3
Basic Research Using Genome Editing
The recent remarkable advances in methods for editing the DNA of genes and genomes have engendered much excitement and activity and had a major impact on many areas of both basic and applied research. It has been known for 60 years that all life on Earth is encoded in the sequence of DNA, which is inherited in each succeeding generation, but accelerating advances have greatly enhanced understanding of and the ability to manipulate DNA.
This chapter reviews the various types of and purposes for basic laboratory research involving human genome editing. It begins by describing the basic tools of genome editing and the rapid advances in genome-editing technology. The chapter then details how genome editing can be used in basic laboratory research aimed at advancing understanding of human cells and tissues; of human stem cells, diseases, and regenerative medicine; and of mammalian reproduction and development. Ethical and regulatory issues entailed in this research are then summarized. Throughout the chapter, key terms and concepts germane to basic research involving genome editing are defined; Box 3-1 defines the most foundational of these terms.
THE BASIC TOOLS OF GENOME EDITING
All living organisms, from bacteria to plants to humans, use similar mechanisms to encode and express genes, although the sizes of their genomes and their numbers of genes differ greatly. Hence, understanding of
any form of life is immensely informative with respect to understanding all other forms, and provides insights and applications that obtain across species—a fact that has been particularly invaluable in the development of methods for editing genes and genomes.
The earliest studies in molecular biology were on bacteria and their viruses. Their relative simplicity and ease of analysis were key in establishing the basis of the genetic code and the expression of genes. Parallel research on more complex organisms built on the advances in these studies of bacteria, and by the mid-1960s, it was clear that bacteria, plants, and animals shared many fundamental molecular mechanisms. Key discoveries in bacteria uncovered some of their mechanisms for protection against viruses, including so-called restriction endonucleases, proteins bacteria use to cleave
the DNA of infecting viruses and “restrict” their growth. This discovery allowed scientists to cut DNA in predictable and reproducible ways and to reassemble the cut pieces into recombinant DNA.
By the mid-1970s, it was evident that recombinant DNA offered a powerful means of combining DNA in productive ways, with promising applications in biotechnology. However, this potential also raised questions about whether the application of these novel methods might entail some risk. In light of those concerns, a group of scientists and others convened a meeting at Asilomar in 1975 to consider what precautions might be needed to oversee this new technology and established a set of guidelines to regulate the containment and conduct of the research. The descendants of those guidelines still regulate recombinant DNA research to this day, some of them incorporated into official regulatory systems. In practice, the most extreme concerns did not eventuate. Today, the use of recombinant DNA methods is widespread worldwide and has yielded enormous benefits to humankind in terms of scientific understanding and medical advances, including many valuable drugs and treatments, and the biotechnology industry is now a thriving part of the world economy.
Among methods developed through the use of recombinant DNA technology is the ability to introduce DNA into cells where it can be expressed—a so-called transgene. This method is widely used in fundamental laboratory research (see Appendix A for more detail). When such exogenous DNA is introduced into a cell, it can insert into the DNA of the cell’s genome largely at random and, depending on how and where it is inserted, can be expressed as RNA and protein, although this overall process is not very efficient. A key advance was the development of techniques for generating molecular tools that could be used to cut the DNA of genes and genomes in specific places to allow targeted alterations in the DNA sequence. It was found that double-strand breaks (DSBs) could be deliberately generated by nucleases that cut DNA at defined sites (homing nucleases, sometimes also called meganucleases, originally discovered in yeast) (Choulika et al., 1995; Roux et al., 1994a,b). In the succeeding 20 years, based on these groundbreaking discoveries, several additional types of nucleases that can be targeted to specific sites were developed and adapted for use in targeted DNA cleavage (Carroll, 2014).
Such double-strand breaks also occur naturally during DNA replication or through radiation or chemical damage, and cells have evolved mechanisms for repairing them by rejoining the ends (a process known as nonhomologous end joining [NHEJ]). However, this rejoining often is not perfect, and small insertions and deletions can be introduced during the repair. Such insertions and deletions (indels) can disrupt the sequence of the DNA and often inactivate the gene that was cut. This targeted cleavage and inaccurate repair through NHEJ provide a means of inactivating genes
or gene-regulatory elements. Although the resulting indels are usually one or a few nucleotides long, in some cases they can consist of thousands of base pairs. Genome editing through NHEJ can also be harnessed to create defined chromosomal deletions or chromosomal translocations by simultaneously creating two double-strand breaks at different sites, followed by rejoining at those two sites. These sites can be either on the same chromosome (producing a deletion) or on different chromosomes (producing a translocation).
More precise editing can be achieved if, during the breakage-repair process in the cell, an extra piece of DNA is provided that shares sequence (i.e., is homologous) with the cleaved DNA. Such homologous repair also is used by normal cellular repair mechanisms. These mechanisms can be exploited to make precise changes. If homologous DNA slightly different in sequence from the cleaved sequence is introduced into the cell, that difference can be inserted into the sequence of the gene or genome, a process termed homology-directed repair (HDR). HDR can also be used to insert a novel sequence (e.g., one or more genes) of variable length at a precise genomic location. In contrast to NHEJ, HDR-mediated genome editing allows scientists to predict both where the edit will occur and the size and sequence of the resulting change. Thus, HDR-mediated editing is very much like editing a document because precise changes in the characters can be made.
Two types of targeted nucleases that have been widely developed for use in editing genes and genomes are zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs). Both rely on proteins whose normal function is to bind to specific relatively short DNA sequences. Zinc fingers are segments of proteins used by multicellular organisms to control the expression of their genes by binding to DNA (they also typically bind zinc as part of their structure; hence their name). They can be engineered by molecular biologists to recognize different short DNA sequences and can be joined to nucleases that cleave DNA. Thus, the zinc fingers target specific sequences in genes and genomes, and the attached nucleases cleave the DNA to generate a double-strand break by cleaving both strands of the DNA. ZFNs have been developed for gene editing and are in clinical trials—for example, in attempts to confer resistance to the HIV virus in AIDS patients (Tebas et al., 2014). TALENs work similarly to ZFNs, also using DNA recognition proteins (transcription activator-like effectors or TALEs) originally identified in bacteria that infect plants. The DNA recognition sequences of TALE proteins are made of repeating units, each of which recognizes a single base pair in the DNA. TALEs are simpler and easier to engineer than are zinc fingers and can similarly be joined to DNA-cleaving nucleases to yield TALENs. The preclinical application of TALENs to engineer lymphocytes for the treatment of acute lymphoblastic leukemia was recently reported (Poirot et al., 2015).
Thus, these tools are already well-established approaches to the use of genome editing for applications in gene therapy, and many of the associated safety and regulatory issues have already been addressed (see Chapter 4). However, the protein engineering required to design site-specific versions of TALENs and, even more so, of ZFNs, remains technically challenging, time-consuming, and expensive.
The past 5 years have seen the development of a completely novel system, known as CRISPR/Cas9 (CRISPR stands for clustered regularly interspaced short palindromic repeats) (Doudna and Charpentier, 2014; Hsu et al., 2014). Short RNA sequences modeled on the CRISPR system, when paired with Cas9 (CRISPR associated protein 9, an RNA-targeted nuclease), or alternatively with other similar nucleases, can readily be programmed to edit specific segments of DNA. The CRISPR/Cas9 system is simpler, faster, and cheaper relative to earlier methods and can be highly efficient. CRISPR/Cas9, like TALEs, was originally discovered in bacteria, where it functions as part of an immunity system to protect bacteria from invading viruses (Barrangou and Dudley, 2016; Doudna and Charpentier, 2014). The key distinguishing feature of CRISPR/Cas9 is that it uses RNA sequences instead of protein segments to recognize specific sequences in the DNA by complementary base pairing.
As first reengineered in 2012 (Jinek et al., 2012), the bacterial nuclease Cas9 binds a single RNA sequence known as a guide RNA tailored to recognize any sequence of choice. This two-component system can bind to the chosen site in DNA via the guide RNA and cleave the DNA using the Cas9 nuclease. Since it is simple to synthesize RNA of any desired sequence, generation of CRISPR/Cas9 targeting nucleases is straightforward—the system is readily programmed to target any sequence in any genome. Programs exist for choosing suitable guide RNAs, and while not all guides work equally well, testing a number of guides to find effective ones is not difficult or expensive. This ease of design, together with the remarkable specificity and efficiency of CRISPR/Cas9 has revolutionized the field of genome editing and has major implications for advances in fundamental research, as well as in such applications as biotechnology, agriculture, insect control, and gene therapy.
Figure 3-1 provides a summary of the ZFN, TALEN, and CRISPR methods of genome editing. As mentioned, these genome-editing methods are being widely applied across a broad range of biological sciences, from fundamental laboratory research on cells and laboratory animals; to applications in agriculture involving improvements in crop plants and farm animals; to applications in human health, both at the research level and, increasingly, in clinical applications. Agricultural applications have been addressed in other studies by the U.S. National Academies of Sciences, Engineering, and Medicine (see Chapter 1) and potential clinical applica-
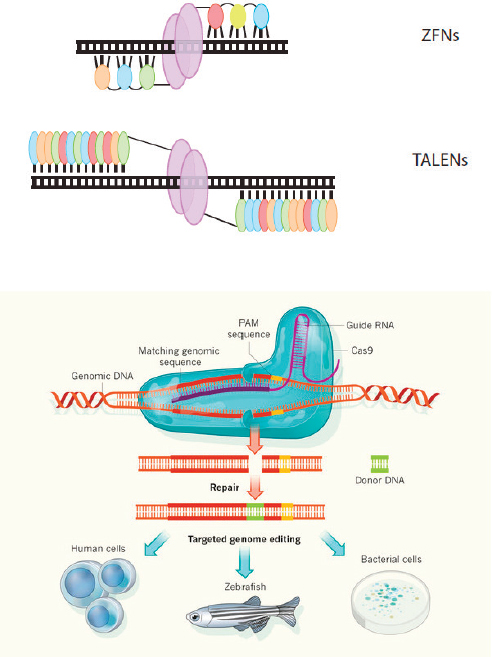
Top: Zinc finger nucleases (ZFNs): The colored modules represent the Zn fingers, each engineered to recognize three adjacent base pairs in the DNA; these modules are coupled to a dimer of the FokI nuclease that makes a double-stranded cut in the DNA.
Middle: Transcription activator-like effector nucleases (TALENs). The colored modules each recognize a single base pair in the DNA; these modules are coupled to a dimer of the FokI nuclease that makes a double-stranded cut in the DNA.
Bottom: CRISPR/Cas9. Two components derived from the clustered regularly interspaced short palindromic repeat (CRISPR) region are needed. A nuclease such as Cas9 (blue) is targeted to a specific site on the DNA by the guide RNA (purple), which binds a 20-base sequence in the genome adjacent to a short protospacer adjacent motif (PAM) sequence (yellow) and targets a double-stranded cut in the DNA.
NOTE: In all three cases, the DNA cut can be repaired by nonhomologous end joining of the ends or by repair directed by a stretch of homologous DNA (green), producing alterations in the genome of the target organism, which can be from any species. (For more detail, see Appendix A.)
SOURCES: Top and middle (Beumer and Carroll, 2014); bottom (Charpentier and Doudna, 2013).
tions are the subject of subsequent chapters of this report. The focus in this chapter is on basic laboratory research using genome editing.
This research addresses fundamental questions concerning the use and optimization of genome-editing methods both in cultured cells and in experimental multicellular organisms (e.g., mice, flies, plants). Such basic discovery research is essential for improving any future applications of genome editing. Applications of genome editing in laboratory research also have added powerful new tools that are contributing greatly to understanding of basic cellular functions, metabolic processes, immunity and resistance to pathological infections, and diseases such as cancer and cardiovascular disease. These laboratory studies are overseen by standard laboratory safety mechanisms. In addition to these applications, this chapter reviews the potential for using similar approaches in basic research on human germline cells, not for the purposes of procreation but solely for laboratory research. This work will provide valuable insights into the processes of early human development and reproductive success, and could lead to clinical benefits, directly as a result of work with human embryos and germline cells or through improvements in the derivation and maintenance of stem cells in vitro.
RAPID ADVANCES IN GENOME-EDITING TECHNOLOGY
The development of CRISPR/Cas9 has revolutionized the science of gene and genome editing, and the basic science is advancing extremely rapidly, with additional CRISPR-based systems being developed and deployed for multiple different purposes. Different species of bacteria use somewhat different CRISPR systems, and although the CRISPR/Cas9 system is currently the most widely used because of its simplicity, alternative systems being developed will provide increased flexibility in methodology (Wright et al., 2016; Zetsche et al., 2015).
Among the issues that need to be addressed going forward are the specificity and efficiency of the DNA cleavage mediated by CRISPR-guided nucleases. While the roughly 20-base sequence recognized by the guide RNA provides a great deal of specificity (an exact match should occur by chance in approximately 1 × 1012 base pairs—1 in a trillion—the equivalent of several hundred mammalian genomes), there is some small potential for so-called off-target events, in which the nucleases make cuts in unintended places, especially if the guide RNA binds to DNA sequences that are slightly different from the intended target. Some early experiments suggested that off-target events might occur at a significant rate, but as the methods have been improved and as their application has increasingly been in normal cells rather than cultured cell lines, the frequency of off-target cleavages appears to be very low. Advances have been achieved in the specificity of Cas9
cleavage (Kleinstiver et al., 2016; Slaymaker et al., 2016), and methods have been developed for monitoring the frequency of off-target cleavage. (See Appendix A for more detail.)
Another significant advance has occurred in the development of methods for modifying the CRISPR/Cas9 system so that DNA cleavage is avoided. For example, the nuclease function of Cas9 can be inactivated so that a complex of guide RNA and such a “dead” Cas9 (dCas9) will target a specific site via the guide RNA but will not cleave the DNA (Qi et al., 2013). By coupling other proteins with different activities to the dCas9, however, different sorts of modifications can be made to the DNA or its associated proteins. Thus, it is possible to design variants of CRISPR/Cas9, ZFN, or TALE that will turn on or turn off adjacent genes, make single-base changes, or modify the chromatin proteins that associate with DNA in chromosomes and thus modify the epigenetic regulation of genes (Ding et al., 2016; Gaj et al., 2016; Konerman et al., 2015; Sander and Joung, 2014). All of these noncleaving variants fail to cleave DNA, thus reducing the potential for deleterious off-target events, and many other modifications are being introduced to enhance specificity and reduce off-target events (see Appendix A for further detail). Most recently, CRISPR/C2c2, a programmable RNA-guided, RNA-cleaving nuclease, has been described (Abudayyeh et al., 2016; East-Seletsky et al., 2016) that could be used to knock down specific RNA copies of genes without affecting the gene itself. This development raises the future possibility of nonheritable or reversible editing.
As can be seen from this brief survey, the rapidly developing versatility of these RNA-guided genome-editing systems is opening up numerous means of manipulating the expression and function of genes. A recent report of methods for inducibly knocking down or knocking out genes in a multiplex fashion in many cell types, including human pluripotent stem cells, as well as in mice (Bertero et al., 2016) further expands the potential of these methods. These and other advances have rapidly rendered these methods basic tools of molecular biology worldwide, adding to the existing toolkit assembled over the past 40 years. These methods are now being applied to study with unprecedented ease the functions of genes in cells and in experimental animals, such as yeast, fish, mice, and many others, to enhance understanding of life. They also are being used to investigate the derivation and differentiation of stem cells, providing fundamental insights relevant to regenerative medicine, and to develop culture models of human disease both to advance understanding of disease processes and to enable testing of drugs on human cells ex vivo.
BASIC LABORATORY RESEARCH TO ADVANCE UNDERSTANDING OF HUMAN CELLS AND TISSUES
Basic biomedical research aimed at discovering more about the mechanisms and capabilities of genome editing offers significant opportunities to advance human medicine. Genome-editing research conducted on human cells, tissues, embryos, and gametes in the laboratory offers important avenues for learning more about human gene functions, genomic rearrangements, DNA-repair mechanisms, early human development, the links between genes and disease, and the progression of cancer and other diseases that have a strong genetic basis. Manipulation of genes and gene expression by genome editing allows one to understand the functions of genes in the behavior of human cells, including why they malfunction in disease. For example, editing of cultured human cells to model the changes that arise in cancer or in genetically inherited diseases provides culture models of those diseases with which to understand the molecular basis of the resulting defects. Such laboratory studies also allow the development of means of combating those defects, such as the testing of potential drugs in cell culture. All of those approaches are much easier now than they were just a few years ago.
Certain cells derived from an early embryo, after fertilization but prior to the developmental stage at which it would implant in a woman’s uterus, are referred to as embryonic stem (ES) cells. These ES cells have scientific advantages because they can reproduce in cell culture and have the potential to form all the different body cell types while lacking the potential themselves to develop into a fetus. It is now also possible to create stem cells by manipulating adult somatic cells to convert them to a state in which they, too, have the ability to form multiple cell types, reducing the need to take stem cells from an early embryo. These are referred to as induced pluripotent stem (iPS) cells. Such pluripotent stem cells can be cultured in vitro and induced to develop into many different cell types, such as neurons, muscle or skin cells, and many others. Advances over the past several decades in understanding stem cells and how they can be used form the foundation for the field of regenerative medicine, which seeks to repair or replace damaged cells within human tissues or to generate new tissues after disease or injury. Although these are increasingly areas of clinical practice, and the application of genetically altered cells in humans is not covered in this chapter (see Chapter 4), there are nevertheless a number of important reasons why scientists aim to undertake basic investigations in human and animal stem cells in the laboratory.
Genome-editing methods have been extremely useful in generating a variety of genetic modifications in human ES and iPS cells. Before the advent of efficient genome-editing tools, these cells had proven resistant to
genetic modification with the standard tools of homologous recombination that had been used effectively in mouse ES cells. Using those tools in human cells resulted in very low frequencies of targeted recombination. Improvements in efficiency resulting from the use of CRISPR/Cas9 have enabled rapid generation of tagged reporter cell lines, making it possible to follow differentiation pathways, look for interacting proteins, sort appropriate cell types, and investigate the functions of individual genes and pathways in cells, among many other applications (Hockemeyer and Jaenisch, 2016). For example, the ability to make precisely targeted mutations or corrections in specific genes has made possible the generation of human ES lines with different specific disease alleles on the same genetic background (Halevy et al., 2016) for use in research on the consequences of such disease genes. Conversely, genome editing also allows the targeted correction of disease mutations in patient-specific iPS cell lines to generate genetically matched control lines. Such modified stem cell lines are used primarily to conduct experimental and preclinical studies, to investigate specific disease processes, and to test drugs that could be used to treat such diseases. In the future, such edited stem cell lines could be used for various forms of somatic cell–based therapies (see Chapter 4).
BASIC LABORATORY RESEARCH TO ADVANCE UNDERSTANDING OF MAMMALIAN REPRODUCTION AND DEVELOPMENT
Germline cells are cells with the capacity to be involved in forming a new individual and to have their genetic material passed on to a new generation. They include precursor cells that form eggs and sperm, as well as the eggs and sperm cells themselves. When fertilization occurs to create an embryo, the earliest stages of this embryo, referred to as the zygote (fertilized egg) and blastocyst, have the potential to divide and form all the cells that will make up the future individual, including somatic (body) cells and new germ cells. As the embryo continues to develop, its cells differentiate into specific cell types that become increasingly restricted in their functions (e.g., to form specialized cells such as those in the nervous system, skin, or gut).
During reproduction and development, genetic changes made directly in gametes (egg and sperm), in egg or sperm precursor cells, or in very early embryos would be propagated throughout the future cells of an organism and may therefore be heritable by subsequent generations. As emphasized above, this chapter focuses exclusively on the use of genome-editing technologies in the laboratory, and not on clinical applications in humans or in embryos for the purposes of implantation to initiate pregnancy. Nevertheless, it is important to understand which cell types are involved in human development and their functions, because this information informs
researchers’ decisions about how to study particular scientific questions and informs ethical, regulatory, and social discussions around when and why it may be useful to use human cells, including embryos, in basic laboratory research.
Genome Editing of Germline Stem Cells and Progenitor Cells
It is already possible in mice to genetically modify the genome in a fertilized egg (the zygote), in individual cells of the early embryo, in pluripotent ES cells, or in spermatogonial stem cells, just as in somatic cells. In all these cases, the effects of the genetic modifications can be studied directly in the embryo or in cells in culture. There are a number of ways to undertake these genetic manipulations and a number of cell types in which they can be conducted. The cell types below are all considered part of the germline or have the capacity to contribute to the germline:
- embryonic stem cells derived from normal early embryos (blastocyst stages)
- cells from early embryos produced after somatic cell nuclear transfer (SCNT)1
- iPS cells obtained by reprogramming somatic cells into an ES celllike state
In mice, these cell types can all be manipulated experimentally through genome editing. Stem cells of the types listed above can contribute to the germline in vivo after they are introduced into mouse embryos at the morula or blastocyst stage. This process generally creates an embryo that is a chimera, in which some cells are derived from the stem cells introduced into the embryo, and some are formed from the initial embryonic cells. Mouse or rat spermatogonial stem cells can be cultured and their genomes edited, and the cells can then be introduced into recipient mouse or rat testes, where they can give rise to sperm able to fertilize oocytes, at least in vitro (see Appendix A and Chapman et al., 2015). In all of these cases, when the resultant embryos are transferred back into the uterus to complete pregnancy, it is possible to establish lines of mice carrying the genetic alterations. These approaches provide unprecedented opportunities to explore the functions of all the genes in the genome and to develop rodent models of human diseases. Proof-of-principle experiments also have been
___________________
1 SCNT is a technique in which the original nucleus of an egg cell is removed and replaced with a “donor” nucleus taken from another cell (e.g., from a somatic cell that has undergone genome editing). This is the technique that was used to create Dolly, the first cloned mammal obtained from an adult cell.
reported in which disease-related genetic mutations have been corrected in mouse zygotes (Long et al., 2014; Wu et al., 2013), embryonic stem cells, or spermatogonial stem cells (Wu et al., 2015) and then transmitted though the germline to produce genetically corrected mice.
The application of genome-editing technologies to the equivalent human cell types holds considerable potential value for fundamental research without any intent to use such manipulated cells for human reproductive purposes. Improved knowledge of how an early human embryo develops also is valuable in its own right, and because such knowledge can help answer questions about humans’ own early development, as well as facilitate understanding and potential prevention or treatment of a wide range of clinical problems. A number of these applications are described below.
Improvements in Assisted Reproductive Technology
The success of human reproductive technologies and preimplantation genetic diagnosis (PGD) of inherited diseases has been, and continues to be, dependent on in vitro fertilization (IVF) and on culturing of human embryos from the zygote to the blastocyst stage. However, tools for ensuring that an individual embryo in culture is normal and capable of completing pregnancy remain limited. Most embryo research has been conducted on mouse embryos, which are similar to human embryos in certain respects but significantly different in others (see Box 3-2). Even the conditions in which human embryos are kept in culture are based largely on those established for mouse embryos. High rates of aneuploidy2 are found in cultured human embryos relative to other species. This aneuploidy is often mosaic—that is, it varies among cells in the embryo (Taylor et al., 2014)—but how it arises and how it relates to in vitro culture conditions are not well understood. There is also concern that epigenetic3 abnormalities might occur in human embryos in vitro (Lazaraviciute et al., 2014), which might compromise development or health, even later in life. Research on early-stage human embryos in culture should enable scientists to better understand the cellular and molecular pathways that control early human embryo development and the conditions under which human embryos in culture can develop successfully. This knowledge could in turn help improve IVF outcomes.
All of the differences between humans and mice discussed above mean that it is not possible to accurately infer developmental events in human embryos from studying mice. This limitation has practical consequences for
___________________
2 Having a chromosome number that is not an exact multiple of the usual haploid number.
3 The term “epigenome” refers to a set of chemical modifications to the DNA of the genome and to proteins and RNAs that bind to DNA in the chromosomes to affect whether and how genes are expressed.
the development of improved IVF technologies, as well as for the ability to derive the best pluripotent or other stem cells for modeling of human disease and for future regenerative therapies. Thus, there is considerable interest in experimental investigation of preimplantation human development in culture, in jurisdictions where such research on human embryos is permitted. The goals of this work are to understand the fundamental events of fertilization, activation of the embryonic genome, cell lineage development, epigenetic events such as X-inactivation, and others, and how these events compare and contrast with what is understood from studying mice.
Similar research also could provide insights into the reasons for the high rates of early pregnancy loss in natural human pregnancies (10 to 45 percent, depending on the age of the mother), as well as the causes of infertility. Better understanding of sperm development would be crucial in addressing issues of male infertility. Pluripotent stem cells arise from the early embryo, and these cells can generate ES cells in culture. Better understanding of human embryonic development would provide insights into the origins and regulation of pluripotency and how to translate that knowledge into improved stem cells for regenerative medicine. The potential benefits of such research are not limited to embryonic stem cells. Cell types that give rise to the yolk sac and the placenta also are determined in the early embryo prior to implantation. The yolk sac and placenta establish the crucial links with the mother during pregnancy and provide nutrients and other factors that enable the embryo to survive. Defects in these tissues can compromise a pregnancy, leading to miscarriage, premature birth, or postnatal abnormalities. Better understanding of how the yolk sac and placenta originate would help in improving techniques for overcoming infertility and preventing early miscarriage, as well as understanding and preventing congenital malformations. These extraembryonic cell types also provide cues that pattern the early postimplantation embryo, although almost nothing is known about these processes in humans. These possibilities and others discussed in this chapter are summarized in Table 3-1.
Understanding of Human Development
Genome editing by CRISPR/Cas9 and similar techniques has a key place in the tool set needed to undertake such experiments. CRISPR/Cas9guided activation or inactivation of specific target pathways could be used to understand overall gene regulation in development. Indeed, as the efficiency of CRISPR/Cas9 continues to increase, it should be possible to use genome editing to knock out4 genes in zygotes and study the effects directly
___________________
4 A gene is said to be “knocked out” when it is inactivated because the original DNA sequence has either been replaced or disrupted.
in genetically altered embryos. None of these experiments would involve human pregnancies, so none could result in heritable germline modifications. They would all be in vitro experiments, with results being analyzed primarily at the blastocyst stage in the first 1-6 days of development.
In some cases, there could be interest in exploring the effects of altering specific genes at the next stages of human development, notably the early stages after the embryo would implant in a uterus. At present, culture of human embryos up to the stage just prior to germ-layer formation (at 14 days after fertilization or the formation of the “primitive streak”) is permitted in many countries. Improved culture systems that allow human embryos to develop in culture during the implantation period are being developed. Recent results suggest that these systems could be used to study the elaboration of extraembryonic structures and of the epiblast into an “embryonic disc”—processes that occur in humans in ways not found in mice (Deglincerti et al., 2016; Shahbazi et al., 2016). These improved cell
culture systems, combined with better ways of analyzing gene function using genome editing, can be expected to lead to better understanding of the fundamental processes of early human development. Already at least two research groups (in the United Kingdom and Sweden) have received regulatory permission to carry out CRISPR/Cas9 experiments on human embryos, aimed at addressing these kinds of fundamental biological questions.
Knowledge gained from such studies is expected to inform and improve IVF procedures and embryo implantation rates and reduce rates of miscarriage. Conversely, the same studies may lead to novel methods of contraception. Such research also should lead to better ways of establishing and maintaining stem cells from these early embryonic stages, which could facilitate efforts to derive cell types for studies and treatments of disease and traumatic injury. Knowledge gained from these laboratory studies using genome-editing methods in early human embryos should also provide information about the suitability of these methods for any eventual
TABLE 3-1 Reasons for Laboratory Studies of Human Embryos
| In Vitro Studies | Clinical Outcomes |
|---|---|
| Studies of fertilization in vitro | Improvements in in vitro fertilization (IVF) and preimplantation genetic diagnosis (PGD) Possible improvements in contraception |
| Improved culture of early human embryos | Improvements in IVF and PGD Insights into reasons for miscarriages and congenital malformations |
| Development of extraembryonic tissues (yolk sac and placenta) | Insights into reasons for failures in implantation and for miscarriages |
| Isolation and in vitro differentiation of pluripotent stem cells | In vitro models for human diseases for experimental testing of drugs and other therapies Improved cells for somatic gene/cell therapies and for regenerative medicine |
| Investigations of sperm and oocyte development | Possible novel approaches to infertility |
potential clinical use. That is, basic research can be expected to inform an understanding of the feasibility of making heritable, and preferably non-mosaic, changes in the genome (see Chapter 5). Because human embryos that can be used in research are a valuable and relatively scarce resource, it will be important to ensure that the most efficient methods are used for these laboratory studies of their basic biology. Thus, it is likely that in the course of this research, various technical issues associated with improving the use of genome-editing methods in human embryos will be addressed. Relevant questions include
- the type and form of genome-editing components to be introduced;
- whether to use Cas9 or an alternative nuclease;
- what method to use to introduce the genome-editing components—for example, as DNA, mRNA, protein, or ribonucleoprotein complex;
- whether to use single guide RNAs, pairs, or multiple guide RNAs as part of the editing machinery;
- the size of the DNA template and whether such a template is required;
- the optimal timing for genome editing, that is, whether information can be obtained by using two-cell embryos, whether it is necessary to use one-cell embryos, or whether it is best to introduce the reagents along with the sperm during in vitro fertilization;
- whether mosaicism can be tolerated, keeping in mind that it may be an advantage for certain experiments, as when cell fate is to be followed, but may need to be avoided in other cases, such as when investigating a gene whose product is a secreted protein; and
- how to test and improve modified versions of nucleases such as Cas9 or inhibitors of certain repair mechanisms (e.g., an effective inhibitor of nonhomologous end joining may be needed if the experiment demands homology-directed repair [Howden et al., 2016]).
Understanding of Gametogenesis and Infertility
In mice, the generation of spermatogonial stem cell (SSC) lines from the adult testes has provided a rich source of cells with which to study the process of spermatogenesis in vitro and in vivo, after regrafting to the testes. It is possible to alter these cells genetically and study the impact of the changes on the process of spermatogenesis itself or, in mice, the impact on the offspring. It is also possible to correct genetic mutations in the stem cells in vitro using CRISPR/Cas9. Proof of principle for such an approach has been published (Wu et al., 2015). This work used CRISPR/Cas9 editing in mouse SSCs to correct a gene mutation that causes cataracts in mice. The edited SSCs were transferred back to mouse testes, and round spermatids were collected for intracytoplasmic sperm injection (ICSI), a form of IVF, to create embryos. Resulting offspring were correctly edited at 100 percent efficiency. Similar experiments have been conducted using SSCs from other species, including macaques (Hermann et al., 2012). Stable human SSC lines have not yet been reported, but would clearly be an important tool for understanding male infertility and for exploring such issues as the higher rate of mutations associated with age. This is an active area of research because it may enable restoration of fertility in male cancer patients after radiation or chemotherapy. The ability to grow and manipulate human SSCs would, however, raise the possibility of generating human germline alterations if the cells were grafted back to the testes or used in IVF.
Related issues arise from experiments in which both oocytes and sperm progenitors have been generated from mouse ES cells. ES-derived oocytes can be fertilized by normal sperm, and ES-derived spermatids can fertilize eggs by ICSI (Hayashi et al., 2012; Hikabe et al., 2016; Saitou and Miyauchi, 2016; Zhou et al., 2016). Human gametes have not yet been generated successfully from pluripotent stem cells, although two recent papers report the generation of early germ cell progenitors from human ES cells (Irie et al., 2015; Sasaki et al., 2015). Through the use of genome-editing methods, this work also highlighted significant differences between mice and humans in the genes involved in specification of primordial germ cells. There is evidence as well that knowledge gained from studying later
stages of spermatogenesis in mice may not always be applicable to the same process in humans. These findings reflect the role of research on human cells in answering questions about human biology. If human haploid gametes could be generated from human pluripotent cells, as they can be in mice, it would open up new avenues for understanding gametogenesis and the causes of infertility. It would also open up possibilities for using heritable genome modifications to address health problems that originate from genetic causes.
ETHICAL AND REGULATORY ISSUES IN BASIC RESEARCH
As described in more detail in Chapter 2, basic science research performed in the laboratory on somatic cells will be subject to regulation focused on safety for laboratory workers and the environment, including special review by institutional biosafety committees for work involving recombinant DNA. Few new ethical issues are raised, although if the cells and tissues come from identifiable living individuals, donor consent and privacy will be a concern, and in most cases the protocols will be subject to at least some review by institutional review boards.
Research with embryos is more controversial. As noted earlier, research using viable embryos is illegal in a small number of U.S. states (NCSL, 2016), and while permitted in most states, research that exposes embryos to risk generally may not be funded by the U.S. Department of Health and Human Services (HHS); this is due to the Dickey-Wicker Amendment,5 which has been adopted repeatedly since the 1990s as part of the HHS appropriations process, including in the bills introduced for 2017 funding (see Chapter 2).6 It states
- None of the funds made available in this Act may be used for—
- the creation of a human embryo or embryos for research purposes; or
- research in which a human embryo or embryos are destroyed, discarded, or knowingly subjected to risk of injury or death greater than that allowed for research on fetuses in utero under 45 CFR 46.204(b) and section 498(b) of the Public Health Service Act (42 U.S.C. 289g(b)).
- For purposes of this section, the term “human embryo or embryos” includes any organism, not protected as a human subject under 45 CFR 46 as of the date of the enactment of this Act, that is derived by fertilization, parthenogenesis, cloning, or any other means from one or more human gametes or human diploid cells.
___________________
5 Public Law No. 114-113, Division H, Title V, § 508.
6 § 508(a) in both S. 3040 and H.R. 5926.
The effect of this combination of state and federal law is to make embryo research legal in most of the United States but generally not eligible for HHS funding.
Additional, extralegal oversight of laboratory research using human embryos comes from the stem cell research oversight committees that were widely adopted pursuant to recommendations of the National Academies regarding embryonic stem cell research (IOM, 2005; NRC and IOM, 2010). Recently, the International Society for Stem Cell Research, whose membership includes investigators from around the world as well as the United States, adopted guidelines calling for the transformation of these voluntary stem cell research oversight committees into human embryo research oversight (EMRO) committees that would oversee “all research that (a) involves preimplantation stages of human development, human embryos, or embryo-derived cells or (b) entails the production of human gametes in vitro when such gametes are tested by fertilization or used for the creation of embryos” (ISSCR, 2016a, p. 5). The review would include details of the proposal and the credentials of the researchers under the auspices of these independent, multidisciplinary committees of scientists, ethicists, and members of the public. The proposed committees would assess research goals “within an ethical framework to ensure that research proceeds in a transparent and responsible manner. The project proposal should include a discussion of alternative methods and provide a rationale for employing the requested human materials, including justification for the numbers of preimplantation embryos to be used, the proposed methodology, and for performing the experiments in a human rather than animal model system” (ISSCR, 2016a, p. 6).
CONCLUSIONS AND RECOMMENDATION
Laboratory research involving human genome editing—that is, research that does not involve contact with patients—follows regulatory pathways that are the same as those for other basic laboratory in vitro research with human tissues, and raises issues already managed under existing ethical norms and regulatory regimes. This includes not only work with somatic cells, but also the donation and use of human gametes and embryos for research purposes, where this research is permitted. While there are those who disagree with the policies embodied in some of those rules, the rules continue to be in effect. Important scientific and clinical issues relevant to human fertility and reproduction require continued laboratory research on human gametes and their progenitors, human embryos, and pluripotent stem cells. This research is necessary for medical and scientific purposes that are not directed at heritable genome editing, though it will also pro-
vide valuable information and techniques that could be applied if heritable genome editing were to be attempted in the future.
RECOMMENDATION 3-1. Existing regulatory infrastructure and processes for reviewing and evaluating basic laboratory genome-editing research with human cells and tissues should be used to evaluate future basic laboratory research on human genome editing.






















