have been consumed by reaction with molecular hydrogen:
H2 + OH → H2O + H.
So the lifetime of CH4 would have been much longer. Photochemical calculations by Brown predict that the CH4 lifetime would have been of the order of 104 years.58,59 Thus, a methane flux equivalent to the present biological source, 550 Tg of CH4 per year,60 could have produced an atmospheric CH4 mixing ratio in excess of 10−3 (Figure 2).
Methane is a good gas to look for in an extrasolar planet atmosphere because it has a strong absorption band
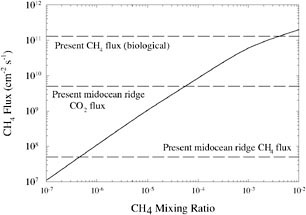
FIGURE 2. CH4 destruction rate in an anoxic atmosphere as a function of CH4 concentration. SOURCE: After J.F. Kasting and L.L. Brown, “Setting the Stage: The Early Atmosphere as a Source of Biogenic Compounds,” in The Molecular Origins of Life: Assembling the Pieces of the Puzzle, A. Brack (ed.), Cambridge University Press, New York, 1998, pp. 35-56.
at 7.7 µm that is within the proposed spectral range for TPF.61 It should be observable if present at a concentration ≥100 ppmv.62 Thus, it could be seen on an early-Earth-type planet if methane was being produced there at the same rate that it is on Earth today. Therein lies ambiguity, however. First, we do not know what the methane production rate would have been on early Earth. Methanogenic bacteria are thought to be evolutionarily ancient,63 but the anaerobic ecosystem on early Earth would have been very different from the modern ecosystem. All methanogens can generate methane from the reaction
CO2 + 4 H2 → CH4 + 2 H2O.
Since both CO2 and H2 should have been abundant on the early Earth,64 methanogens could presumably have flourished in a variety of surface environments. Still, we cannot say precisely what biogenic methane flux would be expected.
A further complication is that methane also has appreciable abiotic sources. (We do not include methane generated from fossil fuels, since these sources are ultimately biogenic.) Today, about 1 percent of the carbon emanating from midocean ridge vents is in the form of CH4.65 The other 99 percent of the carbon is outgassed as CO2. On an early Earth with a more reduced mantle, however, most of the carbon outgassed at the ridges might have been released as CH 4. The abiotic source would then be within a factor of 25 of the present biological source (Figure 2). But the intensity of midocean ridge volcanism might also have been much higher in the past as a consequence of higher geothermal heat flow. At 4 Ga, geothermal heat flow should have been four to five times higher than today.66,67 The rate of volcanism ought to scale somewhere between linearly and quadratically with heat flow.68 Thus, at the higher end, the abiotic CH4 flux could have equaled the present biological CH4 flux. Although these crude estimates for abiotic CH4 generation may be somewhat high, it is clear that we will have to be careful in using methane as a bioindicator. Further research on anoxic atmospheres and anaerobic ecosystems is needed to try to identify other gases that might be used as bioindicators in early-Earth-type atmospheres.
Acknowledgment
This work was supported by a grant from NASA's Exobiology Program.
REFERENCES FOR PAPERS IN SESSION 1
1. F.M. Harold, The Vital Force: A Study of Bioenergetics, W.H. Freeman and Co. , New York, 1986.
2. T. Gold, The Deep Hot Biosphere, Springer-Verlag/Copernicus, New York, 1998.
3. C.R. Woese, “Interpreting the Universal Phylogenetic Tree,” Proc. Natl. Acad. Sci. USA 97:8392-8396, 2000.
4. N.R. Pace, “A Molecular View of Microbial Diversity and the Biosphere,” Science 276:734-740, 1997.
5. S.A. Benner and C.Y. Switzer, “Chance and Necessity in Biomolecular Chemistry: Is Life as We Know It Universal?” in Simplicity and Complexity in Proteins and Nucleic Acids, H. Frauenfelder, J. Deisenhofer, and P.G. Wolynes (eds.), Dahlem Workshop Report, Dahlem University Press, Berlin, 1999, pp. 335-359.
6. S.A. Benner, “The Molecular Origins of Life: Assembling Pieces of the Puzzle,” Science 283:2026, 1999.
7. K. Johnsson, R.K. Allemann, and S.A. Benner, “Designed Enzymes: New Peptides That Fold in Aqueous Solution and Catalyze Reactions,” in Molecular Mechanisms in Bioorganic Processes, C. Bleasdale and B.T. Golding (eds.), Royal Society of Chemistry, Cambridge, U.K., 1990, pp. 166-187.
8. K. Johnsson, R.K. Allemann, H. Widmer, and S.A. Benner, “Synthesis, Structure and Activity of Artificial, Rationally Designed Catalytic Polypeptides,” Nature 365:530-532, 1993.
9. C. Richert, A.L. Roughton, and S.A. Benner, “Nonionic Analogs of RNA with Dimethylene Sulfone Bridges,” Journal Am. Chem. Soc. 118:4518-4531, 1996.
10. D.A. Brant and P.A. Flory, “The Configuration of Random Polypeptide Chains. I. Experimental Results, ” Journal Am. Chem. Soc. 87:2788-2791, 1965.
11. S.A. Benner, A.D. Ellington, and A. Tauer, “Modern Metabolism as a Palimpsest of the RNA World,” Proc. Natl. Acad. Sci. USA 86:7054-7058, 1989.
12. E. Szathmary, “What Is the Optimum Size for the Genetic Alphabet,” Proc. Natl. Acad. Sci. USA 89:2614-2618, 1992.
13. M.J. Lutz, H.A. Held, M. Hottiger, U. Hübscher, and S.A. Benner, “Differential Discrimination of DNA Polymerases for Variants of the Non-standard Nucleobase Pair Between Xanthosine and 2,4-Diaminopyrimidine, Two Components of an Expanded Genetic Alphabet,” Nucleic Acids Research 24:1308-1313, 1996.
14. S.A. Benner, A.D. Ellington, and A. Tauer, “Modern Metabolism as a Palimpsest of the RNA World,” Proc. Natl. Acad. Sci. USA 86:7054-7058, 1989.
15. S.A. Benner, “How Small Can a Microorganism Be?” in Space Studies Board, National Research Council, Size Limits of Very Small Microorganisms: Proceedings of a Workshop, National Academy Press, Washington, D.C., 1999, pp. 126-135.
16. A. Moradian and S.A. Benner, “A Biomimetic Biotechnological Process for Converting of Starch to Fructose: Thermodynamic and Evolutionary Considerations in Applied Enzymology,” Journal Am. Chem. Soc. 114:6980-6987, 1992.
17. K. Biemann, J. Oro, P. Toulmin III, L.E. Orgel, A.O. Nier, D.M. Anderson, P.G. Simmonds, D. Flory, A.V. Diaz, D.R. Rushneck, J.E. Biller, and A.L. Lafleur, “The Search for Organic Substances and Inorganic Volatile Compounds in the Surface of Mars,” Journal of Geophysical Research 82:4641-4658, 1977.
18. S.A. Benner, K.G. Devine, L.N. Matveeva, and D.H. Powell, “The Missing Organic Molecules on Mars,” Proc. Natl. Acad. Sci. USA 97:2425-2430, 2000.
19. H.J. Morowitz, Beginnings of Cellular Life, Yale University Press, New Haven, Connecticut, 1992.
20. A.D. Bangham, “The Liposome as a Membrane Model,” in Permeability and Function of Biological Membranes, L. Bolis, A. Katchalsky, R.D. Keynes, W.R. Lowenstein, and B.A. Pethica (eds.), North-Holland, Amsterdam, 1970, pp. 195-206.
21. W.R. Hargreaves and D.W. Deamer, “Liposomes from Ionic, Single-Chain Amphiphiles,” Biochemistry 17:3759-3768, 1978.
22. P. Walde, R. Wick, M. Fresta, A. Mangone, and P.L. Luisi, “Autopoietic Self-reproduction of Fatty Acid Vesicles,” Journal Am. Chem. Soc. 116:11649-11654, 1994.
23. S. Paula, A.G. Volkov, A.N. Van Hoek, T.H. Haines, and D.W. Deamer, “Permeation of Protons, Potassium Ions and Small Polar Molecules Through Phospholipid Bilayers as a Function of Membrane Thickness,” Biophys. J. 70:339-348, 1996.
24. D.W. Deamer, “The First Living Systems: A Bioenergetic Perspective,” Microbiol. Mol. Biol. Rev. 61:239-262, 1997.
25. P.L. Luisi, “About Various Definitions of Life,” Orig. Life Evol. Biosph. 28:613-622, 1998.
26. A. Chakrabarti, G.F. Joyce, R.R Breaker, and D.W. Deamer, “RNA Synthesis by a Liposome-encapsulated Polymerase,” Journal Mol. Evol. 39:555-559, 1994.
27. P. Walde, A. Goto, P.A. Monnard, M. Wessicken, and P.L. Luisi, “Oparin's Reactions Revisited: Enzymatic Synthesis of Poly(adenylic acid) in Micelles and Self-reproducing Vesicles, Journal Am. Chem. Soc. 116:7541-7547, 1994.
28. R. Shew and D. Deamer, “A Novel Method for Encapsulating Macromolecules in Liposomes,” Biochim. Biophys. Acta 816:1-8, 1983.
29. G.F. Joyce, A.W. Schwartz, S.L. Miller, and L.E. Orgel, “The Case for an Ancestral Genetic System Involving Simple Analogues of the Nucleotides,” Proc. Natl. Acad. Sci. USA 84:4398-4402, 1987.
30. A.A. Beaudry and G.F. Joyce, “Directed Evolution of an RNA Enzyme,” Science 342:255-257, 1992.
31. C. Wilson and J.W. Szostak, “In Vitro Evolution of a Self-alkylating Ribozyme,” Nature 374:777-782, 1994.
32. J.J. Kasianowicz, E. Brandin, D. Branton, and D.W. Deamer, “Characterization of Individual Polynucleotide Molecules Using a Membrane Channel,” Proc. Natl. Acad. Sci. USA 93:13770-13773, 1996.
33. M. Akeson, D. Branton, J.J. Kasianowicz, E. Brandin, and D.W. Deamer, “Microsecond Time-scale Discrimination Among Polycytidylic Acid, Polyadenylic Acid, and Polyuridylic Acid as Homopolymers or as Segments Within Single RNA Molecules,” Biophys. J. 77:3227-3233, 1999.
34. A. Meller, L. Nivon, E. Brandin, J. Golovchenko, and D. Branton, “Rapid Nanopore Discrimination Between Single Polynucleotide Molecules, ” Proc. Natl. Acad. Sci. USA 97:1079-1084, 2000.
35. H. Bayley, “Triggers and Switches in a Self-assembling Pore-forming Protein,” Journal Cell. Biochem. 56:177-182, 1994.
36. L. Song, M.R. Hobaugh, C. Shustak, S. Cheley, H. Bayley, and J.E. Gouaux, “Structure of Staphylococcal α-Hemolysin, a Heptameric Transmembrane Pore,” Science 274:1859-1866, 1996.
37. J.J. Kasianowicz, E. Brandin, D. Branton, and D.W. Deamer, “Characterization of Individual Polynucleotide Molecules Using a Membrane Channel,” Proc. Natl. Acad. Sci. USA 93:13770-13773, 1996.
38. M. Akeson, D. Branton, J.J. Kasianowicz, E. Brandin, and D.W. Deamer, “Microsecond Time-scale Discrimination Among Polycytidylic Acid, Polyadenylic Acid, and Polyuridylic Acid as Homopolymers or as Segments Within Single RNA Molecules,” Biophys. J. 77:3227-3233, 1999.
39. A. Meller, L. Nivon, E. Brandin, J. Golovchenko, and D. Branton, “Rapid Nanopore Discrimination Between Single Polynucleotide Molecules, ” Proc. Natl. Acad. Sci. USA 97:1079-1084, 2000.
40. C. Sagan, W.R. Thompson, R. Carlson, D. Gurnett, and C. Hord, “A Search for Life on Earth from the Galileo Spacecraft,” Nature 365:715-721, 1993.
41. J. Lederberg, “Signs of Life: Criterion-System of Exobiology,” Nature 207:9-13, 1965.
42. J.E. Lovelock, “A Physical Basis for Life Detection Experiments,” Nature 207:568-570, 1965.
43. J.R.P. Angel, A.Y.S. Cheng, and N.J. Woolf, “A Space Telescope for Infrared Spectroscopy of Earth-like Planets , Nature 322:341-343, 1986.
44. A. Leger, M. Pirre, and F.J. Marceau, “Search for Primitive Life on a Distant Planet: Relevance of O2 and O3 Detections,” Astron. Astrophys. 277:309-313, 1993.
45. J.R.P. Angel and N.J. Woolf, “Searching for Life on Other Planets,” Scientific American 274:60-66, 1996.
46. J.F. Kasting, “Habitable Zones Around Low Mass Stars and the Search for Extraterrestrial Life,” Origins of Life 27:291-307, 1997.
47. J. Rosenqvist and E. Chassefiere, “Inorganic Chemistry of O2 in a Dense Primitive Atmosphere,” Planet. Space Sci. 43:3-10, 1995.
48. M.I. Ratner and J.C.G. Walker, “Atmospheric Ozone and the History of Life,” Journal Atmos. Sci. 29:803-808, 1972.
49. P.E. Cloud, “A Working Model of the Primitive Earth,” Am. J. Sci. 272:537-548, 1972.
50. J.C.G. Walker, C. Klein, M. Schidlowski, J.W. Schopf, D.J. Stevenson, and M.R. Walter, “Environmental Evolution of the ArchaeanEarly Proterozoic Earth,” in Earth's Earliest Biosphere: Its Origin and Evolution, J.W. Schopf (ed.), Princeton University Press, Princeton, New Jersey, 1983.
51. J.F. Kasting, “Earth's Early Atmosphere,” Science 259:920-926, 1993.
52. H.D. Holland, “Early Proterozoic Atmospheric Change,” in Early Life on Earth, S. Bengtson (ed.), Columbia University Press, New York, 1994, pp. 237-244.
53. H. Ohmoto, “Evidence in Pre-2.2 Ga Paleosols for the Early Evolution of Atmospheric Oxygen and Terrestrial Biota,” Geology 24:1135 1138, 1996.
54. H. Ohmoto, “When Did the Earth's Atmosphere Become Oxic?” Geochemical News 93(Fall):13, 1977.
55. J.C.G. Walker, C. Klein, M. Schidlowski, J.W. Schopf, D.J. Stevenson, and M.R. Walter, “Environmental Evolution of the ArchaeanEarly Proterozoic Earth,” in Earth's Earliest Biosphere: Its Origin and Evolution, J.W. Schopf (ed.), Princeton University Press, Princeton, New Jersey, 1983.
56. J.F. Kasting, “Earth's Early Atmosphere,” Science 259:920-926, 1993.
57. J.F. Kasting and T.M. Donahue, “The Evolution of Atmospheric Ozone,” Journal of Geophysical Research 85:3255-3263, 1980.
58. L.L. Brown, Photochemistry and Climate on Early Earth and Mars, Ph.D. dissertation, Pennsylvania State University, State College, Pa., 1999.
59. J.F. Kasting and L.L. Brown, “Setting the Stage: The Early Atmosphere as a Source of Biogenic Compounds,” in The Molecular Origins of Life: Assembling the Pieces of the Puzzle, A. Brack (ed.), Cambridge University Press, New York, 1988, pp. 35-56.
60. M. Prather, R. Derwent, D. Ehhalt, P. Fraser, E. Sanhueza, and X. Zhou, “Other Trace Gases and Atmospheric Chemistry,” in Climate Change 1994: Radiative Forcing of Climate Change and an Evaluation of the IPCC IS92 Emission Scenarios, J.T. Houghton et al. (eds.), Cambridge University Press, Cambridge, U.K., 1995, pp. 72-126.
61. C.A. Beichman, N.J. Woolf, and C.A. Lindensmith, The Terrestrial Planet Finder (TPF): A NASA Origins Program to Search for Habitable Planets (JPL Publication 99-3), NASA Jet Propulsion Laboratory, Pasadena, California, 1999, Chapter 4.
62. T.L. Schindler and J.F. Kasting, “Synthetic Spectra of Simulated Terrestrial Atmospheres Containing Possible Biomarker Gases,” Icarus 145:262-271, 2000.
63. C.R. Woese and G.E. Fox, “Phylogenetic Structure of the Prokaryotic Domain: The Primary Kingdoms, ” Proc. Natl. Acad. Sci. USA 74:5088, 1977.
64. J.F. Kasting, “Earth's Early Atmosphere,” Science 259:920-926, 1993.
65. J.A. Welhan, “Origins of Methane in Hydrothermal Systems,” Chem. Geol. 71:183-198, 1988.
66. D.L. Turcotte, “On the Thermal Evolution of the Earth,” Earth Planet. Sci. Lett. 48:53-58, 1980.
67. G.F. Davies, G.F. “Heat and Mass Transport in the Early Earth,” in Origin of the Earth, H.E. Newsom and J.H. Jones (eds.), Oxford University Press, New York, 1990, pp. 175-194.
68. N. Sleep, private communication.


























